Significance
Eukaryotic nuclear genomes store epigenetic information independent of DNA sequence, for example, in the form of 5-methylcytosine (5mC). In organisms as diverse as mammals and flowering plants, removal of 5mC profoundly impacts transcription and reproductive development. We found that the highly conserved protein DRE2 in Arabidopsis controls maternal gene expression and seed development by reducing DNA methylation in the maternal, central cell gamete. An antiapoptotic factor, essential from yeast to human, Dre2 is best known as an assembly component for iron and sulfur into cofactors termed Fe-S clusters and has continually emerging biological roles and functions. To our knowledge, we demonstrate the first epigenetic role for Dre2 in any organism.
Abstract
On fertilization in Arabidopsis thaliana, one maternal gamete, the central cell, forms a placenta-like tissue, the endosperm. The DNA glycosylase DEMETER (DME) excises 5-methylcytosine via the base excision repair pathway in the central cell before fertilization, creating patterns of asymmetric DNA methylation and maternal gene expression across DNA replications in the endosperm lineage (EDL). Active DNA demethylation in the central cell is essential for transcriptional activity in the EDL of a set of genes, including FLOWERING WAGENINGEN (FWA). A DME-binding motif for iron-sulfur (Fe-S) cluster cofactors is indispensable for its catalytic activity. We used an FWA-GFP reporter to find mutants defective in maternal activation of FWA-GFP in the EDL, and isolated an allele of the yeast Dre2/human antiapoptotic factor CIAPIN1 homolog, encoding an enzyme previously implicated in the cytosolic Fe-S biogenesis pathway (CIA), which we named atdre2-2. We found that AtDRE2 acts in the central cell to regulate genes maternally activated in the EDL by DME. Furthermore, the FWA-GFP expression defect in atdre2-2 was partially suppressed genetically by a mutation in the maintenance DNA methyltransferase MET1; the DNA methylation levels at four DME targets increased in atdre2-2 seeds relative to WT. Although atdre2-2 shares zygotic seed defects with CIA mutants, it also uniquely manifests dme phenotypic hallmarks. These results demonstrate a previously unidentified epigenetic function of AtDRE2 that may be separate from the CIA pathway.
Iron-sulfur (Fe-S) clusters are ancient, ubiquitous, and versatile cofactors. They can perform catalytic reactions, accept or donate single electrons, and stabilize protein conformation (1–3). A myriad of proteins functioning in different compartments of the cell require Fe-S clusters. Such proteins are abundant in plastids and mitochondria, which are the sites of the sulfur mobilization (SUF) and iron-sulfur cluster (ISC) pathways, respectively, that mediate the biogenesis of Fe-S clusters. Cytosolic and nuclear proteins in eukaryotes derive Fe-S clusters from the cytosolic iron-sulfur assembly (CIA) pathway. The CIA pathway is dependent on the ISC (1–3).
In each compartment, Fe-S biogenesis follows two steps: First, S and Fe are combined on an appropriate scaffold protein by means of dedicated donors for sulfur, iron, and electrons, and then the Fe-S cluster is transferred to recipient apoproteins, assisted by specialized carrier proteins. The key genes involved in Fe-S biogenesis have been identified in bacteria and yeast and are highly conserved and essential in eukaryotes. The diflavin reductase Tah18 and Derepressed for Ribosomal protein S14 Expression 2 (Dre2) form a short electron transfer chain; their interaction is essential in organisms from yeast to plants (orthologs in Arabidopsis thaliana: AtATR3 and AtDRE2, respectively) to mammals (4–6). The soluble P-loop NTPases Cfd1 and Nbp35 form a scaffold complex, present in plants as an Nbp35 homodimer (7), which transfers Fe-S clusters to Nar1, a protein with sequence similarity to iron dehydrogenases (8). Nar1 functions as an adapter for the targeting complex containing Cia1, a WD40 repeat protein (9), and Met18/Mms19 (10). The spectrum of CIA or non-CIA functions of each of these proteins in eukaryotes remains to be defined.
A decade ago, four CIA proteins, including Dre2, were shown to be involved in processes related to DNA replication (11). Today, numerous enzymes involved in the maintenance of genome integrity, including DNA polymerases, primases, and base excision repair (BER) DNA repair enzymes, are known to be Fe-S–dependent enzymes (3, 10, 12–14). To initiate BER, bacterial MutY and EndoIII glycosylases search cooperatively for DNA lesions based on their redox-active Fe-S cofactors (14). After detection, a base lesion is excised by cleavage of the sugar-phosphate DNA backbone and then replaced by repair DNA polymerases and DNA ligases. In Arabidopsis, the four BER glycosylase homologs—DEMETER (DME), ROS1, DML2 and DML3 (15, 16)—are specialized in excising 5-methylcytosine (5mC) rather than damaged bases, thus catalyzing active DNA demethylation. The activity of DME is dependent on a conserved protein domain for Fe-S cluster binding (17). The gene expression of two DME target genes and the DNA methylation levels of two ROS1 target genes are affected in mutants of CIA proteins, AtNAR1 (18) and ASYMMETRIC LEAVES1/2 ENHANCER7 (AE7) (19), respectively. To our knowledge, these reports represent the first links between CIA proteins and the epigenetic process of active DNA demethylation.
ROS1, DML1, and DML2 demethylate genes in sporophytic tissues (20). DNA demethylation by DME is restricted to a single-cell gamete, the central cell (Fig. 1A, shown in blue), which divides numerous times after fertilization to form the placenta-like tissue, the endosperm (Fig. 1D, shown in blue). The excision of 5mC by DME in the central cell is coupled to active transcription and precedes fertilization, resulting in asymmetric patterns of DNA methylation and gene transcription at maternal and paternal alleles in the endosperm. The active transcription state of genes targeted by DME is epigenetically controlled; once initiated in the progenitor central cell, it is inherited across DNA replications in the endosperm based on DNA methylation levels in a process termed here maternal activation in the endosperm lineage (EDL). The sole mechanism known to activate the DME target, FLOWERING WAGENINGEN (FWA), is maternal activation in the EDL. No developmental phenotypes have been detected in loss-of-function fwa plants (21); however, in maternal dme mutants, seeds abort at the end of endosperm development, and the mutant allele does not transmit to the next generation. dme homozygotes are lethal (15); thus, DNA demethylation of at least some DME target genes is a biologically relevant, albeit not completely understood, epigenetic phenomenon.
Fig. 1.
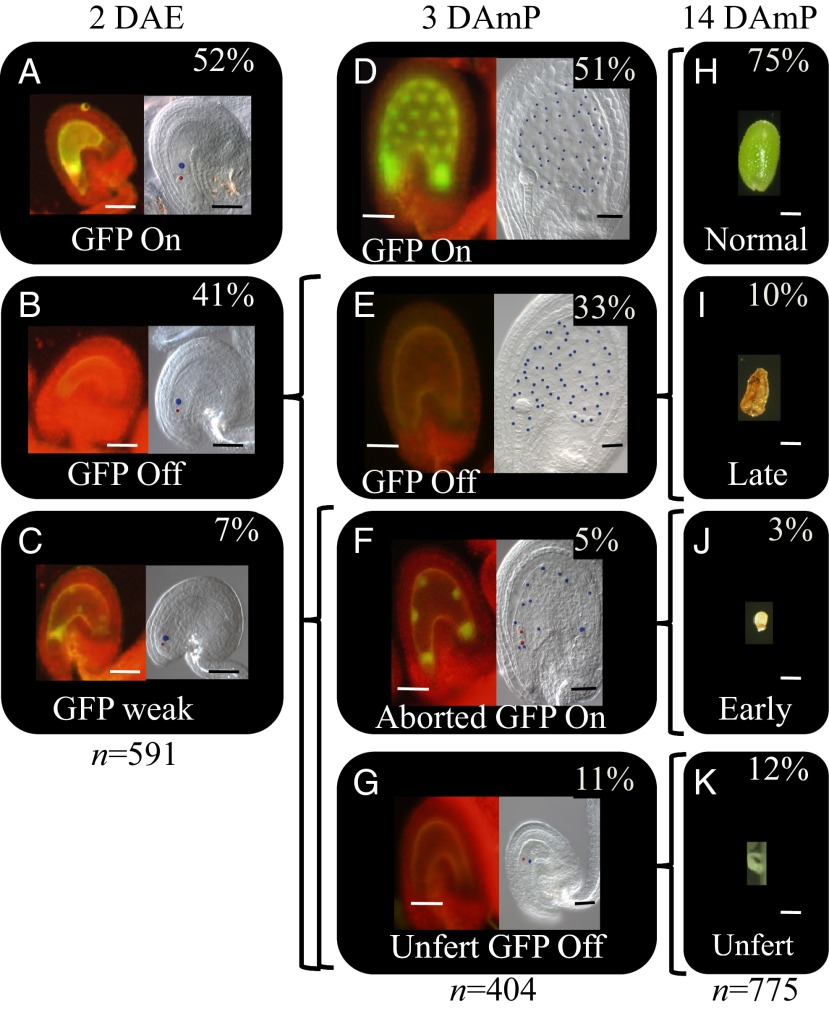
The gametophytic function of AtDRE2 and developmental fate of FWA-GFP and seed phenotype classes in atdre2-2+/−;pFWA-GFP. (A–G) Representative images of FWA-GFP classes (A–C and D–G, Left) and the corresponding Nomarski cleared ovules/seeds (A–C and D–G, Right), in unfertilized ovules at 2 DAE (A–C), in seeds at 3 DAmP with WT pollen (D–G), and at 14 DAmP (H–K). The ovules/seeds cleared separately based on the GFP class. Percentages represent the proportion of each segregating GFP class out of a total number (n) of ovules/seeds. Curly brackets represent developmental fate, and the text presents phenotypic nomenclature. Unfert, unfertilized. Egg cell (A–C and F–G) and embryo (F) nuclei are shown in red, and central cell (A–C) and endosperm (D–G) nuclei are in blue. (Scale bars: 50 μm.)
A pFWA-GFP reporter can easily monitor maternal activation of FWA in the EDL (21) (Fig. 1 A and D, Left). Following ethyl methanesulfonate (EMS) mutagenesis of an Arabidopsis pFWA-GFP line, we isolated a mutant allele in the ortholog of the yeast CIA protein Dre2/human CIA protein, antiapoptotic factor CIAPIN1 (anamorsin), which has reduced expression of pFWA-GFP. Dre2 has an N-terminal domain with a typical overall S-adenosylmethionine (SAM) structure (22, 23) and two C-terminal Fe-S cluster motifs (4). Dre2 can localize to the cytosol, mitochondria, and nucleus in a reactive oxygen species-dependent manner in yeast (24). In mice, CIAPIN1 is an antiapoptotic molecule essential for definitive hematopoiesis (25). In Arabidopsis, the Dre2 ortholog AtDRE2 has been confirmed as the cytosolic partner of AtATR3 and shown to be essential for early embryo development (26).
We found that AtDRE2 is broadly expressed during the reproductive phase, but it is in the central cell that AtDRE2 expression is responsible for maternal activation of FWA, placing AtDRE2 function in the spatiotemporal window of DME action. Moreover, the FWA-GFP expression defect in atdre2-2 was partially suppressed genetically by a mutation in the maintenance DNA methyltransferase MET1; at least four DME target genes were not activated and their methylation levels were not decreased in the atdre2-2 endosperm, and maternal seed defects were detected. These results implicate AtDRE2 in DNA methylation control related to maternal activation in the EDL, an epigenetic function hitherto unrecognized for this protein in any organism.
Results
ALAC4 Encodes a Highly Conserved Fe-S Cluster Biogenesis Protein.
Removal of 5mC from the 5′ region of FWA is required for FWA activation and can be monitored by a pFWA-GFP reporter (21). After mutagenesis of a pFWA-GFP line, several mutants with gametophytic defects, termed alac (alarm clock for FWA imprinting), were isolated based on a 1:1 ratio of segregating “GFP on” and “GFP off” fluorescence phenotypes both before and after fertilization (18). Here we map-based cloned the alac4 mutation (Fig. S1) and identified a single nucleotide deletion at position 1525 from the ATG of At5g18400 that encodes the ortholog (6) of yeast Dre2 (11) and human CIAPIN1 (25), which is involved in Fe-S cluster biogenesis and other processes. The deletion in alac4 is predicted to cause a frame shift and stop codon after another 16 amino acids. The AtDRE2 protein has a generic SAM domain at the N terminus and several conserved cysteines, grouped in two motifs, C202X2CXC207 and C233X2CX7CX2C247. A T-DNA insertion allele, atdre2-1 (6), was introgressed into pFWA-GFP and had similar phenotypes as the EMS allele (Fig. S1 B and C). (alac4 was renamed atdre2-2.) We reintroduced a genomic fragment starting 703 bp upstream of the AtDRE2 ATG and fused with EGFP (pAtDRE2:AtDRE2-EGFP) or with an EGFP and a nuclear localization signal (pAtDRE2:AtDRE2-EGFP-NLS) into atdre2-2+/−;pFWA-GFP. In the progeny of four atdre2-2+/−;pAtDRE2:AtDRE2-EGFP and two atdre2-2+/−;pAtDRE2:AtDRE2-EGFP-NLS independent lines, we identified atdre2-2−/−. These lines were found to complement the seed phenotype (Fig. S2). Taken together, these results confirm that ALAC4 (AtDRE2) is At5g18400 and show that pAtDRE2:AtDRE2-EGFP and pAtDRE2:AtDRE2-EGFP-NLS are functional constructs.
Mutation of AtDRE2 Impairs pFWA-GFP Maternal Activation in the EDL.
We investigated the maternal gametophytic nature of the atdre2-2 mutation. Before fertilization, at 2 d after emasculation (DAE), 52% of ovules were classified as GFP on (Fig. 1A), 41% were GFP off (Fig. 1B), and in 7% the GFP appeared diminished and/or localized into foci, termed “GFP weak” (Fig. 1C). The nature of the latter class is unknown. The central cell and egg cell appeared normally differentiated in all ovule classes (Fig. 1 A–C, Right). After fertilization, at 3 d after manual pollination (DAmP), in crosses between atdre2-2 +/−;pFWA-GFP as female and WT as pollen, 51% of seeds developed normally and were GFP on (Fig. 1D). This class likely represents seeds that have inherited a WT AtDRE2 allele. The phenotype of the remaining seeds, expected to inherit the maternal mutant allele, was found to be heterogeneous and categorized into three classes based on images of seeds cleared after initial observation of their fluorescence. In the largest class, 33% of GFP off seeds, the developmental stage of the embryo and endosperm was comparable to that in normal WT seeds (Fig. 1E). Approximately 5% of the seeds, termed “aborted GFP on,” were smaller, had fewer but enlarged endosperm nuclei compared with WT, and demonstrated GFP fluorescence. The abortion stage for this class was not synchronized, and both the embryo and endosperm divisions arrested early (Fig. 1F), suggesting the existence of primary defects in each lineage. Finally, 11% of ovules remained unfertilized and were GFP off (Fig. 1G).
Based on the ∼1:1 segregation of the WT (Fig. 1 A and G) and abnormal mutant phenotypes (Fig. 1 B, C, H–J) before and after fertilization, we conclude that atdre2-2 has a gametophytic defect in activating pFWA-GFP. In addition to the unfertilized ovules and early-aborted seeds, atdre2-2 +/− also manifested later seed defects (Fig. 1I); see also Gametophytic Function of AtDRE2 below.
We also monitored the reporter gene WT:mutant ratio in ovules expressing reporters for two other genes that are maternally activated in the EDL, MEDEA (15) and FIS2 (27) (Fig. S3). In both cases, the ratio was close to 1:1, confirming the gametophytic effect in atdre2-2 and suggesting that AtDRE2 is essential for the common mechanisms that regulate maternal activation in the EDL.
AtDRE2 Is Broadly Expressed During the Reproductive Phase.
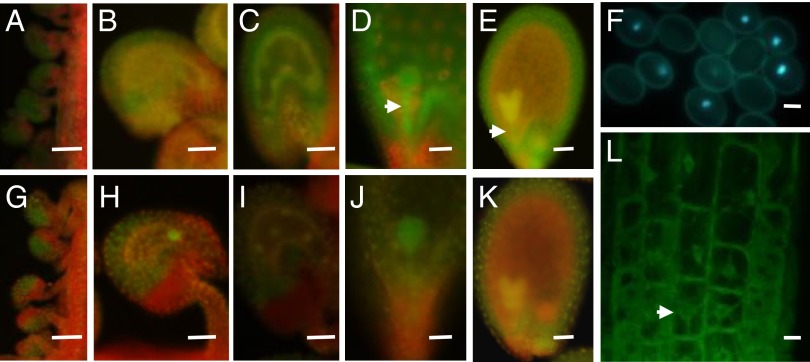
To better understand AtDRE2 function during the reproductive phase, we first characterized its expression in the two types of complementing transgenic lines (Fig. S2). Compared with pAtDRE2:AtDRE2-EGFP, the pAtDRE2:AtDRE2-EGFP-NLS lines allowed better visualization of the nuclei of the female gametophyte, which is embedded within the ovule integument. We found that AtDRE2 is expressed in the central cell and in up to eight endosperm nuclei (Fig. 2 B, C, H, and I) but also has a broader expression (Fig. 2 A–E, G–K), including in the ovule and seed integuments throughout flower and seed development (Fig. 2 A–E, G–K), in the early stage of the embryo, and in the suspensor (Fig. 2 D, E, J, and K). In the male gametophyte, pAtDRE2:AtDRE2-EGFP-NLS expression was found in the vegetative cell of the pollen (Fig. 2F). AtDRE2:AtDRE2-EGFP expression appeared cytosolic. We closely monitored the subcellular localization of pAtDRE2:AtDRE2-EGFP in 4-wk-old roots (Fig. 2L) using confocal microscopy, and found AtDRE2-EGFP in the cytosol as well as in the nucleus. These results indicate that AtDRE2 may function in the cytosol and nucleus.
Fig. 2.
AtDRE2 expression monitored by EGFP fusion constructs. Expression in representative transgenic lines of pAtDRE2:AtDRE2-EGFP without a nuclear localization signal (A–E and L) and with a nuclear localization signal (F–K). (A and G) Expression in ovule integuments from immature floral buds. (B and H) Expression in ovule integuments and in the central cell at 2 DAE. (C and I) Expression in seed integument and in up to eight endosperm nuclei at 1 day after fertilization. (D, E, J, and K) In the embryo and suspensor (arrow), expression is strongest from the globular to heart shape/early cotyledon embryo stages. (F) Fluorescence in the vegetative nucleus of the pollen in a heterozygote transgenic pAtDRE2:AtDRE2-EGFP–NLS. (L) Confocal microscopy of root cells in a transgenic pAtDRE2:AtDRE2-EGFP showing cytosolic and nuclear (arrow) fluorescence. (Scale bars: 50 μm in A–K; 10 μm in F and L.)
AtDRE2 Acts in the Central Cell to Activate FWA-GFP.
To investigate which cells required AtDRE2 for pFWA-GFP activation, we set up complementation tests in which we restricted the expression of AtDRE2 to prefertilized atdre2-2+/− gametophytic cells using two promoters. The At4g12250 (pFM1 promoter) drives specific expression during megagametogenesis (28), and the At2g24840 (pDIANA) promoter drives expression exclusively in the unfused polar nuclei and the central cell (29). We recovered several lines with each promoter that rescued the atdre2-2+/−;pFWA-GFP phenotype at 2 DAE and 3 DAmP (Fig. 3A and Fig. S4), indicating that in the female gametophyte, AtDRE2 most likely functions in the central cell and is sufficient to activate pFWA-GFP. This result also indicates that the atdre2-2 mutation is not dominant with regard to maternal gene activation in the EDL.
Fig. 3.
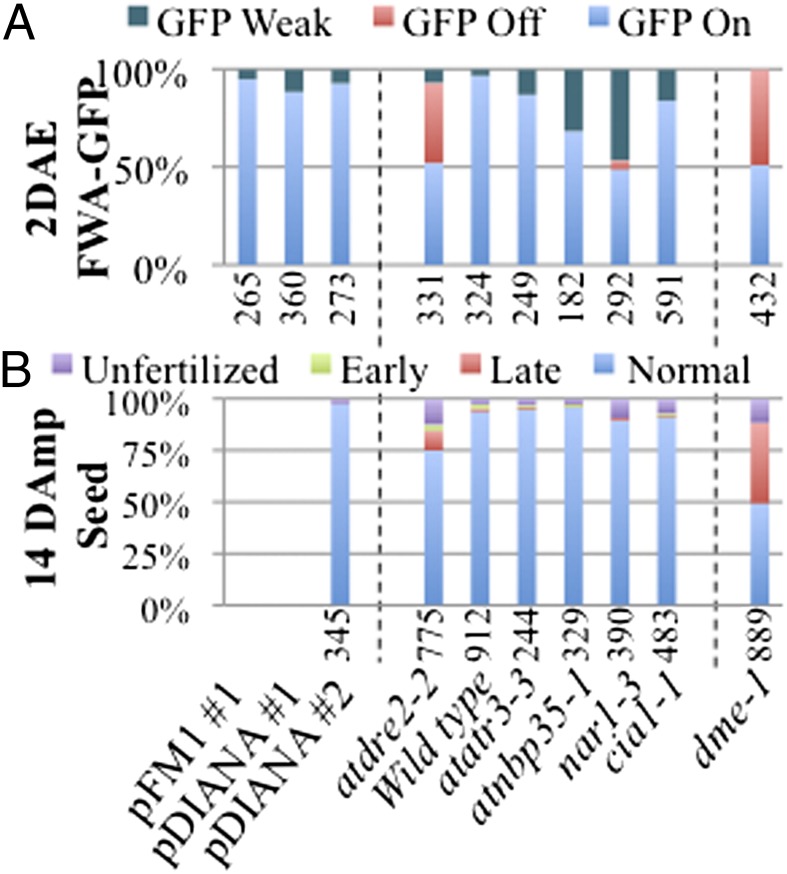
FWA-GFP and seed phenotypes in transgenic lines and CIA mutants. (A) FWA-GFP in atdre2-2+/−;pFM1:AtDRE2#1, atdre2-2+/−;pDIANA:AtDRE2#1, atdre2-2+/;pDIANA:AtDRE2#2, and CIA and DME mutants (atdre2-2, atatr3-3, atnbp35-1, nar1-3, cia1-1, and dme-1) at 2 DAE. (B) Seed phenotype at 14 DAmP for the same genotypes. Data for dme-1 are at 14 DAP. The numbers of ovules/seeds analyzed are indicated below each graph.
Gametophytic Function of AtDRE2.
Although atdre2-2 has been reported to be embryo-lethal (6), we also found a maternal function of AtDRE2 in the central cell. What is the seed phenotype for each GFP seed class when atdre2-2 is present maternally? To answer this question, we directly compared all of the seed classes between the latest stage at which FWA-GFP can be monitored, 3 DAmP (Fig. 1 D–G), and the end of seed development, 14 DAmP (Fig. 1 H–K), in atdre2-2+/− × WT crosses. Because FWA-GFP fluorescence diminishes by 3 DAmP, it was not possible to monitor GFP activity after this stage. The proportion of unfertilized GFP off seeds (11%; (Fig. 1G) corresponds well with that of seeds that appear small and white at 14 DAmP (12.1%; Fig. 1K). The 5% of aborted GFP on seeds (Fig. 1F) most likely represent the 3% of seeds that appear brown and shrunk and abort early (Fig. 1J).
What is the developmental fate of the 33% class of GFP off seeds (Fig. 1E)? The remaining mutant seeds at 14 DAmP, albeit only approximately 10% (Fig. 1I), were larger than early-aborted seeds, contained liquid endosperm and a small embryo, or had a collapsed brown appearance. Therefore, not all GFP off seeds at 3 DAmP manifest a mutant seed phenotype at 14 DAmP. In the reciprocal cross, pFWA-GFP × atdre2-2+/−;pFWA-GFP, we found no GFP off seeds at 3 DAmP and, at 14 DAmP, only the early-aborted class differed from the control (Table S1), indicating that the gametophytic effect of atdre2-2 on seed development is maternal-specific.
Because the maternal mutant atdre2-2 allele appeared to reduce seed viability, we measured the efficiency of atdre2-2 transmission by genotyping seedlings obtained from reciprocal crosses between atdre2-2+/− and WT. We found that the maternal transmission was reduced to 22% (Table S2), in close agreement with the total nonviable seed percentage; however, the findings that atdre2-2 is maternally transmitted and that less than 50% of seeds manifest a phenotype indicate that the gametophytic function of AtDRE2 in the central cell results in a low-penetrance seed phenotype. The paternal transmission of atdre2-2 was also reduced, to 17% (Table S2). These results demonstrate previously unrecognized female and male gametophytic roles of AtDRE2.
AtDRE2 Activates at Least Six DME Target Genes.
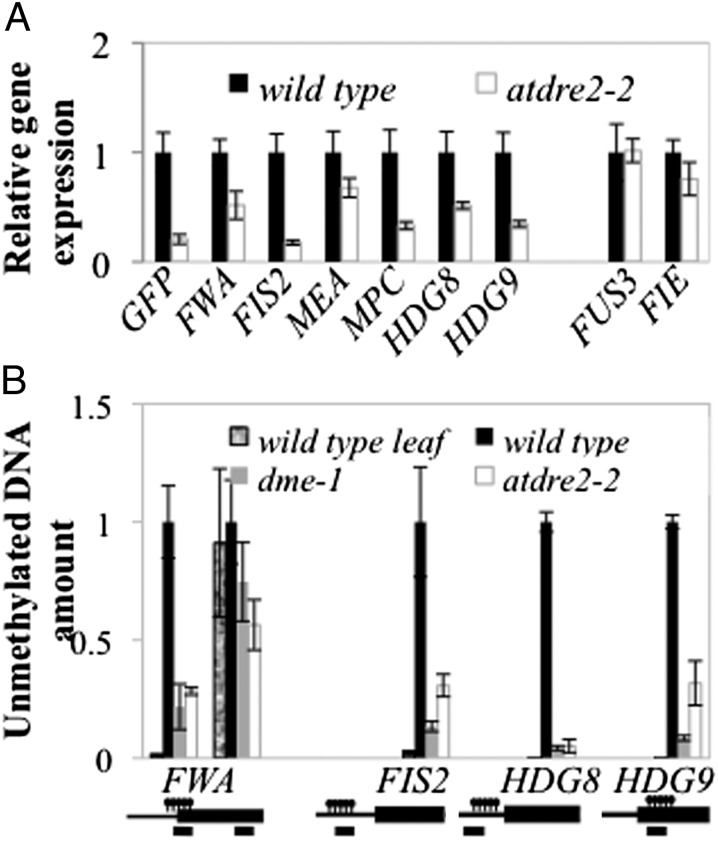
The results from transgenic reporters for FWA, MEA, and FIS2 (Fig. 1 and Fig. S3) suggest that AtDRE2 influences general maternal gene activation in the EDL. To test whether native genes representing DME targets are normally activated, we dissected GFP off seeds at 3 DAmP from atdre2-2 and GFP on seeds from pFWA-GFP. No other mutant seed class was analyzed. We selected six DME target genes (FWA, MEA, FIS2, MPC, HDG8, and HDG9) and two other genes expressed in the endosperm but not regulated by DNA methylation (FIE and FUS3). Using RT-qPCR, we found that the expression of all six DME target genes was reduced in the atdre2-2 mutant compared with WT (Fig. 4B), wheres the expression of DME nontargets did not change. These results indicate that AtDRE2 is essential for maternal activation in the EDL and strengthen the possibility that AtDRE2 acts to reduce DNA methylation.
Fig. 4.
DNA methylation and gene expression defects in atdre2-2. (A) Gene expression of selected DME targets measured by RT-qPCR in 3 DAmP seeds dissected under a fluorescence microscope from pFWA-GFP and atdre2-2. (B) Estimation of endosperm DNA methylation levels from WT, dme-1, and atdre2-2 in 3 DAmP seeds. Equal amounts of seed DNA were digested with McrBC, and undigested genomic DNA was used as a standard for absolute quantification of specific regions. The leaf sample was used as a control for the digest. Promoters and genes are represented by black lines and boxes; the positions of methylated regions and PCR amplicons are indicated above and below, respectively. Values in A and B are mean ± SE from three or four biological replicates, normalized to UBIQUITIN10 (A) and an unmethylated region (B).
AtDRE2 Reduces the DNA Methylation Level of at Least Four DME Target Genes.
We next optimized a quantitative assay for estimating the level of DNA methylation in endosperm from 3 DAmP seed total DNA (Fig. S5), dissected as above, using the DNA methylation-specific enzyme McrBC. AtDRE2 is expected to reduce DNA methylation in the endosperm, and DME is also confirmed to perform this role. Therefore, in mutant GFP off seeds from atdre2-2;pFWA-GFP and dme-1;pFWA-GFP, more DNA may be methylated and digested by McrBC than in WT seeds. We found that after digestion with McrBC, the amount of DNA recovered from methylated regions of FWA, FIS2, HDG8, and HDG9 was reduced in both mutants compared with WT, but there was no difference in the unmethylated control FWA region (Fig. 4B). These results indicate that AtDRE2 has activity toward DNA demethylation.
Antagonistic Interaction Between AtDRE2 and MET1.
Lack of activation of pFWA-GFP in the central cell, predicted to be associated with a defect in DNA demethylation, was found in atdre2-2. Moreover, pFWA-GFP remained inactive at 3 DAmP in atdre2-2 seeds, despite the fact that AtDRE2 expression is not needed during this period but is needed only before fertilization in the central cell, as indicated by the complementation with pFM1:AtDRE2 and pDIANA:AtDRE2 (Fig. 3 A and B). Thus, AtDRE2 may directly or indirectly reduce DNA methylation in the central cell at FWA, a state that can be inherited in the endosperm. If this were the case, then the GFP off phenotype in atdre2-2;pFWA-GFP ovules and seeds should be suppressed by the lack of DNA methylation, found in, for example, the DNA methylation maintenance mutant met1-3. To test this possibility, we constructed the double-heterozygote mutant atdre2-2;met1-3;pFWA-GFP, in which met1-3 is always maintained as a heterozygote F1 from the pollen. The proportion of GFP on class increased in atdre2-2;met1-3;pFWA-GFP compared with atdre2-2;pFWA-GFP, from 52% to 65% at 2 DAE and from 51% to 70% at 3 DAmP (Fig. S6), indicating that in ∼15% of ovules and 20% of seeds, the presence of met1-3 fully rescues the atdre2-2 defect in activating pFWA-GFP. We confirmed the presence of maternal atdre2-2 and met1-3 in 16% or more of ovules/seeds by estimating the maternal transmission of each allele (Table S2). These results support the hypothesis that atdre2-2 has a DNA demethylation defect, and suggest that MET1 and AtDRE2 have antagonistic DNA methylation effects.
Zygotic Function of AtDRE2.
Siliques of self-pollinating atdre2-2+/− contained nearly 25% early-aborting seeds (Fig. S4B) in most experiments or late-aborting seeds in some experiments (Table S1), indicating a zygotic function. To monitor atdre2-2 defects other than those derived from the central cell function, we analyzed the seed phenotype of transgenic lines complementing the FWA-GFP defect in one atdre2-2+/−;pFM1:AtDRE2 line and two atdre2-2+/−;pDIANA:AtDRE2 lines. When self-pollinated, these three transgenic lines had 25% late-aborted seeds (Fig. S4B), but when pollinated with WT pollen (pDIANA:AtDRE2 line 2), the seeds developed normally (Fig. 3B). These results confirm that AtDRE2 has a zygotic function, as previously suggested by the segregation of a mutant seed phenotype in atdre2-1 (6).
Among CIA Mutants, Three dme Hallmarks Are Unique to atdre2.
DME governs DNA demethylation in the central cell and is Fe-S dependent, as discussed above. AtDRE2 regulates the same process and is also known to bind Fe-S clusters (6). To establish whether the CIA pathway might influence DME and AtDRE2 activities, we analyzed three hallmarks of the dme phenotype in mutants of the following CIA proteins: AtATR3, the binding partner of AtDRE2; the scaffold protein AtNBP35; AtNAR1; and the CIA targeting complex protein CIA1. To monitor the first dme hallmark, activation of pFWA-GFP in the EDL, we introgressed pFWA-GFP into strong alleles for all of the mutants, as suggested by embryo lethality (7, 18, 26). In contrast to atdre2-2 and dme-1, no other CIA mutant had 50% GFP off ovules (Fig. 3A). In atnar1-3, pFWA-GFP was not normally activated in approximately one-half of ovules; 47% of ovules had a GFP weak phenotype, as was found for a small percentage of atdre2-2 ovules (Fig. 1C), and only 5% of ovules were GFP off. In the remaining CIA mutants, the sole mutant ovule class was GFP weak, with 32% in atnbp35-1, 13% in atatr3-3, and less than 5% in cia1-1. At 3 DAmP, no major class of GFP off seeds was seen in any mutant (Fig. S4A). We conclude that pFWA-GFP inactivation in the EDL is strong in atdre2-2, weak in atnar1-3, and not detectable in the other mutants.
The second notorious dme hallmark is maternal seed defects. Most CIA mutant proteins are suggested to be zygotic embryo-lethal. Indeed, in self-pollinated siliques, we found approximately 25% early seed abortion in atatr3-3, atnbp35-1, atnar1-3, and cia1-1 at 3 DAmP (Fig. S4A, aborted GFP on) and 14 DAmP (Fig. S4B), which may correspond to zygotic defects. To monitor maternal seed defects, we pollinated all heterozygote mutants with WT pollen (Fig. 3B). In all mutants except atdre2-2, more than 95% of seeds were normally developed at 14 DAmP, similar to control crosses in which WT was the maternal parent. This contrasts with the 50% seed abortion in penetrant dme-1.
We conclude that among all CIA protein mutants investigated here, only atdre2-2 has detectable maternal defects. These data also predict that the third dme-1 hallmark, maternal transmission defects, might not be present in the six CIA protein mutants, a possibility tested and confirmed for atr3-3, atnbp35-1, and atnar1-3 (18). Our data also demonstrate the essential zygotic function of all CIA proteins, including AtDRE2. Taken together, these data identify strong similarities between atdre2 and dme mutant phenotypes, and suggest that most CIA proteins do not influence the activities of DME or AtDRE2 related to gametophytic function.
Discussion
Dre2 is essential in organisms from yeast to humans. Here we distinguished two AtDRE2 roles during reproductive phase in Arabidopsis: a previously unidentified epigenetic role in maternal activation in the EDL via DNA demethylation and an essential zygotic role, already known and shared with most CIA components.
Epigenetic Role of AtDRE2.
We discovered that AtDRE2 has previously unidentified male and female gametophytic roles (Table S2) and focused on the central cell function of AtDRE2 using pFWA-GFP. Our results demonstrate that the AtDRE2 female gametophytic function is epigenetic, based on the following observations: (i) pFWA-GFP activation requires DNA demethylation in the central cell, and atdre2-2;pFWA-GFP is not active in the central cell (Fig. 1B); (ii) the expression of AtDRE2 driven by pFM1 and pDIANA in the central cell can rescue pFWA-GFP expression in both the central cell (Fig. 3A) and the endosperm (Fig. S4A) without protein expression at this stage; (iii) the defect in GFP activation in atdre2-2;pFWA-GFP can be restored when met1-3 is also present (Fig. S6); and (iv) six genes regulated by DNA demethylation in the central cell (30) cannot be activated when AtDRE2 is not functional (Fig. 4A), and four of these genes exhibit increased DNA methylation in the early endosperm (Fig. 4B). To the best of our knowledge, this represents the first report of an epigenetic function for Dre2 in any organism.
Zygotic Role of AtDRE2.
Once the gametophytic central cell function in atdre2-2 was complemented, early seed development also could be completed normally in all independent transgenic lines analyzed (Fig. 3 A and B), but 25% of seeds aborted late in self-pollinated plants (Fig. 3C). Although it is possible that neither of the two promoters used to express AtDRE2 faithfully mimicked the native AtDRE2 expression domain responsible for normal maternal gene expression in the central cell, several clues favor the view that the lack of late seed development complementation may indicate an additional requirement for AtDRE2, possibly in the suspensor/embryo. First, AtDRE2 is highly expressed at this stage in the embryo, as monitored with pAtDRE2:AtDRE2-EGFP. Second, if the late-aborting seed phenotype were related to a lack of AtDRE2 function in the EDL, then the defect would be expected to be maternal; however, our data indicate that it is zygotic, and the same is true for five CIA protein mutants tested (Fig. 3B). This finding also suggests that AtDRE2 has a CIA function.
Does CIA Assemble Fe-S Clusters for DME and AtDRE2?
As a nuclear Fe-S–dependent enzyme, DME is expected to obtain these cofactors in the cytosol via the CIA. AtDRE2 also binds Fe-S clusters (6), but these clusters’ involvement in AtDRE2 functions is unknown. Surprisingly, the common gametophytic phenotypes of atdre2-2 and dme-1 are distinct from the collective zygotic mutant phenotypes of CIA proteins representing all steps of the pathway. Thus, given the lack of redundancy for the CIA proteins tested, our genetic data do not currently support the existence of a canonical CIA pathway upstream of DME. Nonetheless, the lack of common mutant phenotypes between dme and four CIA mutant genes may indicate that a subset of DME target genes is less sensitive to Fe-S cluster depletion and/or is not associated with the phenotypes under investigation here.
In addition, in the case of AtDRE2, the lack of detectable pFWA-GFP and maternal seed phenotypes of the other CIA pathway mutants makes it unlikely that the CIA influences the gametophytic function. Alternatively, the SAM domain, and not the two Fe-S cluster domains, may be critical for the central cell function of AtDRE2. Our genetic, biochemical, and cytologic data may support a scenario in which the SAM domain of AtDRE2 may inhibit the maintenance of DNA methylation by MET1, possibly creating the preferred substrate for DME, namely hemimethylated templates (31). Consistent with this scenario are the antagonistic genetic interaction between AtDRE2 and MET1 (Fig. S6), the function of AtDRE2 in reducing DNA methylation (Fig. 4B), and the possible AtDRE2 nuclear function (Fig. 2L). It was recently suggested that, owing to subtle structural differences of the SAM binding pocket and an absence of in vitro methyltransferase activity, anamorsin inhibits methyl transfer (22, 23, 32). Interestingly, the SAM domain of another nuclear protein has recently been associated with active DNA demethylation (33). Future work elaborating on these possibilities will expand our knowledge of Fe-S clusters and provide insight into the biological context of DNA demethylation.
Materials and Methods
Plant Material.
The background used was always A. thaliana Columbia-0 (Col-0) except in the McrBC assay, where Landsberg erecta (Ler) served as the paternal genotype. The pFWA-GFP line (25), homozygous for the insertion, was used to introgress dme-1;Ler nine times, the EMS-generated alac4+/−;pFWA-GFP five times, and all complementation lines and T-DNA insertions at least once. The following insertion mutants were obtained from the Arabidopsis Biological Resource Center or the Nottingham Arabidopsis Stock Centre: for AtDRE2, SALK_074261, SAIL_1222, GK-368B03; for nbp35-1, SALK_056204; for atatr3-3, GK-004E05-014807; and for cia1-1, SALK_060584.
Histological Analysis.
Seeds were cleared in a drop of chloral hydrate, glycerol, and water mixture (8 g:1 mL:2 mL) under a cover glass at room temperature for at least 4 h. Bright-field images were captured using a Zeiss Axioimager M1 microscope equipped with a Zeiss AxioCam MRc 5 and Nomarski optics. Images of roots were obtained with an Olympus FluoView FV1000 confocal laser scanning microscope.
Generation of Transgenes.
Vectors were constructed by first cloning amplified PCR fragments into pENTR/dTOPO (Invitrogen). The 35S promoter from the Gateway binary vector series (34) was excised using SacI and SpeI and replaced with PCR-amplified fragments of pFM1 and pDIANA for pFM1:AtDRE2 and pDIANA:AtDRE2, respectively. Genomic or coding sequences were introduced into such modified binary vectors using LR clonase (Invitrogen), and plasmids were electroporated into Agrobacterium tumefaciens EH105. T1 transgenic plants were selected on MS plates with the appropriate antibiotic and were crossed into atdre2-2;pFWA-GFP. Seeds from subsequent generations (up to T4) were reselected for the transgene, and up to 24 individuals were genotyped for the atdre2-2 mutation using PCR (5′-ggcaaagaaaccttcttggaa-3′ and 5′-tgggggttgagtttagttgg-3′) and HindIII digestion of the amplified product. Lines segregating as homozygous for the transgene were selected based on the pFWA-GFP and seed phenotypes in the atdre2-2 background. All primer sequences are listed in Table S3.
Quantitative PCR and McrBC Assay.
Gene expression and McrBC assays were performed using 3 DAmP seeds obtained from pollination of atdre2-2;pFWA-GFP (for mutant) or pFWA-GFP (for WT) with Ler pollen. Pools of GFP on and GFP off seeds were selected under a dissecting fluorescence microscope. Approximately 30–50 seeds were used for RNA extraction using an Arcturus PicoPure Kit following the manufacturer’s instructions, and more than 250 seeds per pool were used for DNA extraction. The RNA and DNA were quantified using a NanoDrop spectrophotometer. Approximately 150 ng of RNA was used for cDNA synthesis using Primescript Reverse Transcriptase (TaKaRa), after treatment with DNase (Promega). A 10-fold dilution of the cDNA was used directly for RT-qPCR, which was performed using SYBR Premix Ex Taq (TaKaRa) and Thermal Cycler Dice (TaKaRa). Relative quantification was used for gene expression. Approximately 1 μg of DNA was digested with 1 μL of McrBC overnight at 37 °C. The reaction was stopped by incubation at 65 °C for 20 min, and DNA cleanup was performed using sodium chloride precipitation. A 10-fold dilution was used for RT-qPCR, using absolute quantification against a 1:1 mixture of genomic DNA extracted from Col-0 and Ler leaves. All primer sequences are listed in Table S3. The optimization of this assay is detailed in Fig. S5.
Supplementary Material
Acknowledgments
We thank Yoko Ikeda for advice with experimental procedures, Yukiko Sugimoto for excellent technical assistance, Yuki Kinoshita for screening and initial mapping of alac4 and excellent technical assistance, Akemi Ono and Ian Smith for suggestions on the manuscript, and Noriko Inada for assistance with confocal microscopy. This work was supported by a Research Fellowship from the Japan Society for the Promotion of Science (to D.M.B.) and Grants-in-Aid for Scientific Research on Innovative Areas (23113001 and 23113003, to T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404058111/-/DCSupplemental.
References
- 1.Lill R, Mühlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: Components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 2.Balk J, Schaedler TA. Iron cofactor assembly in plants. Annu Rev Plant Biol. 2014;65:125–153. doi: 10.1146/annurev-arplant-050213-035759. [DOI] [PubMed] [Google Scholar]
- 3.Netz DJ, Mascarenhas J, Stehling O, Pierik AJ, Lill R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 2014;24(5):303–312. doi: 10.1016/j.tcb.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 2008;28(18):5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Netz DJ, et al. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol. 2010;6(10):758–765. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- 6.Bernard DG, Netz DJ, Lagny TJ, Pierik AJ, Balk J. Requirements of the cytosolic iron-sulfur cluster assembly pathway in Arabidopsis. Philos Trans R Soc Lond B Biol Sci. 2013;368(1622):20120259. doi: 10.1098/rstb.2012.0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bych K, et al. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J Biol Chem. 2008;283(51):35797–35804. doi: 10.1074/jbc.M807303200. [DOI] [PubMed] [Google Scholar]
- 8.Balk J, Pierik AJ, Netz DJ, Mühlenhoff U, Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 2004;23(10):2105–2115. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol Cell Biol. 2005;25(24):10833–10841. doi: 10.1128/MCB.25.24.10833-10841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stehling O, et al. MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science. 2012;337(6091):195–199. doi: 10.1126/science.1219723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chanet R, Heude M. Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Curr Genet. 2003;43(5):337–350. doi: 10.1007/s00294-003-0407-2. [DOI] [PubMed] [Google Scholar]
- 12.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Curr Opin Struct Biol. 2012;22(1):94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Kuo CF, et al. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992;258(5081):434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 14.Boal AK, et al. Redox signaling between DNA repair proteins for efficient lesion detection. Proc Natl Acad Sci USA. 2009;106(36):15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 16.Gong Z, et al. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111(6):803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 17.Mok YG, et al. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci USA. 2010;107(45):19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M, et al. The role of Arabidopsis thaliana NAR1, a cytosolic iron-sulfur cluster assembly component, in gametophytic gene expression and oxidative stress responses in vegetative tissue. New Phytol. 2013;199(4):925–935. doi: 10.1111/nph.12350. [DOI] [PubMed] [Google Scholar]
- 19.Luo D, Bernard DG, Balk J, Hai H, Cui X. The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell. 2012;24(10):4135–4148. doi: 10.1105/tpc.112.102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penterman J, Uzawa R, Fischer RL. Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 2007;145(4):1549–1557. doi: 10.1104/pp.107.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303(5657):521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 22.Soler N, et al. A S-adenosylmethionine methyltransferase-like domain within the essential, Fe-S-containing yeast protein Dre2. FEBS J. 2012;279(12):2108–2119. doi: 10.1111/j.1742-4658.2012.08597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song G, et al. Crystal structure of the N-terminal methyltransferase-like domain of anamorsin. Proteins. 2014;82(6):1066–1071. doi: 10.1002/prot.24443. [DOI] [PubMed] [Google Scholar]
- 24.Park KA, et al. Nuclear translocation of anamorsin during drug-induced dopaminergic neurodegeneration in culture and in rat brain. J Neural Transm. 2011;118(3):433–444. doi: 10.1007/s00702-010-0490-8. [DOI] [PubMed] [Google Scholar]
- 25.Shibayama H, et al. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J Exp Med. 2004;199(4):581–592. doi: 10.1084/jem.20031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varadarajan J, et al. ATR3 encodes a diflavin reductase essential for Arabidopsis embryo development. New Phytol. 2010;187(1):67–82. doi: 10.1111/j.1469-8137.2010.03254.x. [DOI] [PubMed] [Google Scholar]
- 27.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA. 2000;97(19):10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huanca-Mamani W, Garcia-Aguilar M, León-Martínez G, Grossniklaus U, Vielle-Calzada JP. CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102(47):17231–17236. doi: 10.1073/pnas.0508186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC. The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell. 2008;20(8):2088–2101. doi: 10.1105/tpc.108.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324(5933):1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124(3):495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Z, Li X, Qiao T, Fan D. Successful expression and purification of human CIAPIN1 in baculovirus-insect cell system and application of this system to investigation of its potential methyltransferase activity. Int J Biol Macromol. 2008;42(1):27–32. doi: 10.1016/j.ijbiomac.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463(7280):554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.