Significance
Ribosomes are the site of protein synthesis in all cells. Ribosomal polypeptide synthesis involves reading the information encoded in the mRNA and adding the correct amino acid via a tRNA. Elongation of the polypeptide chain proceeds through translocation, where the ribosome couples the movement of the mRNA and its associated tRNAs by precisely one codon. Translocation requires large-scale movement of the small-subunit head domain of the ribosome. By comparing 55 ribosome structures, we show that movement universally results from coupled hinging at two separate loci in the small subunit. This mechanism explains the mode of action of the antibiotic spectinomycin and suggests a means by which the ribosome controls translocation.
Keywords: RNA dynamics, translation, S5, three-way junction
Abstract
During ribosomal translocation, a process central to the elongation phase of protein synthesis, movement of mRNA and tRNAs requires large-scale rotation of the head domain of the small (30S) subunit of the ribosome. It has generally been accepted that the head rotates by pivoting around the neck helix (h28) of 16S rRNA, its sole covalent connection to the body domain. Surprisingly, we observe that the calculated axis of rotation does not coincide with the neck. Instead, comparative structure analysis across 55 ribosome structures shows that 30S head movement results from flexing at two hinge points lying within conserved elements of 16S rRNA. Hinge 1, although located within the neck, moves by straightening of the kinked helix h28 at the point of contact with the mRNA. Hinge 2 lies within a three-way helix junction that extends to the body through a second, noncovalent connection; its movement results from flexing between helices h34 and h35 in a plane orthogonal to the movement of hinge 1. Concerted movement at these two hinges accounts for the observed magnitudes of head rotation. Our findings also explain the mode of action of spectinomycin, an antibiotic that blocks translocation by binding to hinge 2.
Central to the elongation phase of protein synthesis is the process of translocation, in which tRNA moves from the aminoacyl (A) to peptidyl (P) and P to exit (E) sites of the ribosome, coupled with mRNA advancement by exactly one codon. Translocation can be broadly divided into two steps. First, the tRNAs move on the large subunit from their classical A/A and P/P states to their hybrid A/P and P/E states (1, 2), facilitated by intersubunit rotation, which can occur spontaneously and reversibly (3–7). During the second step, which is rate limiting and EF-G–dependent (8, 9), the tRNA anticodon stem-loops (ASLs) and their associated mRNAs move from the A to P and P to E sites of the small subunit, thereby advancing the tRNAs into their classical P/P and E/E states. The latter step is accompanied by the intrasubunit rotation of the small-subunit head domain, which unlocks the steric barrier between the P and E sites on the 30S subunit (10–13) and transports the P-site tRNA into the 30S E site (14–16). In addition to a growing structural database of trapped translocation complexes from X-ray and cryo-EM studies (Table S1), computational approaches are being used to analyze ribosome dynamics and to describe the energy landscape of the translocation process (17–22).
Structures of trapped EF-G–containing translocation intermediates show that, during 30S subunit head rotation, the translocating P-site tRNA precisely maintains contact with the head domain but moves relative to the 30S body domain, forming a chimeric hybrid (pe/E) state (14, 15). (Lowercase letters indicate that the tRNA is bound in a chimeric hybrid state, whereas uppercase letters indicate binding to the canonical A, P, or E sites. For example, “pe/E” is meant to indicate that the ASL of the tRNA is bound between P-site elements of the small subunit head and E-site elements of the small subunit body, while its acceptor end is bound to the large-subunit E site.) These findings provide evidence that small-subunit head rotation plays a central role in the second step of translocation. A full description of the molecular mechanism of translocation will therefore depend on an understanding of the structural basis of head rotation.
As has been widely believed (11, 17, 23–26), we anticipated that the axis of head rotation would pass through helix h28 of 16S rRNA (often called the “neck”), the sole covalent connection between the head and body of the 30S subunit. Using the Euler–Rodrigues (E–R) method (27, 28), we localized the axis of head rotation for structures of 55 ribosome complexes, covering a range of more than 20° of head rotation. Unexpectedly, the axis does not pass through helix h28, but is positioned in the space between h28 and the coaxially stacked helices h35 and h36, which form a second, noncovalent connection to the body. In a second approach, we localized the points of divergence of the conserved core of the 30S head to two hinge points, in h28 (hinge 1) and in the linker connecting h34 and h35 (hinge 2), which coincide with the same two features flanking the calculated helical axis. Assignment of the origins of head rotation to these two hinge points was further validated by the strong correlation between the magnitudes of their deflection angles and the degree of head rotation in the different structures. Finally, our findings explain the inhibition of translocation by the antibiotic spectinomycin, which binds to the pivot point of hinge 2 (24, 25, 29, 30).
Results
Localization of the Axis of 30S Head Rotation.
To characterize rotation of the 30S head (Fig. S1A), defined as the 3′ major domain (residues 921–1396) of 16S rRNA and its associated proteins, we applied the Euler–Rodrigues (E–R) formula (27, 28), as described in Methods, to 55 structures of ribosomes trapped with varying degrees of head rotation (Table S1). The E–R formula treats interdomain movement as a simple rigid-body rotation around a single calculated axis (the E–R axis) (Fig. 1A), irrespective of the complexity of the movement. Further analysis (see below) confirmed that the 30S head indeed behaves essentially as a rigid body, in agreement with earlier studies (11, 26). We measured the magnitude and direction of rotation with respect to a reference crystal structure of a nonrotated classical-state ribosome complex (10) [Protein Data Bank (PDB) ID code 2J00]. Positive rotation is defined as counterclockwise, as viewed from the top of the 30S head domain (Fig. 1B and Fig. S1A).
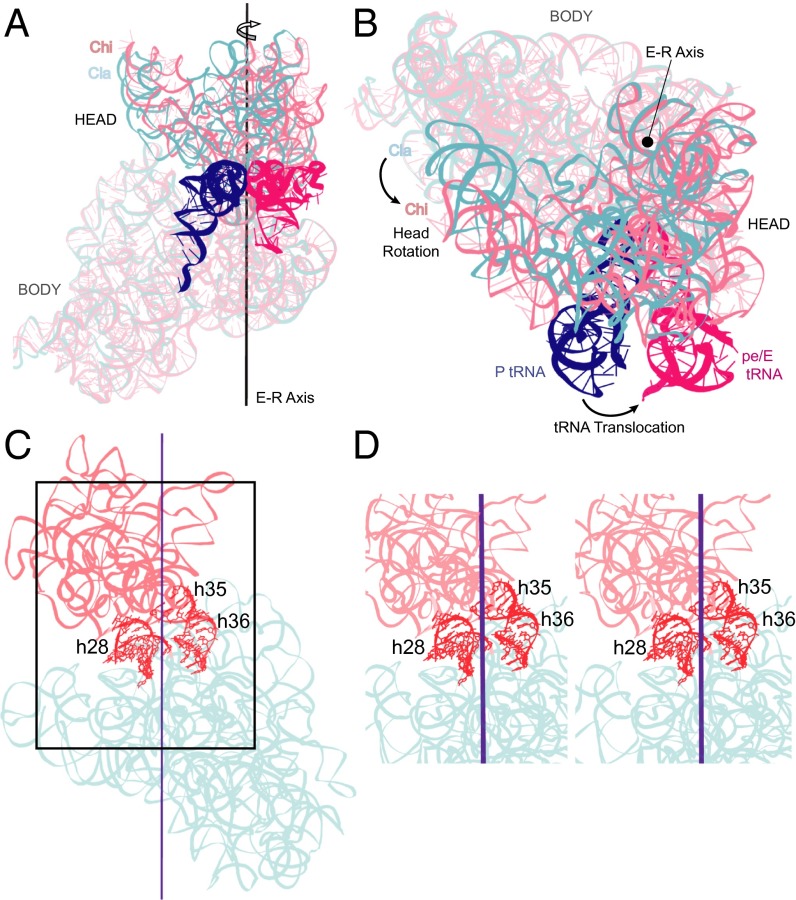
Fig. 1.
Locating the axis of 30S head rotation. Position of the Euler–Rodrigues (E–R) axis for 30S head rotation viewed from (A) the subunit interface and (B) the top of the 30S subunit, showing a 21° rotation of the head in the pe/E chimeric hybrid state (Chi) complex (15) (light pink) relative to the nonrotated classical-state (Cla) reference structure (10) (cyan). Coupling of 30S head rotation to tRNA translocation from the classical P/P (blue) to the chimeric pe/E state (magenta) is shown. (C) Solvent view of the position of the E–R axis, showing the positions of helices h28, h35, and h36. (D) Stereoview showing the position of the E–R axis between h28 and the coaxial h35/h36.
Given that the head and body domains of the 30S subunit are covalently linked by the neck or helix h28 (Fig. 2A), we anticipated that the E–R axis would pass through h28. Surprisingly, the E–R axis does not coincide with h28, but lies between h28 and the coaxially stacked helices h35 and h36 (Fig. 1C). The E–R axis is similarly situated between h28 and h35/h36 in all 28 crystal structures showing significant head rotation in our dataset (Fig. S2). This unexpected finding indicates that the observed motion of the 30S subunit head is not the result of a simple rotation around the neck of the 30S subunit.
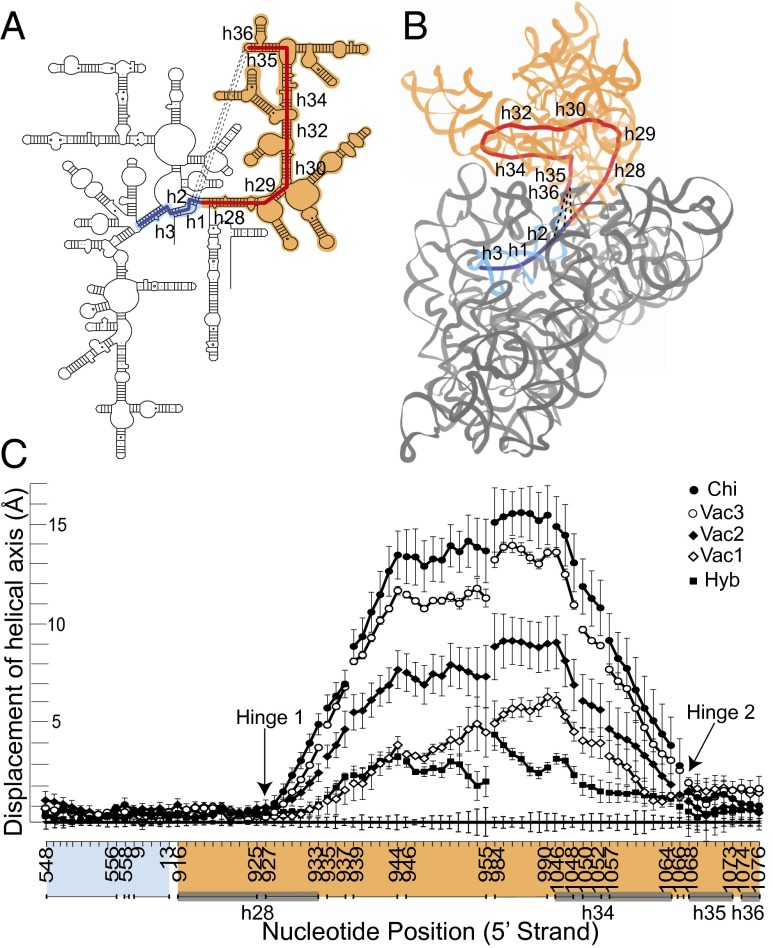
Fig. 2.
Origins of 30S head movement. (A) The 16S rRNA core of the head of the 30S subunit comprises the series of contiguous helices h28 to h36 (red). Dotted lines represent conserved A-minor interactions between the h36 tetraloop and helix h2 in the 30S body. Helices h2, h1, and h3 of the body domain (blue) are coaxial with the core helices of the head. (B) Path of the calculated helical axis (33) (CUR axis) for the RNA core (red), shown for the Thermus thermophilus 70S ribosome crystal structure (10). The head (domain III) is in light brown and the body (domains I, II, and IV) is in gray. (C) Displacement at each position along the CUR axis from the classical-state axis (baseline). Averaged CUR axes were calculated for different structural classes showing head rotation. Chi, chimeric hybrid state; Hyb, hybrid state; Vac1, Vac2, Vac3, vacant ribosome subgroups 1–3. (Compare with Table 1.) The sharp inflection points at positions 927 and 1068 identify the positions of hinge 1 (in helix 28) and hinge 2 (in the single-stranded linker between h34 and h35). Error bars represent SDs. The head domain itself moves essentially as a rigid body (Fig. S3).
Values for 30S head rotation, as well as for intersubunit body rotation (relative to the 50S subunit; also calculated by the E–R method) are distributed nearly continuously, from 0° to +21° and −4° to +10°, respectively (Table S1). Although head and body rotation do not show a general correlation (31) (Fig. S1B), the presence of bound full-length tRNAs constrains their values to three well-defined classes, clustered around 2° head, −1.5° body (classical); 6° head, 7.5° body (hybrid); and 19° head, 2° body (chimeric hybrid), corresponding to three main tRNA-binding states (SI Text).
Localization of the Origins of Movement.
We next identified the origins of head movement by localizing the precise boundaries between the static (body) and dynamic (head) structural elements. This was done by measuring differences between positions of 16S rRNA in rotated vs. nonrotated ribosomes. By superimposing the essentially static body domains of different 30S subunits, we identified structural deviations that correlate with head movement, as described below. We restricted the comparison dataset to the 41 bacterial ribosome crystal structures reported at 4 Å resolution or better (Table S1).
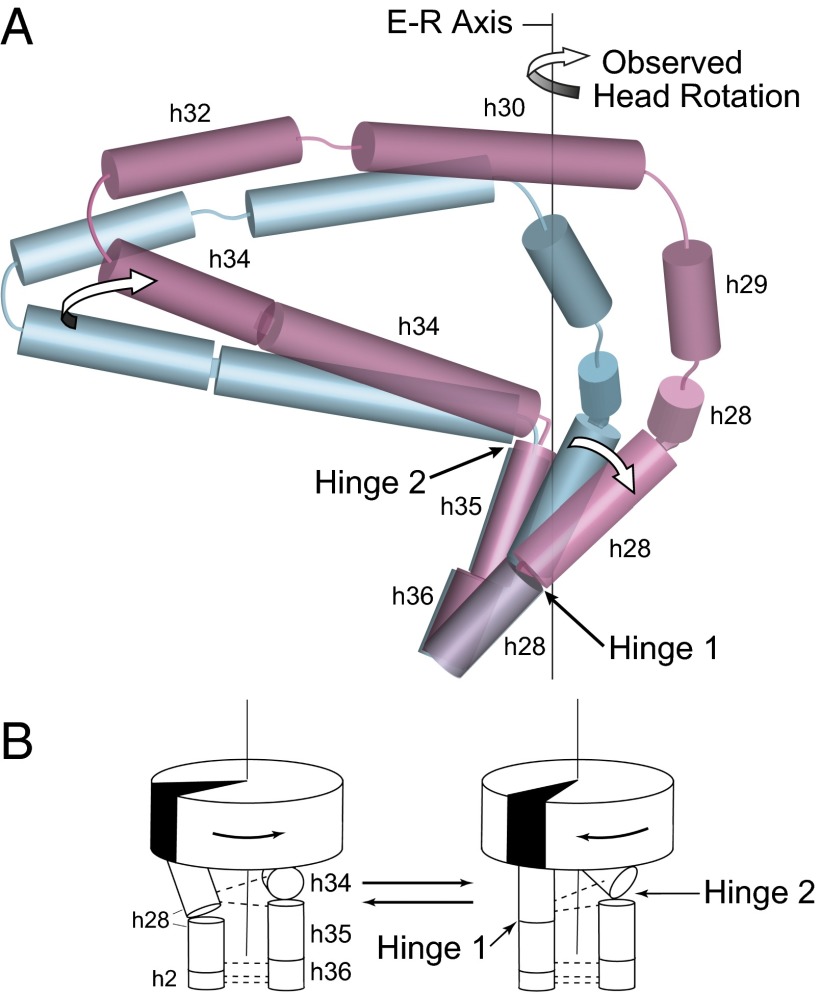
The structural core of the 30S subunit head domain of 16S rRNA comprises a series of contiguous helices stretching in a wide loop from helix h28 to h36, most of which are coaxially stacked (32) (Fig. 2 A and B). At one end, h28 (i.e., the neck) covalently connects the head to the body of the subunit, where it forms a continuous coaxial stack with helices h2, h1, and h3 in the body domain. At the other end of the series, the GUGA tetraloop of h36 forms a noncovalent bridge to the body via A-minor interactions with h2 (32) (Fig. 2B). Within this structural core helices h28–h29, h30–h32, and h35–h36 are coaxially stacked. Thus, the series of helices h28–h29–h30–h32–h34–h35–h36 leads from the body of the 30S subunit into the head, looping back into contact with the body. Based on the positions of these core helices, we calculated a continuous helical axis for the structural core of the 30S head using the program Curves+ (33) for each structure (Fig. 2B).
Deviation of the positions of nucleotides along the helical axis in structures with head rotation compared with those for the unrotated, classical state is plotted in Fig. 2C. A priori, one could imagine, at one extreme, that head movement results from a localized change at a single point in the structure, or alternatively, from the accumulation of multiple smaller changes distributed over a larger segment of the structure. The measurements plotted in Fig. 2C show that head rotation originates in two sharply localized inflection points, adjacent to positions 927 and 1068 of 16S rRNA. Moreover, these same two inflection points are found for all classes of structures (hybrid, chimeric hybrid and three classes of vacant ribosomes grouped according to magnitude of head rotation), despite differences in magnitude of head rotation and in their bound ligands (Fig. 2C). We refer to these inflection points as hinge 1 and hinge 2. The relatively linear behavior of the deviation plot following the sharp inflection at G927 and preceding the inflection at G1068 (Fig. 2C) suggests that the head behaves essentially as a rigid body. Superimposition of the head domains results in an average rmsd of less than 1 Å and a displacement of helical axes of the head domain of less than 2 Å (Fig. S3), showing that the core of the head domain moves essentially as a rigid body.
Hinge 1.
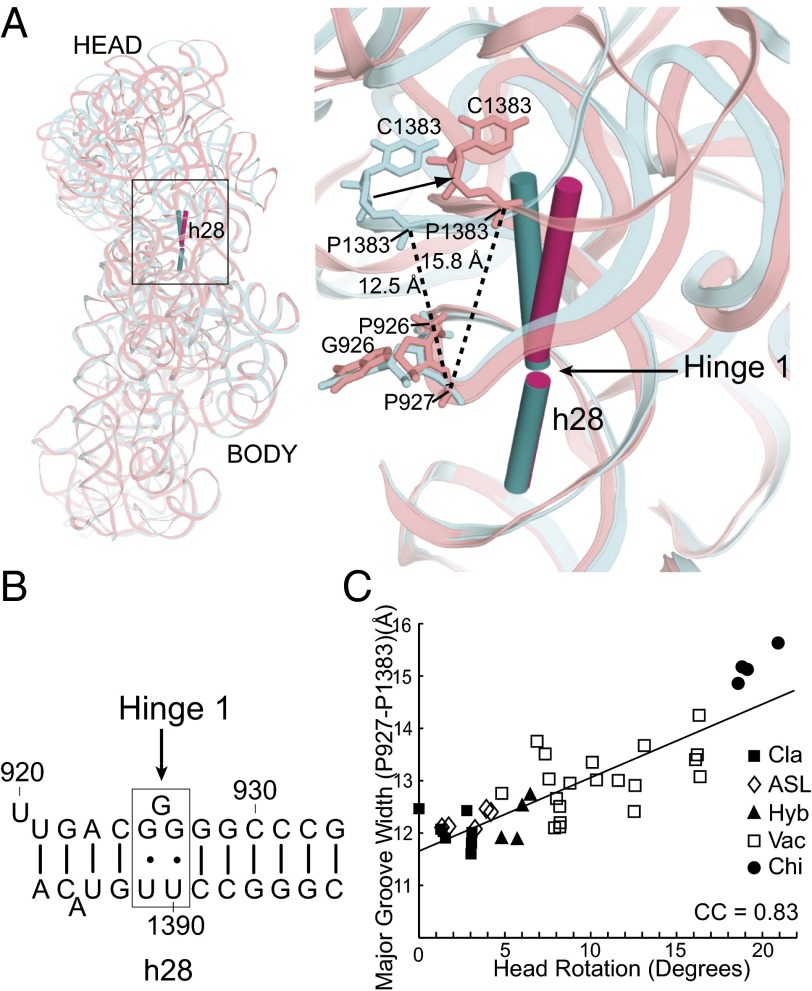
Hinge 1 lies at a weak point within the G-C–rich helix h28 (Fig. 3A) around the universally conserved bulged G926, which is flanked by conserved G⋅U wobble pairs (Fig. 3B). It functions as a simple hinge, which is bent in the classical, nonrotated state at 166° and increasingly straightens during head rotation to 177°. Straightening of hinge 1 (the neck) results in progressive widening of the major groove of h28 as a function of increasing head rotation [correlation coefficient (CC) = 0.83] across the 41 bacterial crystal structures, as measured from the cross-strand distance between P927 and P1383 (Fig. 3 A and C). Additionally, we observe that positions 932 and 1385 show a consistent decrease in their backbone angles (measured between consecutive P atoms) as a function of head rotation, consistent with previous observations (11, 17). These changes are clearly correlated with head movement (CC = −0.81) and occur as a result of structural interactions between hinges 1 and 2 (see below).
Fig. 3.
Hinge 1 lies within the neck (h28) of the 30S subunit. (A) Hinge 1 is positioned in the middle of helix h28, the covalent connection between the head and body domains. The cylinders represent helical axes flanking the articulation point in the classical (10) (0° of head rotation; teal) and chimeric (15) (21.3° of head rotation; magenta) structures. (B) Hinge 1 is located at a weak point in the G-C–rich helix h28 around the bulged G926, which is flanked by conserved G⋅U wobble pairs. (C) The major groove of h28, measured across P927 and P1383, widens by >3 Å as a function of increasing head rotation (CC = 0.83; the correlation is represented with a trendline).
Hinge 2.
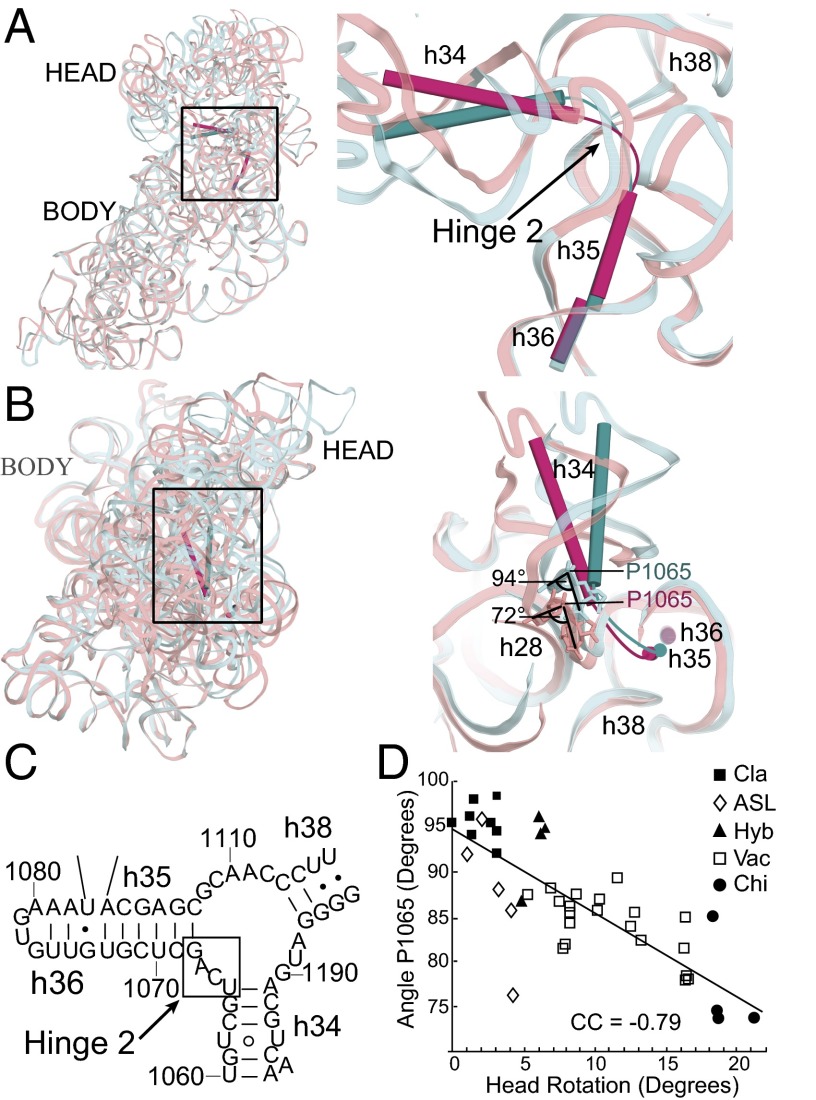
Hinge 2 is linked to the coaxially stacked h35 and h36 (Fig. 4A), which form a noncovalent A-minor connection between the head and body domains (Fig. 2 A and B). Hinge 2 acts as a flag hinge by allowing h34 to swivel around the immobilized h35/h36 (Fig. 4 A and B); its movement is roughly orthogonal to that of hinge 1. It is part of the helical junction formed by h34, h35, and h38 (Fig. 4C), previously identified as a member of the RNA structure motif termed as a Family A three-way junction (34). Helices h35/h36 form a Family C three-way junction with h37 (34) but remain relatively static, with an rmsd of atomic positions of less than 1 Å across the 41 ribosomal crystal structures. The actual hinge point occurs in the loop connecting h34 to h35, between positions 1064 and 1067, where the majority of conformational changes are found. Most prominent is a decrease in the angle subtended by P1064–P1065–P1066 from 94° in the classical-state reference structure to 72° in the highly rotated chimeric hybrid complex (Fig. 4B), mimicking the overall 22° change in head rotation, and decreasing linearly as a function of increasing head rotation (CC = −0.79) (Fig. 4D). This rearrangement directly influences a switch in the interactions between hinge 1 and hinge 2 through G1064, as described below. Most notably, this change in backbone angle is directly centered on the binding site for the antibiotic spectinomycin (30) (Fig. S5), a well-known inhibitor of translocation (35) (Discussion).
Fig. 4.
Hinge 2 is located between h34 and h35. (A) Subunit interface view showing the position of hinge 2 in the single-stranded linker between helices h34 and h35. Helix h34 tilts away from the body in the rotated state (15) (magenta). Helices h35 and h36 remain static between the classical and rotated states. (B) Hinge 2 viewed from the top of the 30S head, showing swiveling of h34 toward the subunit interface in the rotated state (15) structures. Classical (cyan) and chimeric (pink) state 16S rRNAs are shown as ribbons. In the loop connecting h34 and h35 (details shown in Fig. S4), angle P1065 decreases with increasing head rotation by 22°. (C) Secondary structure of Escherichia coli 16S rRNA showing the position of hinge 2 in the three-way junction connecting h34, h35, and h38. (D) Decrease in the P1064–P1065–P1066 backbone angle as a function of increasing head rotation (CC = −0.79; the correlation is represented with a trendline).
Synergy Between Hinges 1 and 2.
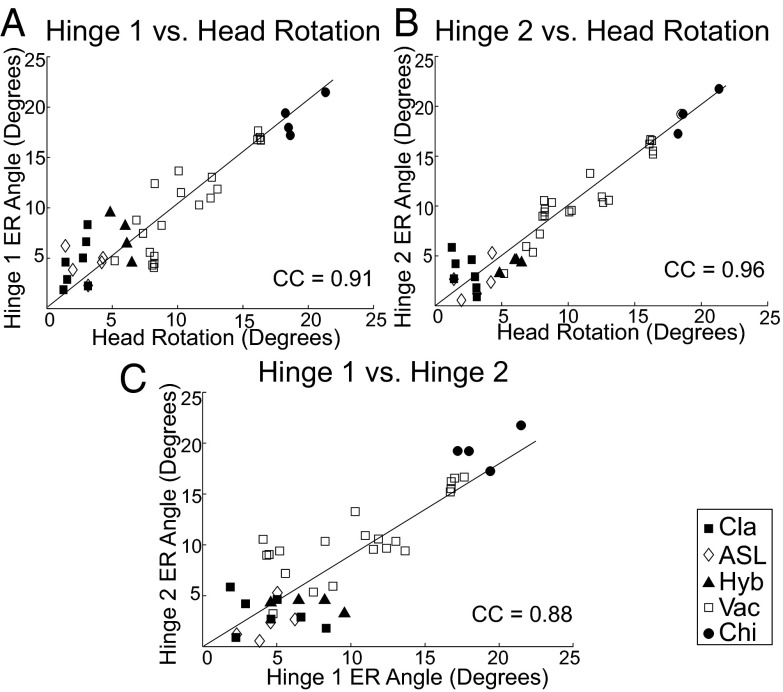
We next assessed how the flexing of each hinge correlates with head rotation and how flexing of the two hinges correlate with each other. We measured flexing by the change in the angle between the helical axes flanking the hinges (compare Figs. 3 and 4), using the E–R method, for each hinge in each crystal structure. Relative to the classical reference state, the angular changes at each hinge correlate with head rotation (CC: hinge 1 = 0.91; hinge 2 = 0.96) (Fig. 5 A and B). Although there is a strong correlation (CC = 0.88) between movement of hinge 1 and hinge 2, it is clear that for individual examples, hinge 1 and hinge 2 are able to move independently from one another (Fig. 5C). We conclude that hinge 1 and hinge 2 effectively combine to result in rotation of the small-subunit head domain, relative to the body domain, around the E–R axis (Fig. 6).
Fig. 5.
Correlation of head rotation with hinge movement. Rotation of the 30S subunit head is strongly correlated with changes in the angles of both (A) hinge 1 (CC = 0.91) and (B) hinge 2 (CC = 0.96). (C) Movement of hinges 1 and 2 are correlated (CC = 0.88). The trendlines represent the correlation of the angles with 30S head rotation. However, the values for several of the structures show that the two hinges can move independently. Symbols indicate the structural class for each point.
Fig. 6.
Rotation of the 30S head results from concerted articulation at hinges 1 and 2. (A) Arrows show movement around hinge 1 and hinge 2 and rotation of the 30S head domain around the E–R axis. During head rotation, movement of hinge 1 is caused by the straightening of the kinked helix h28; the orthogonal movement of hinge 2 is the result of the swiveling of helix h34 around the static h35. Cylinders show positions of helical axes in the 16S rRNA core for the classical (10) (teal) and chimeric (15) (magenta) states. (B) Cartoon representation showing coupling of structural rearrangements at the two hinges to 30S head rotation. Dotted lines represent H-bonded interactions between the two hinges. The backbone contact between h34 and h28 switches between two positions on h28 depending on the degree of head rotation, as described in the text.
Hinge 1 and hinge 2 are connected by RNA–RNA backbone contacts and by interactions mediated by ribosomal protein S5. Of the three RNA hydrogen-bonded contacts, the two between helices h28 in hinge 1 and h35/h36 in hinge 2 are retained throughout head rotation (Fig. S6 A and B). In contrast, phosphate 1064 in h34 switches its interactions as a result of swiveling of h34 toward h28 and straightening of h28 during head rotation (Fig. S6C). In most structures with less than 8° of head rotation, phosphate 1064 H bonds with the 2′-OH of ribose 1385 in h28 (Fig. S6D). However, in structures with greater than 16° of head rotation, this interaction is disrupted, and replaced by a new tertiary contact between phosphate 1064 and the 2′-OH of the neighboring ribose 1386 in h28 (Fig. S6 C and D). This switch in contact between the two hinges influences the backbone conformation of h28. The angle subtended by the consecutive phosphate positions P931–P932–P933 (paired to positions 1386, 1385, and 1384) decreases as a function of increasing head rotation (CC = −0.81), with a maximum decrease of ∼22° between the classical and chimeric hybrid states (Fig. S6E). A further mode of communication between hinges is seen through their mutual interactions with the ribosomal protein S5. The long N-terminal β-hairpin of protein S5 bridges the static elements of hinges 1 and 2, contacting helices h2 and h36 and the static end of h28, whose positions are all maintained during head rotation. However, at the tip of the β-hairpin, Arg25 contacts the mobile helix h34 at positions 1192 and 1193 (Fig. S7A), a contact that is disrupted upon flexing of hinge 2 with high angles of head rotation (Fig. S7B).
Discussion
Localizing the Origin of Head Rotation.
In this study we sought to identify the structural basis for 30S head rotation, the large-scale conformational change in the ribosome that occurs during the rate-limiting, EF-G–dependent second step of translocation (8). By comparing structures of ribosome complexes trapped in different conformational states (Table 1 and Table S1), we used the E–R transform to identify the principal axis of rotation for the 30S head. Although we anticipated that the axis would pass through helix h28 of 16S rRNA (i.e., the neck) (11, 31), the sole covalent connection between the head and body domains of the 30S subunit, we found it positioned instead between h28 and the coaxial helices h35 and h36 (Fig. 1). Next, measuring the deviation of positions along the helical axis describing the core of the head, we localized the origins of head movement to two hinge points, located at the middle of h28 and the end of h35. These results show that h28, which has long been implicated in head rotation (11, 25, 36–39), is indeed involved in head movement, although not as the center of rotation.
Table 1.
30S head and body rotations
| Ribosome state† | 30S head rotation | 30S body rotation‡ | No. of structures | tRNA states |
| Classical (Cla) | 2.0 ± 1.0 | −0.8 ± 0.8 | 8 | Classical |
| P/E Hybrid (Hyb) | 5.9 ± 0.7 | 7.6 ± 1.1 | 4 | P/E Hybrid |
| Vacant 1 (Vac1) | 8.1 ± 0.1 | −3.3 ± 0.1 | 5 | No tRNA |
| Vacant 2 (Vac2) | 11.5 ± 1.3 | 4.9 ± 1.9 | 4 | No tRNA |
| Vacant 3 (Vac3) | 16.3 ± 0.1 | −2.4 ± 0.1 | 4 | No tRNA |
| Chimeric hybrid (Chi) | 19.2 ± 1.3 | 1.8 ± 0.9 | 4 | Chimeric pe/E–hybrid |
The magnitudes of 30S head and body rotation for each ribosome complex were determined by the E–R method relative to 2J00 and 2J01 (10) reference structures (Methods). See Table S1 for the complete list of structures used.
Defined by the tRNA-binding state. Groups were assigned based on the magnitudes of head rotation and conformational states of bound tRNAs (Methods).
Intersubunit rotation.
Hinge 1 is centered on the bulged base G926 within helix h28, the neck helix that covalently connects the head and body domains (Fig. 3). During head rotation, h28, which is kinked in the nonrotated state, becomes straightened. Hinge 2 is positioned in the single-strand linker between h34 and the coaxially stacked h35/h36 (Fig. 4). Hinge 2 swivels around position 1068 (at the tip of h35) in a plane roughly orthogonal to the bending of hinge 1 (Fig. 6). The coaxial h35/h36, although nominally a part of the head domain, form a noncovalent connection to the body and remain static during head rotation (Figs. 2 and 4). Thus, hinge 1 and hinge 2 are identical to the two structural elements that were found to flank the E–R axis (Fig. 1C).
Head rotation thus results from the combined flexing at two separate loci (hinges 1 and 2) in16S rRNA that lie within the two connectors linking the head and body domains, as depicted schematically in Fig. 6B. The angular change at each hinge (hinges 1 and 2) strongly correlates with the degree of head rotation (CC = 0.91 and 0.96, respectively) (Fig. 5 A and B), validating our conclusion that 30S head rotation is based on bending at these two hinge points. Bending of the two hinges is strongly correlated (CC = 0.88), indicating that their movements are generally coupled (Fig. 5C); however, the observed values for several structures show that hinge 1 and hinge 2 can move independently in certain contexts (Fig. 5C), allowing the 30S head to explore a wider range of orientations, such as swiveling or tilting (13, 17, 38, 40).
Functional Implications.
There are indications of functional interactions between hinge 1 and mRNA. Previously, the bulged G926 of helix h28 was found to make a hydrogen-bonded interaction with phosphate +1 of mRNA (41). In addition, the backbone of the Shine–Dalgarno helix around position 1534 was found to contact h28 near position 929 (23). Here, we observe interactions between h28 and the universally conserved A1503, which has been proposed to act as a translocational pawl by intercalation between bases −1 and −2 of the mRNA in the chimeric hybrid state (15). In the classical state, A1503 is held in its retracted position by H-bonding interactions with ribose 927 and G925 in h28 (Fig. S8A). In the highly rotated chimeric state, these interactions are disrupted (Fig. S8B), allowing A1503 to flip into its intercalated state. This appears to be connected to a change in the backbone conformation around position 927. Although the resolutions of these structures are not sufficiently high to distinguish sugar conformations, an increase in the P927–P928 distance from ∼5.9 to ∼7.0 Å between the classical and chimeric states (SI Discussion) is consistent with a change in the ribose sugar pucker from C3′-endo to C2′-endo (42–44), which could help to explain disruption of the contacts between A1503 and h28.
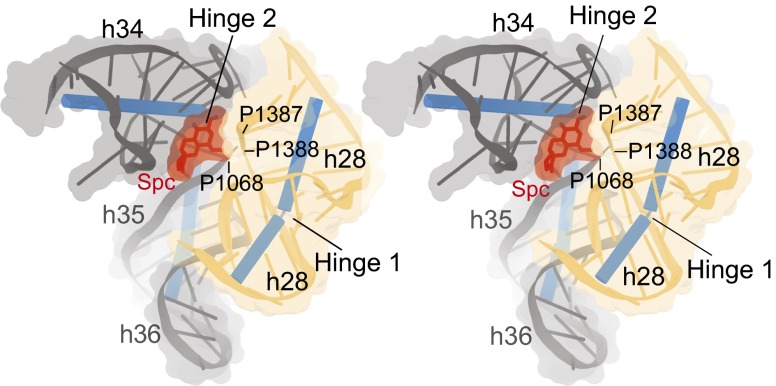
The two-hinge mechanism provides a structural explanation for the inhibition of translocation by the antibiotic spectinomycin. Spectinomycin binds within the 30S head domain (29), trapping the head in a partially swiveled state (24, 25) and drastically reducing the rate of translocation (35). Chemical footprinting (29), mutational analysis (29, 30, 45), and crystal structures of ribosomes (25) or 30S subunits (24) in complex with spectinomycin localized its binding site to G1064, C1066, G1068, A1191, C1192, and G1193 at the h34/h35 junction. Our findings show that these contact positions with spectinomycin are centered on the articulation point of hinge 2 (Fig. 7 and Fig. S5), supporting our conclusion that spectinomycin inhibits translocation primarily by locking hinge 2 and confirming an earlier suggestion by Carter et al. (24). Although spectinomycin also interacts with h28 (Fig. 7 and Fig. S5C), its contact surface is much smaller than that with hinge 2, contacting only the backbone atoms of h28 at G1387 and G1388, some distance away from the articulation point of hinge 1 (Fig. 7). It is nevertheless possible that spectinomycin may indirectly restrict movement of hinge 1, by affecting the interplay between the two hinges. The site of interaction of S5 with h34 at position C1192 is a site of mutation conferring spectinomycin resistance (46) and lies adjacent to hinge 2 at the spectinomycin-binding site (24, 25). The importance of these interactions of S5 with the hinges for ribosome function is reflected in the finding that mutation of the universally conserved Gly23 at the tip of the hairpin to Asp confers cold sensitivity and spectinomycin resistance (47).
Fig. 7.
Spectinomycin binds primarily to hinge 2. Stereoview showing contacts between spectinomycin (Spc) (red) and 16S rRNA in the Escherichia coli 70S ribosome (25). The main contacts made by Spc are in the minor groove between h34 and h35 (grey), at the articulation point of hinge 2, where it forms hydrogen bonds with bases G1064, C1066, G1068, A1191, C1192, and G1193. Spc also contacts h28 (yellow) at the backbone between G1387 and C1388, away from the articulation point of hinge 1 at the bulged G926.
Besides our primary focus on the database of abundant bacterial ribosome crystal structures, preliminary analysis of the vacant yeast 80S ribosome (48) and Tetrahymena (49) 40S ribosomal subunits, which have ∼16° head rotations, suggests that head rotation in eukaryotic ribosomes may be similar to that seen in bacteria. The head and body domains of 18S rRNAs are linked via dual connectors structured identically to those found in bacterial 16S rRNAs. Moreover, the lengths of the core helices in the head domain, as defined above, are precisely conserved between bacterial and eukaryotic ribosomes, despite large differences between the overall sizes of 16S and 18S rRNAs. Analysis of the core helices shows that the location of inflection points for 18S head movement coincides with those found in bacteria. Thus, the two-hinge mechanism may represent a universal strategy for small-subunit head rotation.
Although we can describe the structural basis of 30S head rotation, we understand little about how head rotation is triggered and controlled. The rate-limiting step of mRNA and tRNA ASL movement, which is coupled to head rotation (14, 50), strongly depends on EF-G (8, 51). One possibility is that head rotation is driven or triggered by movement of domain IV of EF-G (14, 15), which contacts h34 of the head domain. However, this possibility seems at odds with the observation (52) that release factor RF3, which has close structural similarities to EF-G but completely lacks domain IV, also induces 16° of head rotation (Table S1). Clearly, additional studies will be needed to shed light on the mechanisms by which head rotation is induced.
Methods
Our analysis was based on the structures of 55 ribosome complexes as listed in Table S1. To calculate 30S head rotation using the E–R formula and to visualize the E–R axis for head rotation, we created a plug-in module for PyMOL (53). The core domain was assigned based on ref. 32. The helical axis for the core helices was calculated using Curves+ (33). Further details are presented in SI Methods.
Supplementary Material
Acknowledgments
We thank Laura Lancaster and Jie Zhou for helpful discussions. This work was supported by Grants GM-17929 and GM-59140 from the National Institutes of Health and by a grant from the Agouron Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413731111/-/DCSupplemental.
References
- 1.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 2.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13(3):234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370(3):530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 5.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13(9):1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32(2):190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 8.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385(6611):37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 9.Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA. 2006;103(37):13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 11.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310(5749):827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 12.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23(5):1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berk V, Cate JHD. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr Opin Struct Biol. 2007;17(3):302–309. doi: 10.1016/j.sbi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340(6140):1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramrath DJF, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110(52):20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock LV, et al. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol. 2013;20(12):1390–1396. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
- 18.Mitra K, Frank J. Ribosome dynamics: Insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct. 2006;35:299–317. doi: 10.1146/annurev.biophys.35.040405.101950. [DOI] [PubMed] [Google Scholar]
- 19.Whitford PC, Sanbonmatsu KY. Simulating movement of tRNA through the ribosome during hybrid-state formation. J Chem Phys. 2013;139(12):121919. doi: 10.1063/1.4817212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanbonmatsu KY. Computational studies of molecular machines: The ribosome. Curr Opin Struct Biol. 2012;22(2):168–174. doi: 10.1016/j.sbi.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penczek PA, Kimmel M, Spahn CMT. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure. 2011;19(11):1582–1590. doi: 10.1016/j.str.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitford PC, Blanchard SC, Cate JHD, Sanbonmatsu KY. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLOS Comput Biol. 2013;9(3):e1003003. doi: 10.1371/journal.pcbi.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korostelev A, et al. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci USA. 2007;104(43):16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 25.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JHD. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2(8):545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues O. On the geometrical laws that govern the displacements of a solid system in space, and on the change of coordinates resulting from these displacements considered independently of the causes that can produce them. J Math Pures Appl. 1840;5:380–440. [Google Scholar]
- 28.Gray J. Olinde Rodrigues’ paper of 1840 on transformation groups. Arch Hist Exact Sci. 1980;21(4):375–385. [Google Scholar]
- 29.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 30.Brink MF, Brink G, Verbeet MP, de Boer HA. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 1994;22(3):325–331. doi: 10.1093/nar/22.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 32.Noller HF. RNA structure: Reading the ribosome. Science. 2005;309(5740):1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 33.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37(17):5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12(1):83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343(5):1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Tsai A, O’Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012;22(6):804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104(50):19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Fredrick K. Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucleic Acids Res. 2013;41(1):565–574. doi: 10.1093/nar/gks1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korostelev A, Noller HF. The ribosome in focus: New structures bring new insights. Trends Biochem Sci. 2007;32(9):434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34(8):390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126(6):1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 42.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 43.Rich A. The double helix: A tale of two puckers. Nat Struct Biol. 2003;10(4):247–249. doi: 10.1038/nsb0403-247. [DOI] [PubMed] [Google Scholar]
- 44.Rana TM. Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8(1):23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 45.Johanson U, Hughes D. A new mutation in 16S rRNA of Escherichia coli conferring spectinomycin resistance. Nucleic Acids Res. 1995;23(3):464–466. doi: 10.1093/nar/23.3.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilina EN, et al. Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front Microbiol. 2013;4:186. doi: 10.3389/fmicb.2013.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirthi N, Roy-Chaudhuri B, Kelley T, Culver GM. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA. 2006;12(12):2080–2091. doi: 10.1261/rna.302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 49.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331(6018):730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 50.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109(50):20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodnina MV, Wintermeyer W. The ribosome as a molecular machine: The mechanism of tRNA-mRNA movement in translocation. Biochem Soc Trans. 2011;39(2):658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Lancaster L, Trakhanov S, Noller HF. Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome. RNA. 2012;18(2):230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DeLano WL. The PyMOL Molecular Graphics System, Version 1.7 (DeLano Scientific LLC, San Carlos, CA)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.