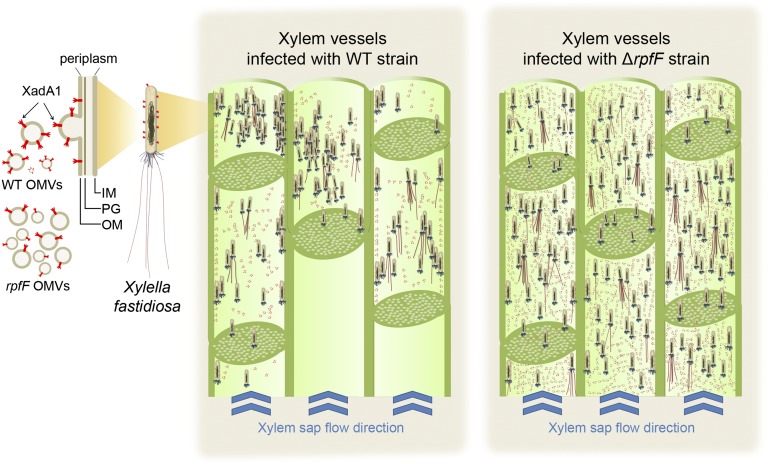
Significance
Release of outer membrane vesicles (OMVs) is a general feature of Gram-negative bacteria. Most studies have addressed the mechanisms of their formation or the cargo they can carry, but other roles remain to be explored further. Here we provide evidence for a novel role for OMVs in Xylella fastidiosa, a bacterial pathogen that colonizes the xylem of important crop plants. OMVs, whose production is suppressed by a quorum-sensing system, serve as an autoinhibitor of cell adhesion to surfaces, thereby blocking attachment-driven biofilm formation that would restrict movement within the xylem and thus colonization of plants. The ubiquity of OMV formation in the bacterial world suggests that these extracellular products may have alternative roles that might modulate movement and biofilm formation.
Keywords: phytopathogen, Pierce disease, XadA, antiadhesiveness
Abstract
Outer membrane vesicles (OMVs) of Gram-negative bacteria have been studied intensively in recent years, primarily in their role in delivering virulence factors and antigens during pathogenesis. However, the near ubiquity of their production suggests that they may play other roles, such as responding to envelope stress or trafficking various cargoes to prevent dilution or degradation by other bacterial species. Here we show that OMVs produced by Xylella fastidiosa, a xylem-colonizing plant pathogenic bacterium, block its interaction with various surfaces such as the walls of xylem vessels in host plants. The release of OMVs was suppressed by the diffusible signal factor-dependent quorum-sensing system, and a X. fastidiosa ΔrpfF mutant in which quorum signaling was disrupted was both much more virulent to plants and less adhesive to glass and plant surfaces than the WT strain. The higher virulence of the ΔrpfF mutant was associated with fivefold higher numbers of OMVs recovered from xylem sap of infected plants. The frequency of attachment of X. fastidiosa to xylem vessels was 20-fold lower in the presence of OMVs than in their absence. OMV production thus is a strategy used by X. fastidiosa cells to adjust attachment to surfaces in its transition from adhesive cells capable of insect transmission to an “exploratory” lifestyle for systemic spread within the plant host which would be hindered by attachment. OMV production may contribute to the movement of other bacteria in porous environments by similarly reducing their contact with environmental constituents.
Many important plant diseases such as Pierce disease of grapes and citrus variegated chlorosis (CVC) are associated with the xylem-limited bacteria Xylella fastidiosa (1). Infected plants exhibit progressive leaf scorching or other foliar symptoms consistent with the water stress that is associated with the occlusion of large numbers of xylem vessels by bacterial cells or by tyloses that are induced by the presence of bacteria within vessels (2–4). The virulence of X. fastidiosa is associated with its ability to migrate widely and proliferate within xylem vessels after its spatially limited introduction by infested sharpshooter vectors during feeding (5). Disease symptoms may be largely an inadvertent effect caused by successful colonization that causes interference with xylem sap flow (6). Cells of X. fastidiosa colonize specific areas of the foreguts of insect vectors, where they multiply and form a biofilm, being firmly attached to the foregut cuticular lining to endure the high fluid flow during sap feeding (7, 8). This turbulent environment may lead to occasional detachment of cells, allowing pathogen inoculation into plants (9). Thus, insect colonization and transmission of X. fastidiosa depends on its ability to attach to the insect’s foregut.
X. fastidiosa uses diffusible signaling factors (XfDSF), a family of related unsaturated fatty acids, to regulate its behavior in a cell density-dependent manner (10, 11). XfDSF-mediated signaling suppresses motility and stimulates the production of cell-surface adhesins, thus increasing cell aggregation, surface attachment, and biofilm formation (10, 12–14). A ΔrpfF mutant of X. fastidiosa, blocked in the production of XfDSF, is hypervirulent to grapevine but is impaired in insect colonization and transmission (11, 12, 15). The accumulation of diffusible signaling factors (DSF) with increasing cell concentration increases the adhesiveness of the cells, presumably better to enable their acquisition by insect vectors, but reduces their ability to move and multiply within plants. These observations support the hypothesis that XfDSF signaling is used in a context-dependent lifestyle switch that enables a subset of the bacterial cells in a plant to become more adhesive, and thus able to be acquired by insects, by inducing a phenotype incompatible with the movement of the more solitary cells throughout the plant (6).
A recent study (16) indicated that an extracellular factor produced by X. fastidiosa attenuated its ability to adhere to surfaces. This extracellular factor, produced by the WT strain and in greater amounts by the ΔrpfF mutant during both plant colonization and growth in broth culture, suppressed transmission by insect vectors and blocked adhesion of X. fastidiosa cells to various surfaces.
Although the nature of this antiadhesive extracellular factor was unclear, it seemed likely that it could be one or more of the surface components overreleased by the ΔrpfF mutant, perhaps related to outer membrane (OM) proteins, because at least 11 OM protein-encoding genes are up-regulated in the ΔrpfF mutant as compared with the WT strain (14). X. fastidiosa OM proteins such as hemagglutinin-like proteins and the autotransporter XatA have been shown to be localized not only in the OM but also extracellularly, both as soluble proteins or in outer membrane vesicles (OMVs) (17, 18).
OMVs are spheroid particles ranging in size from ca. 20 to 250 nm that are produced through the blebbing and pinching off of portions of the OM from all Gram-negative bacteria investigated to date (19–22). OMVs contain integral OM proteins embedded within the glycerophospholipid-LPS double layer along with OM-anchored lipoproteins and entrapped soluble periplasmic proteins (20, 22). OMVs from certain bacteria contain a variety of virulence factors and antigens and thus participate in pathogenesis (23–26). Other functions assigned to OMVs include modulating the immune response (27), trafficking of degradative enzymes against competing bacterial species (28, 29), delivering cargoes to benefit complex microbial communities (30), serving as a response to envelope stress (22, 31), and perhaps even acting as decoys for predators such as viruses (32).
In this study, we demonstrate that X. fastidiosa is a particularly active producer of OMVs that, in turn, are an extracellular antiadhesive factor in this species. Using immunoassays against XadA1, an OM protein found to be solely present in the OM and in OMVs, nanoparticle-tracking analysis (NTA), and fluorescence and electron microscopy, we show that the production of OMVs is suppressed by DSF-mediated quorum sensing in X. fastidiosa both in vitro and within the host plant. We propose that secretion of OMVs by X. fastidiosa cells participates in a strategy to adjust its adhesiveness to surfaces when transitioning from a biofilm-forming stage capable of being vectored to a nonadhesive “exploratory” lifestyle for spreading in xylem vessels. Such a novel function of OMVs might contribute to the behavior of other species in which such planktonic/sessile transitions are necessary.
Results
X. fastidiosa Releases Large Numbers of OMVs in Vitro.
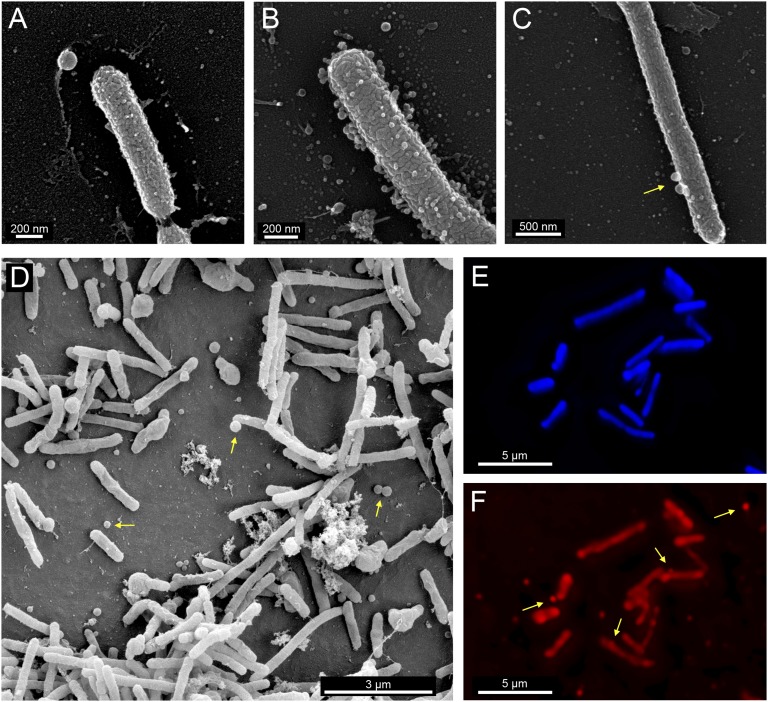
Scanning electron microscopy images of cultures of X. fastidiosa revealed the presence of OMV-like nanoparticles attached to cells as well as unattached particles in samples of planktonic cells (Fig. 1 A–C) or cells that had attached to glass surfaces in PD3 (complex medium) broth cultures (Fig. 1D). The particles usually were rather small, less than 30% the diameter of the bacterial cell. Deconvolution microscopy images of X. fastidiosa cells recovered from broth cultures and stained with both DAPI and FM4-64 revealed the presence of DNA-containing cells that were stained with both materials (Fig. 1 E and F) as well as nanoparticles that were assumed to be OMVs because they were stained only with FM4-64. Although some nanoparticles were attached to cells of X. fastidiosa, others were unattached (Fig. 1F). Such particles were conspicuous in any given image, suggesting that the X. fastidiosa WT strain releases numerous OMVs to the environment.
Fig. 1.
X. fastidiosa produces OMVs in vitro. (A–D) Scanning electron microscopy of the WT strain with OMV-like nanoparticles apparent on and near planktonic cells recovered from broth cultures (A–C) or cells attached to glass surfaces within those cultures (D). (E and F) Deconvolution microscopy images of cells of the WT strain cultured in PD3 stained with either DAPI (E) or FM4-64 (F). Arrows indicate putative OMVs.
The number of OMVs produced by bacterial cells in broth cultures was estimated using NTA. X. fastidiosa WT cells grown in PD3 or PIM6 [a minimal medium developed in the laboratory of Michele Igo, University of California, Davis, CA (10)] broth were removed by low-speed centrifugation, and the OMV concentration in the extracellular medium was determined. The particles released by X. fastidiosa ranged in size from 20 to 400 nm, and sizes were described by a normal distribution with a mean of 110 nm as illustrated in Fig. S1. As seen in other bacterial species (20, 33), the composition of the culture medium clearly affects the production of OMVs by X. fastidiosa. Although the WT X. fastidiosa strain produced 51 ± 20 OMVs per cell in PD3 medium, only 7 ± 1 OMVs/cells were produced in PIM6 broth (Table 1). Escherichia coli, Xanthomonas citri, and Pseudomonas aeruginosa produced three- to 10-fold fewer OMVs per cell than X. fastidiosa under the conditions tested (Table 1). Thus, in comparison with model bacteria commonly used in studies of OMVs, X. fastidiosa produces and releases many more such particles.
Table 1.
Outer membrane vesicle quantification by nanoparticle tracking analysis
| Bacterial species | Growth condition* | OD600† | OMVs/cfu‡ | LSD test§ |
| X. fastidiosa Temecula1 WT | PD3 | 0.59 | 51 ± 20 | b |
| X. fastidiosa Temecula1 WT | PIM6 | 0.15 | 7 ± 1 | c |
| X. fastidiosa ∆rpfF | PD3 | 0.53 | 131 ± 28 | a |
| X. fastidiosa ∆rpfF | PIM6 | 0.14 | 22 ± 4 | b, c |
| Xanthomonas citri 306 | PD3 | 0.84 | 14 ± 2 | b, c |
| E. coli BL21(DE3) | LB¶ | 1.24 | 0.6 ± 0.1 | c |
| E. coli BL21(DE3) | PD3 | 1.07 | 16 ± 1 | b, c |
| P. aeruginosa PA14 | PIM6|| | 1.17 | 0.7 ± 0.2 | c |
Growth of X. fastidiosa was for 4 d in PD3 or for 2 d in PIM6; growth of X. citri, E. coli, and P. aeruginosa was for ca. 4 h.
OD600 at time of harvest for one of the assays.
Counts of OMVs per cfu obtained by NTA are shown as mean ± SD from three biological replicates.
Values of samples marked with the same letter do not differ significantly (P < 0.05) according to Fisher’s LSD test.
E. coli BL21(DE3) does not grow in PIM6.
P. aeruginosa does not grow in PD3.
To investigate further the production of OMVs by X. fastidiosa, we assessed the subcellular localization of XadA1, a trimeric autotransporter adhesin predicted to be localized to the OM (34). Previous work had demonstrated that XadA1 is present not only on the surface of the 9a5c CVC strain of X. fastidiosa but also away from bacterial cells in biofilms formed in vitro and in infected plant tissue (35). As shown in Fig. 2A, the anti-XadA1 antibody raised against XadA1 from the CVC strain (35) also recognized the native XadA1 of the Temecula1 strain of X. fastidiosa used here. Western blot analysis revealed that XadA1 was localized in similar amounts in cell lysates and in the cell-free culture supernatant (Fig. 2B and Fig. S2A). As expected, no XadA1 was detected in cell lysates of a ΔxadA1 mutant (Fig. 2A and Fig. S2A). However, a slightly smaller cross-reacting polypeptide was present in cell lysates of both the WT strain and ΔxadA1 mutant (Fig. 2A and Fig. S2A). The cross-reacting polypeptide was present only in the total cell lysate fractions and was used as a cell marker per se and thus as a measure for fractionation quality. As expected, cell-associated XadA1 is found in the OM protein fraction (Fig. S2B). Because XadA1 from both the cellular and cell-free fractions exhibited similar electrophoretic mobility, it apparently is not cleaved during the release process from the cell (Fig. S2B).
Fig. 2.
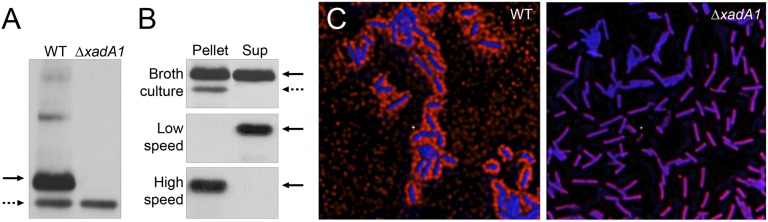
XadA1 localizes to the cell envelope and to OMVs. (A) Anti-XadA1 antibody detects XadA1 in total cell lysates of WT X. fastidiosa (arrow) but not in lysates from a ΔxadA1 mutant. (B) XadA1 is present in both cells and cell-free supernatant of broth cultures (Top). XadA1 present in cell-free supernatant remains in suspension following low-speed (38,000 × g) centrifugation (Middle) but is removed as a pellet during high-speed (150,000 × g) centrifugation (Bottom). The dashed arrow indicates a cross-reacting polypeptide detected only in cells. (C) Deconvolution microscopy of WT cells (Left) and a ΔxadA1 mutant (Right) treated with both an anti-XadA1 antibody and DAPI, following a red-fluorogenic secondary antibody, revealing that XadA1 localizes to both the cell envelope and to the extracellular environment of cells, presumably in OMVs.
To determine whether XadA1 is secreted as a free protein and/or is associated with OMVs, the cell-free culture supernatant was fractionated further by differential centrifugation. As shown in Fig. 2B, extracellular XadA1 was not removed from culture supernatants by low-speed (38,000 × g) centrifugation but was recovered in the pellet by subsequent high-speed (150,000 × g) centrifugation, indicating that it is associated with particles such as OMVs and is not present as a free soluble protein. The high-speed pellet prepared from X. fastidiosa broth cultures has been demonstrated previously to be enriched in hemagglutinin-like adhesins and the autotransporter XatA (17, 18), but the full protein composition of such OMVs remains unknown.
Additional evidence that XadA1 is localized both in the cell envelope and in OMVs was obtained by immunofluorescence localization using deconvolution microscopy of cell preparations treated with polyclonal anti-XadA1 followed by fluorescently labeled secondary antibody and DAPI staining. As shown in Fig. 2C, large amounts of XadA1 surround cells, presumably in OMVs, in addition to that associated with the surface of cells (stained with DAPI). Therefore XadA1 was used as a marker for OMVs produced by X. fastidiosa. As might be expected, OMVs could be detected more readily using anti-XadA1 antibodies than by FM4-64 staining (compare Figs. 1F and 2C).
X. fastidiosa ΔrpfF Strain Overproduces OMVs in Vitro and in Planta.
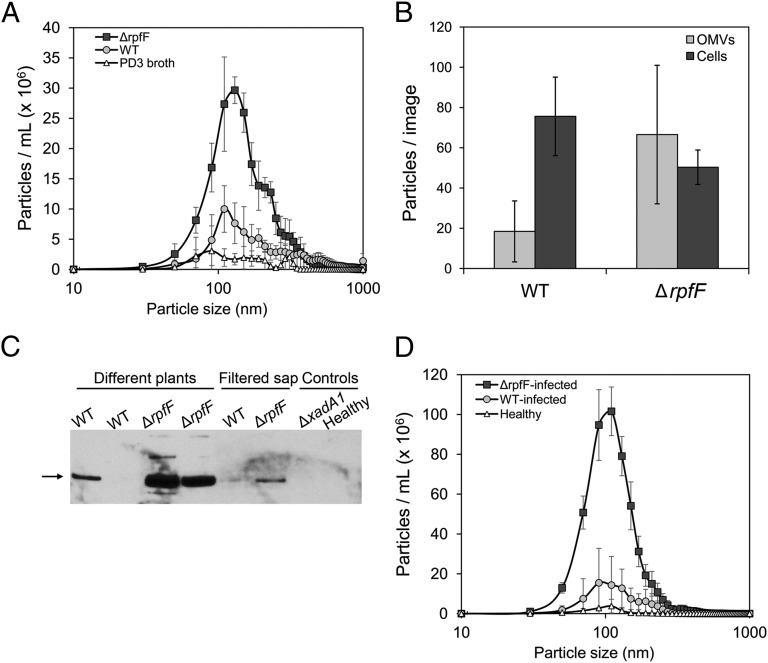
X. fastidiosa mutant strains were scored for the enhanced ability to release XadA1 as an indication of hypervesiculation. Although ΔrpfF mutants express lower levels of XadA1 (Fig. S3A) and xadA1 transcript (Fig. S3B) than the WT strain, the mutants release a much higher proportion of this protein into the culture supernatant than does the WT strain (Fig. S3A). Given that this marker for OMVs was preferentially shed by the ΔrpfF mutant, it seemed likely that the ΔrpfF mutant would release more OMVs than the WT strain. The number of OMVs released into the extracellular medium by the ΔrpfF mutant grown in PD3 or PIM6 medium as assessed by NTA was about threefold higher than the number released by the WT strain, regardless of the growth medium (Table 1). Although much more numerous, the secreted by the ΔrpfF mutant were similar in size to those secreted by the WT strain, exhibiting a normal distribution of sizes with a median size of about 110 nm (Fig. 3A). Estimates of the number of relatively large FM4-64–stained OMVs that could be visualized by deconvolution microscopy also revealed that the ΔrpfF mutant produced higher amounts of such particles: 0.24 ± 0.11 and 1.35 ± 0.87 OMVs per cell (P = 0.02) were produced by the WT and ΔrpfF mutant, respectively (Fig. 3B). These numbers are consistent with the relatively small fraction of large (>200 nm) particles present in the OMV population in culture supernatants observed by NTA (Fig. 3A).
Fig. 3.
The ΔrpfF mutant overproduces OMVs. (A) Size distribution of particles detected in extracellular fractions of the WT strain (○) and ΔrpfF mutant (■) of X. fastidiosa grown in PD3 broth, as well as uninoculated PD3 broth (△) determined by NTA. (B) Numbers of OMVs (light bars) and cells (dark bars) of the WT and the ΔrpfF mutant grown in PD3 broth and stained with FM4-64 were measured using deconvolution microscopy of culture samples. (C) Immunoblot detection of XadA1 in xylem sap extracted from grape plants infected with the WT, ΔrpfF, and ΔxadA1 mutant strains as indicated or from uninfected healthy plants. Where indicated, sap also was filtered through a 0.22-µm filter before analysis. (D) Size distribution of particles detected in filtered xylem sap from plants infected with the WT strain (○) or ΔrpfF mutant (■) of X. fastidiosa or uninfected healthy plants (△) determined by NTA. The error bars represent the SD of the mean of three biological replicates in A and B and two biological replicates in D. Differences between WT and ΔrpfF mutant were determined to be statistically significant with P < 0.05 in two-tailed homoscedastic t tests.
To assess if OMVs were produced by X. fastidiosa in planta, we quantified XadA1 abundance in xylem sap recovered from grape plants infected with either the WT or the ΔrpfF mutant strains. Much more XadA1 was detected in xylem sap of plants infected with the ΔrpfF mutant than from the sap of plants infected with the WT strain (Fig. 3C). As expected, no XadA1 was detected in sap from plants infected with the ΔxadA1 strain or the uninfected control (Fig. 3C). Importantly, XadA1 was detected readily in sap from plants infected with the ΔrpfF mutant from which cells of the pathogen had been removed by filtration through a 0.2-µm filter, suggesting that OMVs are abundant in xylem sap of infected plants. As measured by NTA, filtered xylem sap recovered from plants infected with the ΔrpfF mutant contained fivefold more particles assumed to be OMVs than did sap from plants infected with the WT strain (Fig. 3D). Very few particles were detected in sap from uninfected plants. Clearly, large numbers of OMVs are produced by X. fastidiosa in planta, and the concentration of such particles in plants infected with the ΔrpfF mutant is particularly high. Because the WT and particularly the ΔrpfF mutant release an antiadhesive factor into both culture medium and the xylem of infected plants (16), we posit that OMVs are the agent that blocks attachment of X. fastidiosa to surfaces.
OMVs Impair X. fastidiosa Attachment to Surfaces Under Steady and Flow Conditions.
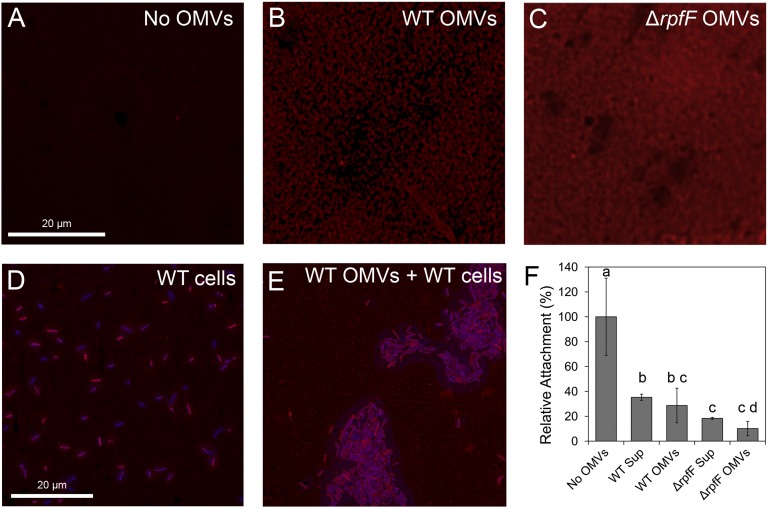
To determine the effects of OMVs on the attachment of X. fastidiosa, we tested the ability of the WT strain to attach to various surfaces in the presence and absence of OMVs isolated from either the WT or the ΔrpfF strain. Because OMVs are stable in suspension (36, 37), it was possible to conduct a variety of in vitro assays. We first performed these assays on a glass surface using pure or crude OMV samples. OMVs present in culture supernatants or more purified OMV preparations recovered from culture supernatants by ultracentrifugation were applied to glass coverslips before the addition of WT cells. Deconvolution microscopy of the treated surfaces that were stained by FM4-64 revealed that the OMVs had attached to the glass surfaces (Fig. 4 A–C). Because OMVs from comparable numbers of X. fastidiosa cells had been applied, this approach did not show whether the more abundant binding of OMVs from the ΔrpfF mutant reflected qualitative differences in their adhesiveness or merely the large number of OMVs in preparations from this mutant; however, the results of NTA analysis supported the latter supposition. Application of OMVs to glass surfaces resulted in dramatic differences in the pattern of attachment of X. fastidiosa WT cells as compared with their attachment on noncoated glass surfaces. The cells were evenly distributed and attached individually to the noncoated surface (Fig. 4D), but attachment occurred at only a few sites on the coated surfaces, and most cells attached at those sites formed discrete aggregates (Fig. 4E). Compared with the noncoated surfaces, much less cell attachment to the OMV-coated surfaces was observed after 1 h of incubation. Deconvolution microscopy revealed that cells attached primarily at those few sites in which OMVs were not present on the treated glass surfaces (Fig. 4E). The antiadhesive effect was equally pronounced when purified OMVs or culture supernatants containing OMVs were applied to glass surfaces (Fig. 4F). Again, perhaps because OMVs were more abundant in the preparations from a given number of cells of the ΔrpfF mutant than in preparations from the same number of cells of the WT strain (Fig. 3), the attachment of X. fastidiosa cells was suppressed more by OMV preparations from the mutant strain (Fig. 4F). Together, these observations indicate that OMVs reduce the accessibility of cells to the glass surface, perhaps by competing with cells for attachment sites.
Fig. 4.
OMVs suppress the attachment of X. fastidiosa cells to the glass surface. (A–C) Deconvolution microscopy images of untreated glass coverslips (A) and coverslips treated with OMVs isolated from WT (B) or ΔrpfF mutant (C) strains, stained with FM4-64. (D and E) Attachment of WT cells to untreated coverslips (D) or to coverslips treated with OMVs from the WT strain (E) when stained with FM4-64 and DAPI. (F) Relative number of sites colonized by WT strain on surfaces treated or untreated with OMVs or with cell-free supernatants (Sup) before cells. The error bars represent the SD of the mean; values of samples marked with the same letter do not differ significantly (P < 0.05) according to Fisher’s LSD test.
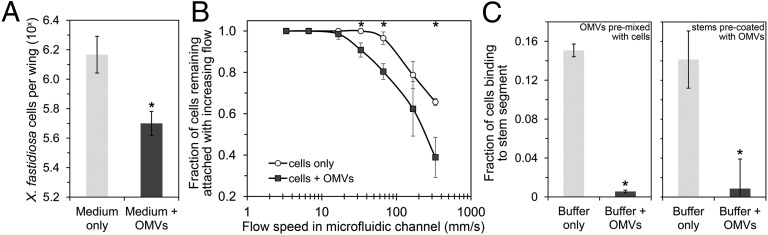
Because the antiadhesive factors present in culture supernatants of X. fastidiosa originally had been observed to block adhesion to chitinaceous surfaces such as the wings of glassy-winged sharpshooter (Homalodisca vitripennis) (16), we assessed the ability of purified OMVs to interfere with adhesion of WT X. fastidiosa on this substrate. Treatment of insect wings with purified OMVs from a ΔrpfF mutant reduced the attachment of WT cells by threefold as compared with that of cells suspended in medium alone (Fig. 5A). It thus appears that OMVs produced by X. fastidiosa can account for the reduced retention and transmission of this pathogen by insect vectors that acquire the pathogen either from laboratory cultures or from the sap of infected plants, especially plants colonized by the ΔrpfF mutant (16).
Fig. 5.
OMVs impair the attachment of X. fastidiosa cells under steady and flow conditions. (A) Number of cells that attached to insect wings pretreated with medium alone (light bar) or containing OMVs (dark bar). (B) Fraction of individually tracked cells remaining attached to a microfluidic channel under increasing flow speeds. (C) Fraction of cells that attached to grape stem segments when introduced in buffer alone (light bar) or when premixed with OMVs (Left) or when OMVs were introduced into the stem segment before the bacterial cells (Right), calculated from dilution plating and cfu counts. The error bars represent the SD of the mean from three biological replicates. Asterisks indicate a significant difference between values with and without OMVs (P < 0.05 in a two-tailed homoscedastic t test).
Although OMVs appear to be capable of blocking the adhesion of X. fastidiosa to insect vectors, this blocking would appear to be an inadvertent function that would not benefit the pathogen, because adhesion to the vectors is needed for the successful retention of the pathogen during the insect’s feeding on infected plants. Instead, the ability to move extensively along and between xylem vessels after inoculation would favor the development of large population sizes and better enable acquisition of the pathogen by insect vectors. Adhesion of X. fastidiosa to xylem vessels would constrain the virulence of the pathogen. It would be expected, therefore, that OMVs would contribute to the virulence of X. fastidiosa by blocking its adhesion to xylem vessels and thus enabling its active and passive movement through the plant. This notion was verified by determining the ability of OMVs to interfere with adhesion of X. fastidiosa cells to xylem vessels by introducing cells together with OMV preparations into microfluidic chambers and grape stem segments and comparing the number of cells retained after flushing events. In the first approach, cells were monitored individually for surface attachment under increasing flow speeds within channels with a cross-section of 5,000 µm2, similar to a xylem vessel with a diameter of 80 µm. Preincubation of WT cells with pure OMV preparations causes more cells to be detached and washed downstream at any given flow speed above 10 mm/s (Fig. 5B). In the other approach, the fraction of cells introduced together with OMVs into stem segments that were retained after flushing was about was 30-fold lower than observed in cells introduced with buffer alone (Fig. 5C).
OMVs strongly blocked the adhesion of X. fastidiosa cells to xylem vessels, regardless of the fluid in which they were cointroduced into the plant. For example, cell attachment was reduced more than fourfold when introduced with an OMV preparation suspended in PIM6 medium as compared with introduction in PIM6 medium alone (Fig. S4A). Furthermore, particulate material, presumably OMVs, was sufficient to account for the blocking of the attachment of cells to xylem vessels. As shown in Fig. S5, approximately a 30-fold higher fraction of X. fastidiosa cells had attached to xylem vessels when they were introduced in cell-free culture supernatants in which OMVs had been removed by ultracentrifugation as compared with cells introduced together with OMVs suspended in PIM6 medium. The presence of OMVs thus is sufficient to explain the blocking of attachment of X. fastidiosa to plant surfaces.
The strong blockage of X. fastidiosa attachment to xylem vessels when introduced together with OMVs might have been caused by the binding of OMVs to the cells of the pathogen or to the plant surface. To distinguish between these possibilities, purified OMVs were introduced into grape stem segments and allowed to interact with the plant tissue for 1 h; then the xylem vessels were flushed with buffer to remove excess OMVs before the X. fastidiosa cells were introduced. As was seen when cells were coincubated with OMVs, the fraction of cells retained in stem segments that were pretreated with buffer alone was 18-fold higher than the fraction retained in xylem vessels in which OMVs had been introduced before the bacterial cells (Fig. 5C). These results support the model in which OMVs bind to sites on the plant where cells of X. fastidiosa might preferentially have bound. Apparently X. fastidiosa cells bind to OMVs less efficiently than to the surfaces to which OMVs had bound, and these particles therefore interfere with adhesion by blocking sites at which the pathogen might attach.
Discussion
It is recognized that bacteria can modify the surfaces of the local habitat in which they reside by secreting extracellular molecules. Examples of such habitat modification include the production of extracellular polysaccharides that protect cells from predators and from some toxicants to which they might otherwise be exposed and that also retain water (38, 39). Likewise, secreted biosurfactants can modulate bacterial motility and modify the chemical environment (40). The work reported here suggests that OMVs also can strongly influence the interaction of bacteria with surfaces. This finding further expands the growing recognition of the many possible roles of OMVs. They not only are carriers of specific cargo or merely a way to shed OM components (20–22, 30, 41); they also can play important roles in the interactions that occur between the cell and its surrounding environment. Although previous work had suggested that X. fastidiosa, like other Gram-negative bacteria, could secrete various surface proteins in insoluble forms (17, 18), the characteristics of the OMVs that appear to be the reservoir of such proteins had not been described previously. It is possible that the OMVs of X. fastidiosa might have still other functions, such as in transporting various proteinaceous and other cargoes, serving as decoys for predators such as viruses, and perhaps even transporting DSF signal molecules, as they do in certain other taxa (32, 42). The strong effect of these OMVs in blocking the attachment of the producing cells suggests that they have evolved for this function. Although this property has not been described previously for the OMVs produced by other taxa, it seems possible that such a trait might be beneficial to at least some of the species capable of producing OMVs. For example, bacterial motility generally is quite restricted in soils because of the strong retention of cells within soil particle pores and the strong interaction of cells with the inorganic components of the soil matrix (43, 44). The mobility of soil-borne bacteria might be enhanced by their production of OMVs with properties similar to those produced by X. fastidiosa that could minimize interactions with soil components. Likewise, rhizosphere bacteria are known to move along the surface of roots (45). Thus, it is possible that their production of OMVs could be modulated to facilitate movement along the roots by interfering with their attachment to the root surface. Although only a few studies have investigated the production of OMVs produced by plant-associated bacteria (17, 18, 46, 47), it seems likely that many such taxa might use OMVs not only to deliver virulence factors but possibly also to modulate attachment to plants and other aspects of such interactions. It is even possible that the development of bacterial biofilms on surfaces could be influenced by the liberation of OMVs. Bacteria might exploit appropriate the temporal or spatial patterns of OMV release to alter attachment to surfaces or even to create the complex 3D structures typical of many biofilms (48). Clearly, more studies are needed to determine better how commonly OMVs that have the antiadhesive properties exhibited by X. fastidiosa’s OMVs are made by bacteria and the contexts in which such particles would play a role in the bacterium’s lifestyle.
DSF-mediated signaling, linked to modulation of the levels of the intracellular signal cyclic di-GMP, strongly controls the production of OMVs in X. fastidiosa. Perhaps it is not surprising that cyclic di-GMP signaling is linked to vesiculation in X. fastidiosa, because this signaling system generally controls surface features of bacteria (49). DSF-mediated signaling controls a large number of genes in X. fastidiosa; the expression of more than 20% of its genes, including 11 transcriptional regulators, is altered in a ΔrpfF mutant, (14). Interestingly, at least 11 genes encoding OM-associated proteins were up-regulated in a ΔrpfF mutant. Thus, enhanced OMV release in this mutant might result from an imbalance in the content of specific envelope proteins. It has been reported that a lack of specific OM proteins in Salmonella typhimurium and Vibrio cholerae triggers the shedding of large numbers of OMVs (50, 51).
It is clear that OMV production is a highly regulated process (22, 41, 52, 53). Several factors that increase vesiculation in other Gram-negative bacteria have been reported (20, 22), but the molecular mechanisms of OMV biogenesis have not been fully deciphered. For instance, deletion of DegP, a periplasmic protease/chaperone that controls envelope stress caused by the presence of misfolded proteins, results in a hypervesiculation phenotype (31, 54). Moreover, disruption of connections between the outer and inner membranes and the peptidoglycan increases OMV production (33, 41, 50, 55, 56). Although X. fastidiosa harbors a degP ortholog (PD0231/XF0285) that is up-regulated upon heat shock (57), its expression does not appear to be RpfF-dependent (14).
The strong DSF-dependent production of OMVs by X. fastidiosa can be explained readily by considering the context-dependent behaviors necessary for such a bacterium to colonize successfully both plants and insect vectors—processes that require apparently incompatible traits. Retention of X. fastidiosa cells in the mouth parts of insect vectors that ingest large volumes of xylem sap through a narrow orifice would require the bacterium to be quite adhesive so as to be retained in a channel with such rapid and turbulent liquid flow (8, 9). DSF-mediated induction of a variety of cell-surface adhesins such as HxfA in those X. fastidiosa cells present in relatively large numbers in xylem vessels would enhance their opportunities to be retained by insects during feeding. However, such adhesive cells might be expected to bind to plant surfaces, thereby hindering their movement along and between xylem vessels. The apparent suppression of OMV production in X. fastidiosa cells experiencing high levels of DSF suggests that those more solitary cells, in which DSF levels should be relatively low, should produce relatively large numbers of OMVs that, in turn, would inhibit the binding of the pathogen to plant surfaces (Fig. 6). Conversely, cells present in relatively high local concentrations in which DSF levels would be elevated would shed few OMVs, and adhesins induced by DSF would be present on the cell surface, thereby facilitating the direct binding of cells to insect vectors. Such a strategy of DSF-regulated production would allow spatially separated release of OMVs in the plant, enabling them to contribute to plant colonization but avoiding the conflict that otherwise would arise in acquisition by insect vectors.
Fig. 6.
X. fastidiosa OMVs fine-tune surface adhesiveness in planta. X. fastidiosa WT releases OMVs containing XadA1 within xylem vessels. Cells are present in relatively low concentrations in xylem vessels where DSF would not have accumulated. A ΔrpfF mutant incapable of DSF production would release higher numbers of OMVs, which, by binding to the walls of xylem vessels, would reduce the ability of X. fastidiosa cells to bind to the plant, thereby facilitating movement through the plant and thus contributing to virulence.
Materials and Methods
Bacterial Strains and Culture Conditions.
The bacterial strains used in this work are described in SI Materials and Methods and are listed in Table S1. X. fastidiosa Temecula1 is the WT strain, and KLN59.3 is its gfp-marked derivative (3). KLN61 (11) is a ΔrpfF strain, and KLN121 (12) is its gfp-marked derivative. The ΔxadA1 mutant was constructed by transforming the WT strain with a construct based on pFXFkan-based suicide vector (13) as described in the SI Materials and Methods. Kanamycin-resistant colonies were screened for deletion of xadA1 (PD0731) by PCR using primers listed in Table S2.
X. fastidiosa strains were routinely grown for 5–7 d on periwinkle wilt (PW) medium (58) prepared with Gelrite (8 g/L) (PWG) at 28 °C and then were collected, suspended in 10 mM KPO4 (pH 7.4), and inoculated into PW, PD3 (58), or PIM6 broths to a desired initial cell density (usually to an OD600 of 0.05). Kanamycin (50 µg/mL) was included as required. X. fastidiosa broth cultures were grown for 4–7 d at 28 °C in a rotary shaker at 120 rotations per minute. E. coli BL21(DE3), P. aeruginosa PA14, and X. citri 306 were grown on lysogeny broth (10 g/L Bacto tryptone, 5 g/L yeast extract, 5 g/L NaCl) at 37 °C and were transferred to either PD3 (E. coli and X. citri) or PIM6 (P. aeruginosa) for OMV determination.
Scanning Electron Microscopy.
X. fastidiosa cells were removed from PWG plates and smeared on borosilicate coverslips or were cultured for 7 d in PD3 or 4 d in PIM6 broths in Falcon 50-mL conical centrifuge tubes (Fisher Scientific). To serve as a surface for cell attachment and biofilm development, a microscope glass slide was inserted inside the tube, and a borosilicate coverslip was fixed at the meniscus. The coverslips were detached carefully, and cell samples were processed as detailed in SI Materials and Methods before being sputter-coated with a thin (10-nm) layer of gold and analyzed in an FEI Quanta FEG 250 scanning electron microscope using 5 or 10 KV.
Deconvolution Microscopy.
For deconvolution microscopy, the lipophilic fluorescent dye FM4-64 was used in combination with DAPI (as a nucleic acids dye) to differentiate between particles consisting of lipids only (OMVs) and particles consisting of both lipids and DNA (cells). X. fastidiosa was collected from PWG plates and smeared over borosilicate coverslips, fixed briefly with fire, and placed in flat-bottomed six-well tissue-culture plates (Becton Dickinson). FM-4-64 and DAPI (Molecular Probes) were diluted in 10 mM KPO4 (pH 7.4) to final concentrations of 1 μg/mL and 0.2 μg/mL, respectively, and 1 mL of the solution was placed on each well. After 5 min of staining, each sample was washed twice with the buffer, and the coverslip was mounted immediately on a microscope slide. Immunolocalization of XadA1 was done using anti-XadA1 antiserum (35) as primary antibody and Alexa Fluor 594 goat anti-rabbit IgG (Life Technologies) antibody as secondary antibody. Briefly, heat-fixed cells on glass coverslips were incubated with a 1:1,000 dilution of the primary antibody in BSA-PBS (0.1% BSA fraction V in PBS, filtered sterilized) for 1 h at 37 °C and then were washed three times with BSA-PBS. The cells then were incubated with the secondary antibody in BSA-PBS for an additional 1 h in the dark, washed again similarly, and then stained with DAPI as described above. DAPI, FM-4–64, and Alexa Fluor 594 fluorescence were captured using a filter-optimized set on an Applied Precision DeltaVision Spectris DV4 deconvolution microscope. Images were analyzed using Zeiss 510 software and subsequently processed using Adobe Photoshop. For deconvolution images, samples were imaged at 0.05-µm (50-nm) z-axis intervals.
Isolation of OMVs.
X. fastidiosa was cultured for 7 d in PD3 or for 4 d in PIM6 broth, and cells were removed by centrifugation at 10,000 × g for 15 min at 4 °C. The supernatant then was centrifuged at 38,000 × g for 1 h at 4 °C. A portion of this supernatant was removed carefully and subjected to centrifugation at 150,000 × g for 4 h at 4 °C. The resulting pellet containing OMVs was resuspended in 10 mM KPO4 (pH 7.4) buffer and stored frozen at 4 °C or −80 °C until used in the attachment assays. A typical OMV preparation derived from 100 mL of bacterial culture was grown for 7 d in PD3; the OMV pellet formed after the ultracentrifugation steps was resuspended in 500 uL phosphate buffer.
OMV Enumeration by NTA.
To determine OMV concentrations in bacterial culture broths and saps, cells and debris were removed by two cycles of centrifugation at 16,000 × g for 20 min each at 4 °C, and the final supernatant was kept on ice. To assure complete cell removal, 5-µL aliquots were spotted onto PD3 plates and allowed to grow for 14 d. The OMV concentration was determined by NTA using a NanoSight LM10 system (NanoSight Ltd.) configured with a 405-nm laser and a high-sensitivity digital camera system. Videos were collected and analyzed using the NTA software (version 2.3), with the minimal expected particle size, minimum track length, and blur setting all set to automatic. Camera sensitivity and detection threshold were set to 14. Ambient temperature ranged from 20 to 22 °C. Samples were diluted in 0.22-µm filtered PBS (pH 7.4) to a concentration of 108–109 particles/mL. For each sample, three videos of 30–60 s duration were recorded, with sample mixing between recordings. Technical and biological replicates were averaged and plotted using Microsoft Excel. The OD600 of aliquots of the starting bacterial culture were measured; 10-fold serial dilutions were plated on PWG medium, and colonies were counted after 7–10 d to determine the number of cfu/mL. Then the numbers of OMV/mL were normalized for the cell concentration to yield the number of OMVs per cell.
Immunodetection of XadA1.
Immunodetection of XadA1 on cell lysate fractions separated by SDS/PAGE was performed as described in SI Materials and Methods using anti-XadA1 antiserum (35) and HRP-conjugated anti-rabbit IgG secondary antibody (1:5,000; Promega) as a secondary antibody followed by chemiluminescent detection.
Quantitative Reverse-Transcription PCR.
XadA1 transcript levels of WT and of ΔrpfF (KLN61) X. fastidiosa strains were determined by quantitative RT-PCR performed as described in SI Materials and Methods.
Attachment Assays.
Borosilicate glass coverslips previously sterilized in 80% ethanol were placed in six-well microtiter plates and submersed in 1-mL suspensions of OMVs diluted 1:1 in PIM6 broth (or in PIM6 alone as control) for 1 h before rinsing; then a suspension of X. fastidiosa Temecula1 WT cells (108 cells/mL) was added. After 1-h incubation at 28 °C, coverslips were rinsed three times with PIM6 to remove unbound cells; then cells were stained and mounted on microscopy slides and inspected by deconvolution microscopy using various magnifications, as described above. Staining with FM4-64 (1 µg/mL) was used to verify surface coverage by OMVs (magnification, 1,000×). Cells were visualized by staining with DAPI. Attachment parameters of cells to coverslips were quantified by analyzing the microscopic images taken at 20× magnification using ImageJ software (59). The amount of attachment of X. fastidiosa cells to glass surfaces was calculated as the percentage of the total pixels in a field of view exhibiting DAPI fluorescence; coverage of the glass surfaces treated with OMVs was normalized by that by cells on paired non-pretreated glass surfaces. The number of sites in a field of view in which one or more X. fastidiosa cells had adhered to the glass surface was counted and normalized for the number of such sites on non-pretreated glass slides. Fisher’s least significant difference (LSD) test was performed to compare samples, evaluated at P < 0.05.
Hindwings of Homalodisca vitripennis were washed twice in 80% ethanol, dried, and stored at 4 °C until use. Hindwings were submersed for 1 h in 1-mL suspensions of OMVs prepared as above (or in PIM6 alone as a control); then a suspension of gfp-marked X. fastidiosa KLN59.3 cells was added. After 2-h incubation at 28 °C, wings were rinsed three times with PIM6 to remove unbound cells. Wings then were mounted on microscopy slides, and the number of GFP fluorescent cells was determined by epifluorescence microscopy as described previously (9). When a non-gfp WT strain was used, cells were stained after the washing step with Syto 9 (Molecular Probes). Appropriate dilutions of wing macerates also were plated on PWG to determine the number of adhering cells (16).
Attachment of X. fastidiosa to plant tissues was assessed by measuring the retention of cells introduced into grape stems (Fig. S4B). OMV suspensions (25 µL) in 10 mM KPO4 buffer (pH 7.4) or PIM6 medium or control solutions of buffer or PIM6 alone were introduced into the end of fresh, detached stem segments (4 cm long and 0.6 cm in diameter) of Thompson Seedless grape (Vitis vinifera) using a 5-mL syringe attached to Nalgene 180 PVC tubing (diameter ca. 0.6 cm) that connected the syringe barrel to the basal end of the stem segment. The stem segments and attached syringes were incubated at 28 °C for 2 h. After incubation the segments were flushed gently with 5 mL of KPO4 buffer to remove unattached OMVs, and 25 µL of a suspension of strain KLN59.3 (108 cells per mL) was introduced, as described above. After 2 h bacterial cells were flushed from the stem segment as described above, and the number of cells removed by flushing was enumerated by plating appropriate dilutions on PWG medium as described above. The stems then were surface sterilized with 80% ethanol and macerated in 5 mL of KPO4 buffer using a Pro Scientific PRO200 homogenizer, and appropriate dilutions of macerates were plated on PWG containing kanamycin to determine the numbers of attached bacterial cells. The fraction of attached bacterial cells was calculated from the total number of cells introduced into the stem segment (sum of unattached and attached cells). In some assays, cells of X. fastidiosa were mixed with and introduced simultaneously with 25-µL aliquots of OMV suspension rather than being introduced after the introduction of OMVs. All other subsequent procedures were performed as described above.
Extraction of Xylem Sap.
Xylem sap was extracted from mature plants of Thompson seedless grapevine showing symptoms of Pierce disease 10 wk after inoculation with the WT, ΔrpfF (KLN61), or ΔxadA1 X. fastidiosa strains or from uninoculated plants using a pressure chamber as described previously (16).
Construction and Use of a Microfluidic Chamber.
A spin-coated photoresist wafer mold was purchased from the McGill University Advanced Nano Design Applications Facility. A poly(dimethylsiloxane)-borosilicate glass chamber containing observation channels of 50 × 100 µm were built as described in ref. 60. For cell-surface attachment assays, a modification in the procedure described in ref. 61 was performed. The chamber was filled initially with a flow medium consisting of a 1:1 mixture of PW broth medium and PBS (pH 8). The chamber then received 100 µL of a suspension (107 cells/mL) of strain PKLN59.3 previously incubated for 30 min with the same volume of a pure OMV suspension obtained as described above or with PBS alone. Unbound cells were washed away by slow-flowing medium at 3 mm/s, and 18–46 cells were left in the observation field in each run. After 10 min under the initial flow condition, the remaining bound cells were photographed and tracked individually during subsequent 10-min steps in which flow speeds were increased from 3 to 300 mm/s. At the end of each interval cells were photographed and counted. Cell counts were plotted in Microsoft Excel, and statistical significance between differences of treatment and control was determined at P < 0.02.
Supplementary Material
Acknowledgments
We thank Dr. Alessandra A. de Souza for providing anti-XadA1 polyclonal antibody, Dr. Vilma R. Martins for allowing us to use the NanoSight equipment, and Dr. Denise Schichnes for help with the deconvolution microscopy. This research was funded by the Pierce’s Disease Control Program of the California Department of Food and Agriculture. M.I. was supported by Vaadia-BARD Postdoctoral Fellowship Award FI-427-09 from the United States–Israel Binational Agricultural Research and Development Fund (BARD). P.A.Z. was supported by Postdoctoral Fellowship Award 2011/09409-3 from the São Paulo Research Foundation. A.M.d.S. was supported in part by Research Fellowship Award 306709/2009-0 from the National Council for Scientific and Technological Development.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414944111/-/DCSupplemental.
References
- 1.Purcell AH, Hopkins DL. Fastidious xylem-limited bacterial plant pathogens. Annu Rev Phytopathol. 1996;34:131–151. doi: 10.1146/annurev.phyto.34.1.131. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, Wistrom C, Lindow SE. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc Natl Acad Sci USA. 2008;105(7):2670–2675. doi: 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman KL, Almeida RPP, Purcell AH, Lindow SE. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microbiol. 2003;69(12):7319–7327. doi: 10.1128/AEM.69.12.7319-7327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q, Sun Y, Walker MA, Labavitch JM. Vascular occlusions in grapevines with Pierce’s disease make disease symptom development worse. Plant Physiol. 2013;161(3):1529–1541. doi: 10.1104/pp.112.208157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redak RA, et al. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol. 2004;49:243–270. doi: 10.1146/annurev.ento.49.061802.123403. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–271. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 7.Purcell AH, Finlay AH, McLean DL. Pierce’s disease bacterium: Mechanism of transmission by leafhopper vectors. Science. 1979;206(4420):839–841. doi: 10.1126/science.206.4420.839. [DOI] [PubMed] [Google Scholar]
- 8.Almeida RPP, Purcell AH. Patterns of Xylella fastidiosa colonization on the precibarium of sharpshooter vectors relative to transmission to plants. Ann Entomol Soc Am. 2006;99(5):884–890. [Google Scholar]
- 9.Killiny N, Almeida RPP. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl Environ Microbiol. 2009;75(2):521–528. doi: 10.1128/AEM.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaulieu ED, et al. Characterization of a diffusible signaling factor from Xylella fastidiosa. MBio. 2013;4(1):e00539–e12. doi: 10.1128/mBio.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman KL, Almeida RPP, Purcell AH, Lindow SE. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci USA. 2004;101(6):1737–1742. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Newman KL, Lindow SE. Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol Plant Microbe Interact. 2008;21(10):1309–1315. doi: 10.1094/MPMI-21-10-1309. [DOI] [PubMed] [Google Scholar]
- 13.de Souza AA, Ionescu M, Baccari C, da Silva AM, Lindow SE. Phenotype overlap in Xylella fastidiosa is controlled by the cyclic di-GMP phosphodiesterase Eal in response to antibiotic exposure and diffusible signal factor-mediated cell-cell signaling. Appl Environ Microbiol. 2013;79(11):3444–3454. doi: 10.1128/AEM.03834-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Li JL, Lindow SE. RpfF-dependent regulon of Xylella fastidiosa. Phytopathology. 2012;102(11):1045–1053. doi: 10.1094/PHYTO-07-12-0146-R. [DOI] [PubMed] [Google Scholar]
- 15.Ionescu M, et al. Diffusible signal factor (DSF) synthase RpfF of Xylella fastidiosa is a multifunction protein also required for response to DSF. J Bacteriol. 2013;195(23):5273–5284. doi: 10.1128/JB.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baccari C, Killiny N, Ionescu M, Almeida RPP, Lindow SE. Diffusible signal factor-repressed extracellular traits enable attachment of Xylella fastidiosa to insect vectors and transmission. Phytopathology. 2014;104(1):27–33. doi: 10.1094/PHYTO-06-13-0151-R. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto A, Huston SL, Killiny N, Igo MM. XatA, an AT-1 autotransporter important for the virulence of Xylella fastidiosa Temecula1. Microbiologyopen. 2012;1(1):33–45. doi: 10.1002/mbo3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voegel TM, Warren JG, Matsumoto A, Igo MM, Kirkpatrick BC. Localization and characterization of Xylella fastidiosa haemagglutinin adhesins. Microbiology. 2010;156(Pt 7):2172–2179. doi: 10.1099/mic.0.037564-0. [DOI] [PubMed] [Google Scholar]
- 19.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181(16):4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schertzer JW, Whiteley M. Bacterial outer membrane vesicles in trafficking, communication and the host-pathogen interaction. J Mol Microbiol Biotechnol. 2013;23(1-2):118–130. doi: 10.1159/000346770. [DOI] [PubMed] [Google Scholar]
- 22.Schwechheimer C, Sullivan CJ, Kuehn MJ. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry. 2013;52(18):3031–3040. doi: 10.1021/bi400164t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berlanda Scorza F, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol Cell Proteomics. 2008;7(3):473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Altindis E, Fu Y, Mekalanos JJ. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc Natl Acad Sci USA. 2014;111(15):E1548–E1556. doi: 10.1073/pnas.1403683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32(1):1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 26.Bomberger JM, et al. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5(4):e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74(1):81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans AGL, et al. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158(Pt 11):2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol. 1998;180(20):5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24(1):40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63(2):545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biller SJ, et al. Bacterial vesicles in marine ecosystems. Science. 2014;343(6167):183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 33.Bager RJ, et al. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis. Vet Microbiol. 2013;167(3-4):565–572. doi: 10.1016/j.vetmic.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Lyskowski A, Leo JC, Goldman A. In: Structure and Biology of Trimeric Autotransporter Adhesins. Bacterial Adhesion: Chemistry, Biology and Physics, Advances in Experimental Medicine and Biology. Linke D, Goldman A, editors. Vol 715. Berlin: Springer; 2011. pp. 143–158. [DOI] [PubMed] [Google Scholar]
- 35.Caserta R, et al. Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation. Appl Environ Microbiol. 2010;76(13):4250–4259. doi: 10.1128/AEM.02114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post DM, et al. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J Biol Chem. 2005;280(46):38383–38394. doi: 10.1074/jbc.M508063200. [DOI] [PubMed] [Google Scholar]
- 37.Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology. 1999;145(Pt 8):2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- 38.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 39.Hidalgo-Cantabrana C, et al. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol. 2014;80(1):9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burch AY, Zeisler V, Yokota K, Schreiber L, Lindow SE. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ Microbiol. 2014;16(7):2086–2098. doi: 10.1111/1462-2920.12437. [DOI] [PubMed] [Google Scholar]
- 41.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80(6):1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 43.Männik J, Driessen R, Galajda P, Keymer JE, Dekker C. Bacterial growth and motility in sub-micron constrictions. Proc Natl Acad Sci USA. 2009;106(35):14861–14866. doi: 10.1073/pnas.0907542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vos M, Wolf AB, Jennings SJ, Kowalchuk GA. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol Rev. 2013;37(6):936–954. doi: 10.1111/1574-6976.12023. [DOI] [PubMed] [Google Scholar]
- 45.Danhorn T, Fuqua C. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 46.Sidhu VK, Vorhölter FJ, Niehaus K, Watt SA. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol. 2008;8:87. doi: 10.1186/1471-2180-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury C, Jagannadham MV. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta. 2013;1834(1):231–239. doi: 10.1016/j.bbapap.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188(16):5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deatherage BL, et al. Biogenesis of bacterial membrane vesicles. Mol Microbiol. 2009;72(6):1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song T, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70(1):100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon KJ, Castelli ME, García Vescovi E, Feldman MF. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol. 2012;194(12):3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rath P, et al. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110(49):E4790–E4797. doi: 10.1073/pnas.1320118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwechheimer C, Kuehn MJ. Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli. J Bacteriol. 2013;195(18):4161–4173. doi: 10.1128/JB.02192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lappann M, Otto A, Becher D, Vogel U. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J Bacteriol. 2013;195(19):4425–4435. doi: 10.1128/JB.00625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol. 2013;195(2):213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koide T, Vêncio RZ, Gomes SL. Global gene expression analysis of the heat shock response in the phytopathogen Xylella fastidiosa. J Bacteriol. 2006;188(16):5821–5830. doi: 10.1128/JB.00182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis MJ, French WJ, Schaad NW. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr Microbiol. 1981;6(5):309–314. [Google Scholar]
- 59.Schneider CA, Rasband WS, Eliceiri KW. NIH image to imagej: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meng Y, et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187(16):5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De La Fuente L, et al. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl Environ Microbiol. 2007;73(8):2690–2696. doi: 10.1128/AEM.02649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.