Significance
We found that expression of phospholipase Cδ1 (PLCδ1) is down-regulated in colorectal cancer (CRC) cells compared with normal colon epithelium. Ectopic expression of PLCδ1 in CRC cells induced expression of E-cadherin, a tumor-suppressive cell–cell adhesion molecule, whereas knockdown of PLCδ1 repressed E-cadherin expression. Moreover, PLCδ1 overexpression reduced the malignant phenotypes of CRC. We also identified that PLCδ1 expression is repressed by the Kirsten rat sarcoma viral oncogene homolog (KRAS)/mitogen-activated protein kinase kinase (MEK) pathway, which is constitutively activated in many of CRC cells. Furthermore, PLCδ1 expression suppressed the phosphorylation of ERK1/2, which is an MEK target, by E-cadherin induction. These data suggest that PLCδ1 has tumor-suppressive functions in CRC through E-cadherin induction and KRAS/MEK/ERK signal attenuation. These findings could provide a valuable strategy for CRC treatment.
Keywords: phospholipase C delta 1, epithelial-to-mesenchymal transition, tumor suppressor
Abstract
Colorectal cancer (CRC) is one of the most common causes of cancer-related deaths worldwide, and Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in CRC predict the ineffectiveness of EGF receptor-targeted therapy. Previous transcriptional microarray analysis suggests the association between phospholipase Cδ1 (PLCδ1) expression and KRAS mutation status in CRC. However, both the roles and the regulatory mechanisms of PLCδ1 in CRC are not known. Here, we found that the expression of PLCδ1, one of the most basal PLCs, is down-regulated in CRC specimens compared with normal colon epithelium by immunohistochemistry. Furthermore, we examined the roles of PLCδ1 in CRC cell lines that harbor an activating KRAS mutation. Ectopic expression of PLCδ1 in CRC cells induced the expression of E-cadherin, whereas knockdown of PLCδ1 repressed the expression of E-cadherin. Moreover, the overexpression of PLCδ1 suppressed the expression of several mesenchymal genes and reduced cell motility, invasiveness, and in vivo tumorigenicity of SW620 CRC cells. We also showed that PLCδ1 expression is repressed by the KRAS/mitogen-activated protein kinase kinase (MEK) pathway. Furthermore, PLCδ1 suppressed the phosphorylation of extracellular signal-regulated kinase (ERK)1/2 through E-cadherin induction in CRC cells, suggesting the presence of a negative regulatory loop between KRAS/MEK/ERK signaling and PLCδ1. These data indicate that PLCδ1 has tumor-suppressive functions in CRC through E-cadherin induction and KRAS/MEK/ERK signal attenuation.
Colorectal cancer (CRC) is one of the most common cancers and causes of cancer-related deaths worldwide. Although CRC patients with unresectable tumors and metastasis have been treated with chemotherapy, recent advances in molecular research about CRC have resulted in the development of molecular targeted therapies, such as cetuximab and panitumumab. Chemotherapy combined with these targeted therapies improves the prognosis of CRC patients with unresectable tumors to some extent (1, 2). However, some CRC patients with activating Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations are unable to benefit from such drugs, because both cetuximab and panitumumab are epidermal growth factor (EGF) receptor-targeting agents and mutant KRAS constitutively activates the downstream signaling of EGF receptor (3). A more vigorous study using KRAS-mutant CRC is needed to identify novel molecular targets for drugs that will improve the prognosis of CRC patients with unresectable tumors, especially those with KRAS mutations.
Mutations in KRAS are found in about 40% of CRC patients. Constitutively active KRAS mutations lead to the hyperactivation of mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) [mitogen-activated protein kinase (MAPK)] signaling and/or phosphatidylinositol-3 kinase (PI3K) pathways (4). Activation of MEK/ERK (MAPK) signaling results in increased phosphorylation of ERK1/2, which in turn, phosphorylates several proteins related to cell cycle progression and cell motility. The PI3K pathway also promotes aberrant cell growth and survival by phosphorylation of v-akt murine thymoma viral oncogene homolog (AKT) (4). In addition to the aberrant effects of KRAS mutations on cell proliferation and survival, KRAS mutation involvement in the epithelial-to-mesenchymal transition (EMT) has been indicated in several cancer cell types (5, 6). EMT confers cells with stem-like properties, including invasiveness, with a loss of epithelial characteristics, such as E-cadherin expression, and a gain of mesenchymal characteristics. In pancreatic cancer cells, knockdown of mutant KRAS causes a significant decrease in cell motility, invasiveness, proliferation, and metastasis in association with increased E-cadherin expression and decreased ERK1/2 phosphorylation, suggesting the oncogenic KRAS roles through ERK phosphorylation and E-cadherin suppression (6).
Several pieces of evidence have indicated roles of some phospholipase C (PLC) enzymes in CRC progression. PLC is a signaling molecule that hydrolyzes phosphatidylinositol-4,5-bisphosphate to generate inositol-1,4,5-trisphosphate and 1,2-diacylglycerol, which increase the intracellular Ca2+ level and activate protein kinase C (PKC) signaling pathways. These pathways are involved in many biological processes, including tissue differentiation and tumorigenesis (7). Recent meta-analysis of CRC reveals that some PLC isozymes are deregulated in CRC, and low expression levels of PLCδ1 and PLCε1 genes are associated with KRAS mutation status (8). PLCε1 is a ras effector with dual roles in CRC tumor progression (9–11). However, little is known about the function of PLCδ1 in CRC. Moreover, the relationship between PLCδ1 and KRAS has not been elucidated.
Here, we have elucidated the roles of PLCδ1 in KRAS-mutant CRC cell lines. We found that PLCδ1 regulates expression of E-cadherin, suppression of EMT, cell motility, invasiveness, and tumorigenicity. Furthermore, KRAS/MEK signaling repressed PLCδ1 expression, whereas PLCδ1 suppressed the ERK1/2 phosphorylation by E-cadherin. These data indicate that PLCδ1 has tumor-suppressive functions in CRC through E-cadherin induction and KRAS/MEK/ERK signal attenuation and provide a valuable perspective on therapeutic approaches, which are also applicable to KRAS-mutated CRC, by regulating PLCδ1-mediating signals.
Results
PLCδ1 Is Down-Regulated in Colon Adenocarcinoma.
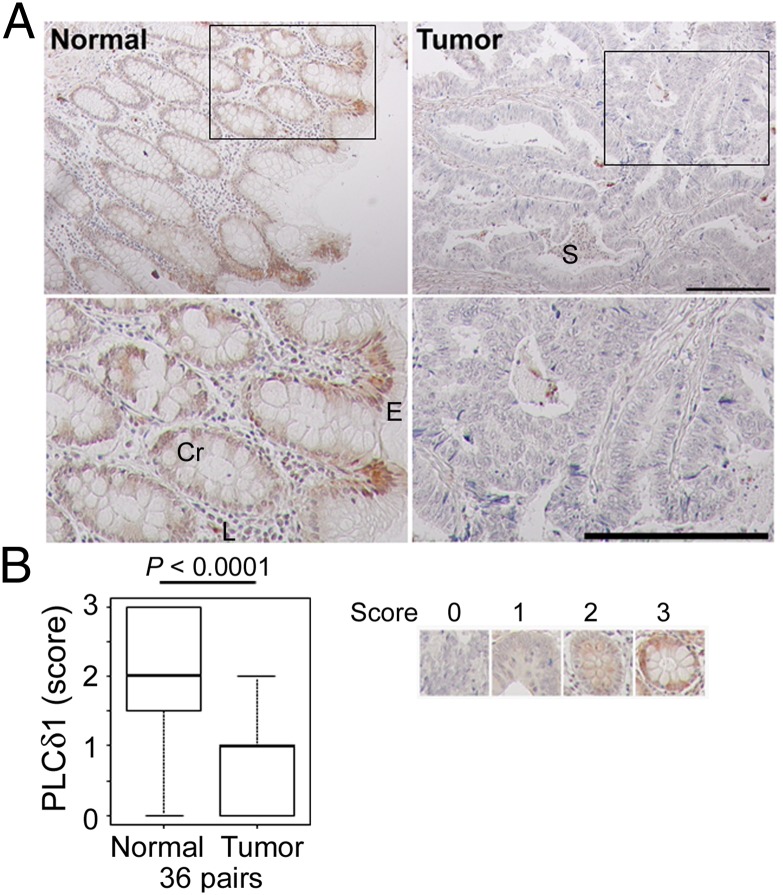
Because the involvement of PLCδ1 in CRC remains virtually unknown, we first examined if PLCδ1 protein expression is down-regulated in colon carcinoma compared with normal colon epithelium. We performed immunohistochemistry with human colon carcinoma tissue arrays, which contain 36 matched normal and adenocarcinoma tissues. PLCδ1 expression was observed strongly in normal colon surface epithelium and moderately in crypt cells as well as in the lamina propria (Fig. 1A). PLCδ1 expression was significantly diminished in CRC cells, but it was maintained in stromal cells (Fig. 1 and Fig. S1A). No clinicopathological features, such as sex, age, differentiation, or stage, were found to be associated with PLCδ1 levels observed in adenocarcinoma tissues (Fig. S1B).
Fig. 1.
PLCδ1 was down-regulated in colon adenocarcinoma. (A) Immunohistochemistry with human colon carcinoma tissue arrays, which contain 36 matched normal and adenocarcinoma tissues, was performed with anti-PLCδ1 antibody. Insets in Upper are shown magnified in Lower. Cr, crypt; E, surface epithelium; L, lamina propria; S, stroma. (Scale bar: 200 µm.) (B) The expression level of PLCδ1 in each sample was scored. As shown in Right, scores of 0–3 indicate complete loss, mild staining, moderate staining, and marked staining, respectively. The scored PLCδ1 levels were assessed between normal and tumor samples (36 pairs) using the Wilcoxon signed rank test.
PLCδ1 Induces the Expression of E-Cadherin.
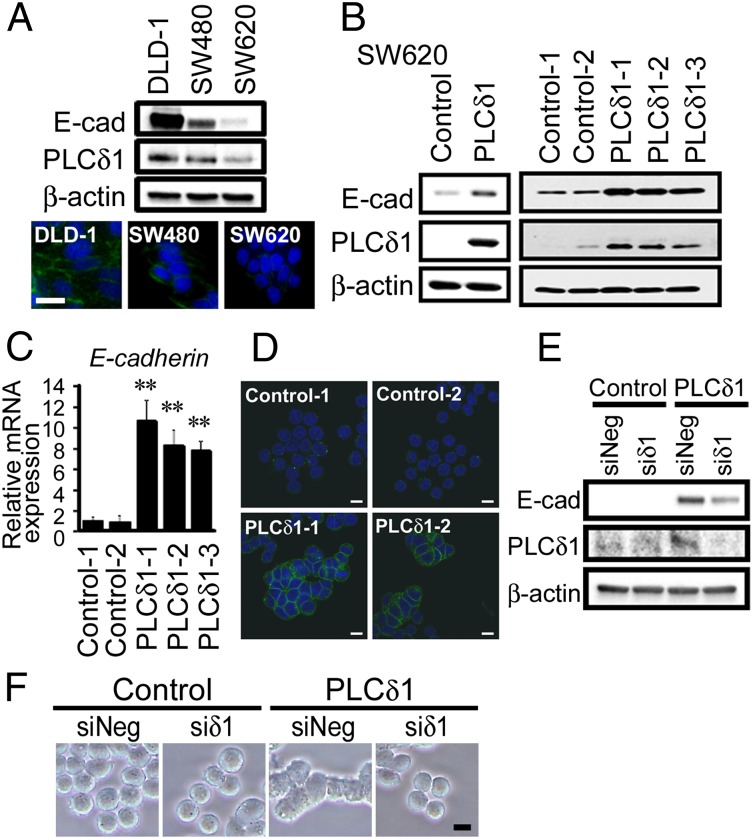
To elucidate the role of PLCδ1 in CRC, we investigated PLCδ1 expression in the CRC cell lines SW620, SW480, and DLD-1. The SW620 cell line was established from the metastatic site of a CRC patient, whereas the SW480 cell line was established from the primary tumor of the same patient. The DLD-1 cell line was established from CRC tissues from a different patient. Previously, SW620 was shown to have very low E-cadherin expression and acquired expression of Vimentin as well as EMT-promoting transcription factors, such as Zeb1 and Snail. These reports suggest that SW620 cells have undergone EMT, whereas SW480 and DLD-1 cells have not (12). Consistent with these reports, we observed the very low expression of E-cadherin in SW620 cells, whereas robust E-cadherin expression was observed in DLD-1 and SW480 cells (Fig. 2A). Interestingly, we found that PLCδ1 expression was very low in SW620 cells, whereas it was relatively high in DLD-1 cells (Fig. 2A). Because of these results, we investigated the role of PLCδ1 in SW620 cells in association with the expression of E-cadherin. We found that PLCδ1 overexpression in SW620 significantly up-regulated E-cadherin protein levels (Fig. 2B). Similar results were also obtained by quantitative real-time PCR (qRT-PCR), showing that PLCδ1 induced E-cadherin expression transcriptionally (Fig. 2C). The E-cadherin and PLCδ1 mRNA expression levels in PLCδ1-overexpressing clones were in the range between levels observed in SW480 and DLD-1 cells (Fig. S2), suggesting that the levels of PLCδ1 overexpression and restored E-cadherin are biologically relevant. Confocal microscopy showed that E-cadherin was localized at the cell–cell junctions in PLCδ1-overexpressing cells, suggesting the functional restoration of E-cadherin by PLCδ1 (Fig. 2D). This restoration of E-cadherin was attenuated by PLCδ1-targeting siRNA (Fig. 2E). Cell morphology was also changed with E-cadherin expression, because individual rounded cells adapted an epithelial morphology with increased cell-to-cell tight adhesions (Fig. 2F).
Fig. 2.
PLCδ1 induced the expression of E-cadherin. (A, Upper) The expressions of E-cadherin, PLCδ1, and β-actin (as a loading control) in DLD-1, SW480, and SW620 cells were determined by Western blotting. (A, Lower) E-cadherin and nuclei were stained with anti–E-cadherin antibody and hoechst33342, respectively (blue, nuclei; green, E-cadherin). E-cad, E-cadherin. (Scale bar: 20 µm.) (B) SW620 cells were transfected with either PLCδ1-expression vector or the relevant empty vector and then selected with G418 treatment for 8 d. The expressions of E-cadherin, PLCδ1, and β-actin (as loading control) in (Left) bulk or (Right) stable clone cells were determined by Western blotting. (C) PLCδ1-overexpressing stable clones (PLCδ1-1, -2, and -3) and the relevant control clones (Control-1 and -2) were assessed for E-cadherin mRNA expression by qRT-PCR analysis. The relative expression levels of E-cadherin, normalized to β-actin (as an internal control), are shown (n = 3), with the value of Control-1 set as one. Statistical analysis was performed using the Tukey multiple comparison of means test. **P < 0.005. (D) E-cadherin (green) and nuclei (blue) were observed by confocal microscopy. (Scale bar: 10 µm.) (E) The PLCδ1-overexpressing stable clone (PLCδ1-1) and the relevant control clone (Control-1) were transfected with siRNA against PLCδ1 (siδ1) or nontarget siRNA (siNeg) and assessed by Western blots for the indicated proteins. (F) The morphology of the cells in E is shown. (Scale bar: 10 µm.)
Endogenous PLCδ1 Contributes to E-Cadherin Expression in a Lipase-Dependent Manner.
Because PLCδ1 overexpression induced E-cadherin expression, we next tried to evaluate the more physiological effects of PLCδ1 on E-cadherin expression in CRC cell lines by silencing experiments. In SW620 and SW480 cells, siRNA-mediated PLCδ1 knockdown reduced E-cadherin expression (Fig. 3A and Fig. S3). In DLD-1 cells, PLCδ1 knockdown also reduced E-cadherin expression and junctional localization (Fig. 3B). Because functional redundancy between PLC isoforms is possible, we also examined whether another PLCδ isoform, PLCδ3, also regulates E-cadherin expression. Although, PLCδ3 expression was significantly down-regulated in CRC (Fig. S4) and ectopic expression of PLCδ3 in SW620 up-regulated E-cadherin expression (Fig. S5A), knockdown of PLCδ3 scarcely affected E-cadherin expression, even in the PLCδ1-knockdown cells (Fig. 3C and Fig. S5 B and C). These results suggest that endogenous PLCδ3 hardly contributes to E-cadherin expression, at least in these cell lines, but our results of PLCδ3 overexpression retain the possibility of PLCδ3 contribution to E-cadherin expression in other cell context.
Fig. 3.
Knockdown of PLCδ1 but not PLCδ3 repressed the expression of E-cadherin in CRC cells. (A) Cells were transfected with negative control siRNA (siNeg) or siRNA for PLCδ1 (siδ1-1 or -2). After 7 d, the expression levels of E-cadherin and PLCδ1 were determined by qRT-PCR (n = 3). The relative mRNA expression, normalized by β-actin, is shown. Western blots for the expression levels of E-cadherin, PLCδ1, and β-actin (as loading control) are shown below. E-cad, E-cadherin. (B) DLD-1 cells transfected with siNeg, siδ1-1, or siδ1-2 were assessed by Western blots for E-cadherin, PLCδ1, and β-actin. Confocal microscope images of these cells stained with E-cadherin antibody (green) and hoechst33342 (blue) are also shown. (Scale bar: 30 µm.) (C) The expression levels of E-cadherin, PLCδ1, and PLCδ3 in SW620 cells transfected with siNeg, siδ1, or siRNA for PLCδ3 (siδ3) as indicated were determined by Western blots. (D) The expression levels of E-cadherin and PLCδ1 (human) mRNA in SW620 cells transduced with siNeg, siδ1 (human), and siδ1 (human) with mouse PLCδ1 (mδ1) or lipase activity-dead PLCδ1 (mδ1LD) expression plasmid were determined by qRT-PCR (n = 3). The relative mRNA expression, normalized by β-actin, is shown. Statistical analysis was performed using the (A) Dunnett or (D) Tukey multiple comparison of means test. n.s., Not significant. *P < 0.05; **P < 0.005.
Moreover, E-cadherin down-regulation by PLCδ1 knockdown was reversed by the coexpression of murine PLCδ1 but not the PLCδ1 lipase activity-dead construct, suggesting the functional redundancy of human and murine PLCδ1 and the importance of lipase activity of PLCδ1. (Fig. 3D).
PLCδ1 Represses the Expression of EMT-Associated Genes.
We next investigated the effect of PLCδ1 on EMT-associated gene expression. Western blots showed that SW620 cells stably overexpressing PLCδ1 express reduced levels of Vimentin, a mesenchymal gene, compared with vector-introduced control cells (Fig. 4A). Down-regulation of Vimentin in PLCδ1-overexpressing cells was also observed by immunofluorescence microscopy (Fig. 4B) and qRT-PCR analysis (Fig. 4C). To investigate the expression of other EMT-related genes, qRT-PCR analysis was performed and showed that ectopic PLCδ1 expression suppressed TGF-β, Zeb1, Slug, and Snail1 mRNA expression (Fig. 4D). Previous reports show that increased E-cadherin expression results in the relocalization of β-catenin, a key factor in CRC progression (13, 14), from the nucleus to the membrane adherens junctions and also, causes a reduction in β-catenin-T-cell factor/lymphocyte enhancer factor (TCF/LEF) signaling as well as the expression of some of the transcription factors that promote EMT (15). To investigate if the PLCδ1-induced E-cadherin is associated with the suppression of β-catenin-TCF/LEF–mediated transcription, a TCF4 transcriptional reporter (TOP-/FOP-FLASH; Upstate) assay was performed. PLCδ1-overexpressing cells showed TOP-/FOP-FLASH activity of about 50–60% compared with control cells (Fig. 4E). These results indicate that PLCδ1 suppressed the expression of EMT-promoting factors in association with the down-regulation of β-catenin-TCF/LEF signaling pathways.
Fig. 4.
PLCδ1 repressed EMT-associated gene expression. (A) The expressions of Vimentin and β-actin (as loading control) in SW620 cells that stably overexpress PLCδ1 (PLCδ1-1, -2, and -3) and control cells (Control-1 and -2) were determined by Western blotting. (B) Vimentin in control cells and PLCδ1-overexpressing SW620 cells was stained using an anti-Vimentin antibody and observed by immunofluorescence microscopy. (C and D) The mRNA expressions of Vimentin, TGF-β, Slug, Snail, and Zeb1 in SW620 cells that stably overexpress PLCδ1 (PLCδ1-1 and -2) and control cells (Control-1 and 2) were determined by qRT-PCR (n = 3). The relative expression levels, normalized to β-actin expression, are shown. (E) Control or PLCδ1-overexpressing cells were transfected with TOP- or FOP-FLASH reporter plasmids and pRL-TK control plasmid in triplicate. The relative values of firefly luciferase activity, normalized by Renilla luciferase activity, are shown. Statistical analysis was performed using Tukey multiple comparison of means test. *P < 0.05; **P < 0.005 (vs. Control-1).
Role of PLCδ1 in Proliferation, Motility, and Invasiveness of CRC Cells.
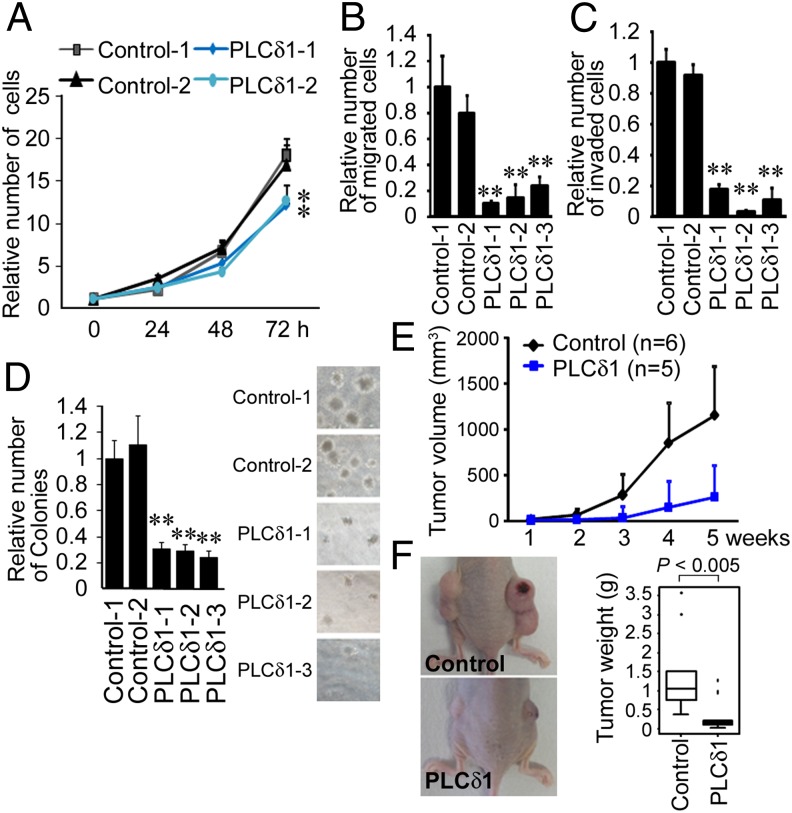
Because E-cadherin is a prognostic molecule and because the loss of E-cadherin has been associated with CRC malignant phenotypes (16), the effects of PLCδ1 on CRC malignant phenotypes were assessed. Because E-cadherin mediates contact inhibition of proliferation (17), we investigated the role of PLCδ1 on CRC cell proliferation. Cells were plated in 24-well plates, and the cell number was counted every 24 h. Induction of PLCδ1 hardly affected the proliferation of SW620 cells at 24 h, but the proliferation rates of PLCδ1-overexpressing cells decreased at 48 and 72 h (Fig. 5A), which could have been caused by E-cadherin–mediated contact inhibition. We next evaluated if PLCδ1 affected cell motility and invasiveness using transwell migration and invasion assays, respectively. The number of migrated SW620 cells, which stably expressed PLCδ1, was about 10–20% of control cells (Fig. 5B). The number of invaded SW620 cells, which stably expressed PLCδ1, was about 3–20% of control cells (Fig. 5C). These results suggest that PLCδ1 repressed the cell proliferation, migration, and invasion of CRC cells.
Fig. 5.
The roles of PLCδ1 on malignant phenotypes of CRC cells. (A) Control or PLCδ1-overexpressing SW620 cells (Control-1 and -2 or PLCδ1-1 and -2) were plated at 10,000 cells/well in 24-well plates. Cells were dissociated by trypsinization, and the total number of cells was determined every 24 h (n = 3). (B and C) Transwell migration or invasion assays were performed with control or PLCδ1-overexpressing SW620 cells as described in Materials and Methods. The relative numbers of migrated or invaded cells are shown (n = 3). (D) Control or PLCδ1-overexpressing SW620 cells were plated in six-well plates with soft agar. After 14 d, the numbers of colonies were counted and are shown in a bar graph (n = 3). The representative images of colonies are shown in Right. (E) Control (n = 6) or PLCδ1-overexpressing SW620 (n = 5) cells were inoculated into the flanks of nude mice. The volume of xenografts was determined as described in Materials and Methods. (F) Representative images of the mice with xenografts are shown in Left. After 5 wk, the weights of the xenografts were assessed. The statistical difference was determined by Mann–Whitney U test. (B–D) Statistical analysis was performed by Tukey multiple comparison of means test. **P < 0.005 (vs. Control-1).
PLCδ1 Suppresses Anchorage-Independent Cell Growth and Tumorigenicity of CRC Cells.
The anchorage-independent growth of cancer cells is one of the hallmarks of malignant phenotypes and promoted by loss of E-cadherin (18). Soft agar assays revealed that PLCδ1 reduced the anchorage-independent cell growth of SW620 cells (Fig. 5D). Anchorage-independent growth in soft agar often relates to the tumorigenic potential of tumor cells (19). We next performed in vivo experiments to evaluate the effect of PLCδ1 overexpression on tumorigenicity. SW620 cells, which stably expressed PLCδ1, and the relevant control cells were inoculated into the flanks of nude mice, and the established xenograft volumes were assessed one time per week. As a result, PLCδ1-overexpressing cells had significantly reduced tumor volume (Fig. 5E). The weights of xenografts from PLCδ1-overexpressing cells were also significantly reduced (Fig. 5F). These results strongly suggest that PLCδ1 functions as a tumor suppressor.
KRAS/MEK Signaling Suppressed the Expression of PLCδ1.
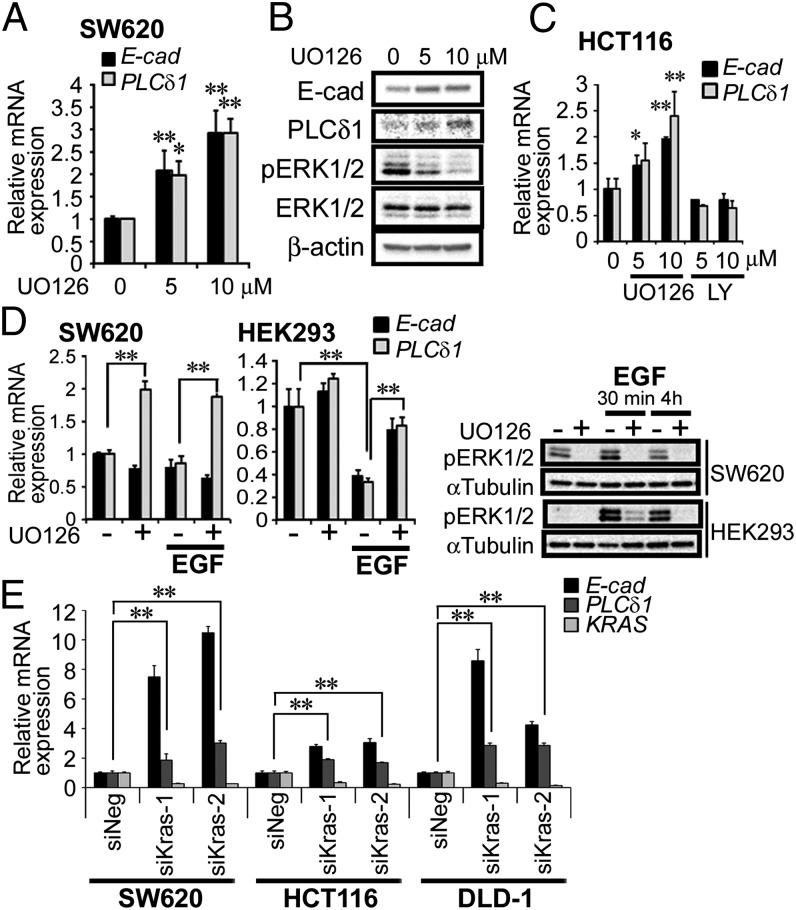
In CRC specimens, low levels of PLCδ1 mRNA expression have been shown to correlate with the presence of KRAS mutations (8). Therefore, we examined if two downstream signaling pathways of KRAS (MEK/ERK and PI3K) affect the expression of PLCδ1 using cells with mutant KRAS (SW620 and HCT116) and cells with WT KRAS (HEK293 and HeLa). SW620 cells (with KRAS G12V) were treated with the MEK inhibitor UO126, and we assessed the expression of PLCδ1. MEK inhibitor treatment increased both PLCδ1 and E-cadherin transcriptional levels about two- to threefold in a dose-dependent manner (Fig. 6A). MEK inhibitor treatment also increased PLCδ1 and E-cadherin protein levels, whereas phosphorylation of the MEK downstream effectors ERK1/2 was decreased in a dose-dependent manner, indicating the effectiveness of UO126 (Fig. 6B). MEK inhibitor treatment also induced PLCδ1 expression about 1.5- to 2.5-fold in HCT116 cells (with KRAS G13D) but not HEK293 (KRAS WT) and HeLa cells (KRAS WT), whereas the PI3K inhibitor LY294002 did not enhance the expression of PLCδ1 in any of these cell lines (Fig. 6C and Fig. S6). The phosphorylation statuses of the downstream effectors of MEK/ERK and PI3K signaling (ERK1/2 and AKT, respectively) were reduced by these inhibitor treatments in these cell lines (Fig. S6C). Furthermore, EGF treatment, which promotes the RAS/MEK pathway and the phosphorylation of ERK1/2 in HEK293 cells, suppressed PLCδ1 expression. The EGF-mediated PLCδ1 suppression in HEK293 cells was recovered by cotreatment with the MEK inhibitor (Fig. 6D). However, EGF did not suppress PLCδ1 expression in SW620 cells in which the KRAS/MEK signal is constitutively active because of the KRAS G13D mutation, whereas UO126 treatment up-regulated PLCδ1 expression (Fig. 6D). Knockdown of KRAS increased PLCδ1 and E-cadherin genes expression in SW620, HCT116, and DLD-1 cells, showing the involvement of KRAS in PLCδ1 repression (Fig. 6E). These results clearly indicated that activation of the KRAS/MEK signaling pathway suppressed PLCδ1 expression. In these experiments, E-cadherin expression was also assessed and shown to be changed similarly to PLCδ1 expression, but the expression of PLCδ1 tended to precede E-cadherin expression, because only PLCδ1 expression was up-regulated with a 4-h treatment of the MEK inhibitor in SW620 cells (Fig. 6D, SW620), although both E-cadherin and PLCδ1 expressions were up-regulated with a 48-h treatment with the MEK inhibitor (Fig. 6A).
Fig. 6.
KRAS/MEK signaling suppressed the expression of PLCδ1. (A) SW620 cells were treated with the MEK inhibitor UO126 (5 or 10 µM) for 48 h. Cells were harvested, and the mRNA expression levels of PLCδ1, E-cadherin, and β-actin (as internal control) were determined by qRT-PCR analysis (n = 3). The relative expression levels of PLCδ1 and E-cadherin, normalized by β-actin, are shown. E-cad, E-cadherin. (B) SW620 cells treated as in A were assessed by Western blots with the indicated antibodies. (C) HCT116 cells were treated with UO126 (5 or 10 µM) or LY294002 (5 or 10 µM) for 24 h, and then, the mRNA expression levels were determined (n = 3). The relative expression levels of PLCδ1 and E-cadherin, normalized by β-actin, are shown. (D) SW620 and HEK293 cells were treated with UO126 (10 µM) or EGF (50 ng/mL) in FBS-free medium at indicated combinations for 4 h; then, cells were harvested, and the mRNA expression levels were determined (n = 3). The relative expression levels of PLCδ1 and E-cadherin, normalized by β-actin, are shown. (D, Right) Cells were also assessed by Western blots for the indicated proteins. (E) SW620, HCT116, and DLD-1 cells were transfected with negative control siRNA (siNeg) or siRNA targeting KRAS (siKras-1 or -2) for 48 h. The relative mRNA expression levels of PLCδ1, E-cadherin, and KRAS, normalized by β-actin, are shown (n = 3). Statistical analysis was performed using Tukey multiple comparison of means test. *P < 0.05; **P < 0.005.
PLCδ1 Suppressed ERK1/2 Phosphorylation Through E-Cadherin.
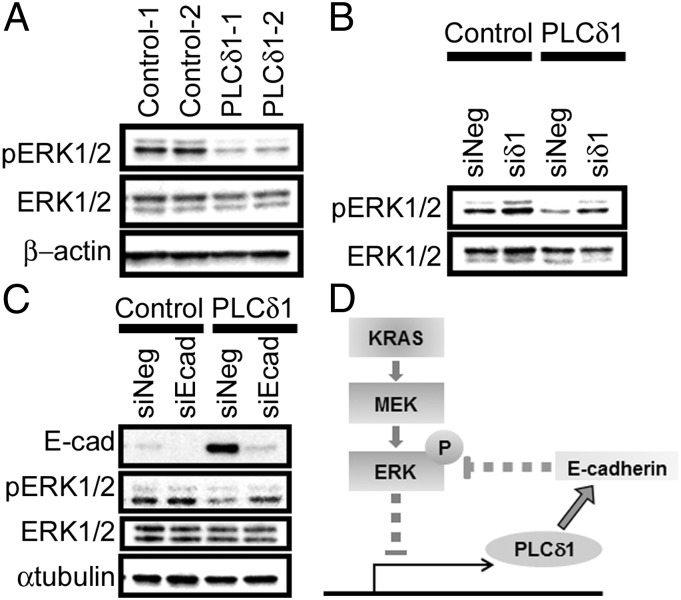
Finally, we examined if PLCδ1 affects the KRAS/MEK/ERK signaling pathway. ERK1/2 phosphorylation levels were assessed by Western blots in control or PLCδ1-overexpressing stable cells. As shown in Fig. 7A, phosphorylated ERK1/2 was reduced in PLCδ1-overexpressing cells. The reduction was mitigated by siRNA-mediated PLCδ1 knockdown (Fig. 7B). Because MEK/ERK signaling is reportedly attenuated by E-cadherin (20), we assessed if suppression of ERK phosphorylation by ectopic PLCδ1 could be mediated by E-cadherin. As shown in Fig. 7C, the decreased ERK1/2 phosphorylation levels in PLCδ1-overexpressing cells were restored by E-cadherin knockdown to the levels of control cells. These results clearly indicate that PLCδ1 suppresses ERK1/2 phosphorylation by restoring E-cadherin expression.
Fig. 7.
PLCδ1 suppressed the phosphorylation of ERK1/2 by E-cadherin. (A) Control or PLCδ1-overexpressing SW620 cells (Control-1 and -2 or PLCδ1-1 and -2) were assessed for the protein levels of phosphorylated ERK1/2, ERK1/2, and β-actin (as a loading control) by Western blots. (B) Control or PLCδ1-overexpressing SW620 cells (Control-1 or PLCδ1-1) were transfected with siRNA for PLCδ1 (siδ1) or negative control siRNA (siNeg). After 5 d, cells were harvested and assessed for protein levels with the indicated antibodies by Western blots. (C) Control or PLCδ1-overexpressing SW620 cells (Control-1 or PLCδ1-1) were transfected with E-cadherin siRNA (siEcad) or siNeg. After 5 d, cells were harvested and assessed for the protein levels with the indicated antibodies by Western blots. E-cad, E-cadherin. (D) The regulatory loop of KRAS/MEK/ERK signaling and PLCδ1 in CRC. KRAS/MEK signaling, which is constitutively promoted by KRAS mutations in many CRC patients, suppressed the expression of PLCδ1. Because PLCδ1 promotes the expression of E-cadherin, one of major tumor-suppressive molecules that also suppresses MEK/ERK signaling, PLCδ1 down-regulation by KRAS mutation accelerates tumor progression. P indicates phosphorylation.
Discussion
Loss of the epithelial adhesive molecule E-cadherin promotes CRC growth and invasiveness and is associated with CRC metastasis and poor prognosis (21, 22). Loss of E-cadherin is promoted by many tumor-promoting factors, including EMT inducers, cytokines, and several tumor-promoting mutant genes, such as p53 (23) or KRAS (24). In this study, we clarified the notable mechanisms of E-cadherin suppression and CRC progression mediated by PLCδ1 down-regulation.
To the best of our knowledge, we show here for the first time that PLCδ1 ectopic expression induced E-cadherin expression, whereas PLCδ1 suppression decreased E-cadherin expression in CRC. To investigate the link between the expressions of PLCδ1 and E-cadherin in other cancer types, we assessed PLCδ1 and E-cadherin levels in hepatocellular carcinoma cell lines HepG2 (with KRAS mutation), SK-Hep-1, and HLE and breast cancer cell lines MDA-MB-231 (with KRAS mutation) and MCF-7. As shown in Fig. S7A, we observed little correlation in these cells. Interestingly, knockdown of PLCδ1 significantly decreased E-cadherin expression in hepatocellular carcinoma cell lines HepG2 and HLE but not MCF-7 (Fig. S7B). These results suggest that, although the total expression level of E-cadherin is regulated by multiple factors in addition to PLCδ1 and the link between total E-cadherin levels and PLCδ1 may be cell-content dependent, E-cadherin is regulated by PLCδ1 in some cancer cell types. Down-regulation of E-cadherin has been reported to be mediated by EMT inducers, and E-cadherin restoration can suppress the expression of EMT inducers (15, 25). Among EMT inducers, we found here that the expressions of TGF-β, Slug, Zeb1, and Snail1 were down-regulated with ectopic PLCδ1 expression. In contrast, transient PLCδ1 knockdown rarely up-regulated the expression of EMT-related genes, whereas E-cadherin expression was reduced by PLCδ1 knockdown. These results suggest that E-cadherin maintenance by PLCδ1 is not always correlated with these EMT inducers.
PLCδ1 is important for the intracellular Ca2+ maintenance in epithelial cells, especially keratinocytes. PLCδ1-KO mice have significantly decreased intracellular Ca2+ levels, resulting in the abnormal differentiation of epidermal and hair follicles (26). Ca2+ and the calcium-sensing receptor are reported to be important for differentiation and E-cadherin expression in colonic epithelial cells, and disruption of calcium-sensing receptor system contributes to abnormal differentiation and malignant progression (27). Therefore, PLCδ1 may contribute to intracellular Ca2+ maintenance, which is necessary for E-cadherin expression in CRC. Ca2+ activates several signaling molecules, including calcineurin/nuclear factor of activated T cells and PKC. Additional study to understand the molecules downstream of PLCδ1 that are necessary for its tumor-suppressive function is needed in future works.
In addition, we clarified that KRAS/MEK signaling represses PLCδ1 expression. Furthermore, we first elucidated that PLCδ1 protein levels were significantly reduced in CRC specimens compared with the normal colonic epithelium by tissue microarray analysis. In this analysis, no association between PLCδ1 expression and tumor grade or stage was observed. This phenomenon is likely caused by KRAS/MEK signaling-mediated PLCδ1 down-regulation. Because KRAS mutation is observed during the early stages in multistep processes of carcinogenesis (28), PLCδ1 down-regulation by KRAS/MEK signaling may also occur in the early stages of colorectal carcinogenesis. We also showed that PLCδ1 suppressed the KRAS/MEK/ERK pathway, suggesting a negative regulatory loop between KRAS/MEK/ERK signaling and PLCδ1 (Fig. 7D). From these results, we speculate that activation of PLCδ1 or PLCδ1 downstream signaling, which restores E-cadherin and suppresses tumor malignancy, could be a novel strategy for CRC treatments. Notably, this strategy may lead to a virtuous cycle of restoring PLCδ1 expression, enhancing E-cadherin expression, and attenuating KRAS/MEK/ERK signal, which may, ultimately, inhibit cancer malignancy.
In this study, we found a significant reduction in PLCδ1 expression in CRC cells in clinical specimens and clarified the roles of PLCδ1 in KRAS-mutant CRC cell lines. We revealed that PLCδ1 was responsible for E-cadherin expression, suppression of EMT, cell motility, invasiveness, and tumorigenicity. Furthermore, the expression of PLCδ1 was repressed by KRAS/MEK signaling, whereas PLCδ1 suppressed the phosphorylation of ERK1/2 through E-cadherin. These data indicate that PLCδ1 has tumor-suppressive functions in CRC through E-cadherin induction and KRAS/MEK/ERK signal attenuation. It is worth noting that KRAS knockdown increased E-cadherin and PLCδ1 expressions in CRC. Because the level of E-cadherin and malignancy of CRC are changeable by PLCδ1, PLCδ1-inducible compounds may be a convincing candidate for CRC drugs. Our results provide a valuable perspective on the therapeutic approaches to KRAS-mutated CRC.
Materials and Methods
Immunohistochemistry.
Human colon carcinoma tissue arrays with matched adjacent normal colon tissue were purchased from Biomax (US Biomax). The immunostaining was performed as described previously (29). Immunohistochemical assays for human PLCδ1 were performed with a Vectastain Elite Rabbit ABC Kit (Vector Laboratories) with anti-PLCδ1 antibody (Sigma) followed by light counterstaining with Mayer’s hematoxylin (Wako). Sections were examined under a BX51 microscope (Olympus).
Cell Culture.
The colorectal adenocarcinoma cell lines SW620 and HCT116 were obtained from the American Type Culture Collection. DLD-1 and HEK293 cell lines were obtained from the Japanese Collection of Research Bioresources Cell Bank (National Institute of Health Sciences). These cells were maintained at 37 °C in a 5% (vol/vol) CO2 humidified atmosphere in RPMI medium 1640 (Invitrogen) supplemented with 10% (vol/vol) FBS. SW480 and HeLa cells were cultured as described previously (30).
Western Blot Analysis.
Western blot analysis was performed as described previously (29) with some modifications. Primary antibodies for E-cadherin (BD Biosciences), GAPDH, phospho-Akt (Ser473), Akt, phospho-ERK1/2 (Thr202/204), ERK1/2 (Cell Signaling), anti–β-actin antibody (Sigma), α-tubulin (GeneTex), Vimentin (Santa Cruz), and PLCδ1 (Santa Cruz) were used.
Migration and Invasion Assay.
Migration assays were performed using cell culture insert with 8-µm-sized pores (BD Biosciences) in a 24-well plate with RPMI Medium 1640 containing 10% (vol/vol) FBS. For invasion assays, the cell culture inserts were added with 60 µL (2.5 mg/mL) BD Matrigel Basement Membrane Matrix Growth Factor Reduced (BD Biosciences) as described previously (31). Materials and methods for plasmids; siRNA and transfection; immunofluorescence microscopy; RNA isolation, cDNA synthesis, and qRT-PCR; luciferase reporter assay; cell proliferation assay; soft agar colony formation assay; animal experiments; and statistical analysis are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Y. Nakamura, and Dr. A. Yoneda for fruitful discussions. This work was supported by the Funding Program for the Next Generation World-Leading Researchers (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.G.R. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405374111/-/DCSupplemental.
References
- 1.Heinemann V, Douillard JY, Ducreux M, Peeters M. Targeted therapy in metastatic colorectal cancer — an example of personalised medicine in action. Cancer Treat Rev. 2013;39(6):592–601. doi: 10.1016/j.ctrv.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 2.Láng I, et al. Quality of life analysis in patients with KRAS wild-type metastatic colorectal cancer treated first-line with cetuximab plus irinotecan, fluorouracil and leucovorin. Eur J Cancer. 2013;49(2):439–448. doi: 10.1016/j.ejca.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 3.Normanno N, et al. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 4.Zenonos K, Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World J Gastrointest Oncol. 2013;5(5):97–101. doi: 10.4251/wjgo.v5.i5.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agbunag C, Bar-Sagi D. Oncogenic K-ras drives cell cycle progression and phenotypic conversion of primary pancreatic duct epithelial cells. Cancer Res. 2004;64(16):5659–5663. doi: 10.1158/0008-5472.CAN-04-0807. [DOI] [PubMed] [Google Scholar]
- 6.Rachagani S, et al. Activated KrasG¹²D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br J Cancer. 2011;104(6):1038–1048. doi: 10.1038/bjc.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang YR, Follo MY, Cocco L, Suh PG. The physiological roles of primary phospholipase C. Adv Biol Regul. 2013;53(3):232–241. doi: 10.1016/j.jbior.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Danielsen SA, et al. Phospholipase C isozymes are deregulated in colorectal cancer—insights gained from gene set enrichment analysis of the transcriptome. PLoS ONE. 2011;6(9):e24419. doi: 10.1371/journal.pone.0024419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert JP, Zhou Y, Hicks SN, Sondek J, Harden TK. Dual activation of phospholipase C-epsilon by Rho and Ras GTPases. J Biol Chem. 2008;283(44):29690–29698. doi: 10.1074/jbc.M805038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Edamatsu H, Kitazawa R, Kitazawa S, Kataoka T. Phospholipase Cepsilon promotes intestinal tumorigenesis of Apc(Min/+) mice through augmentation of inflammation and angiogenesis. Carcinogenesis. 2009;30(8):1424–1432. doi: 10.1093/carcin/bgp125. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, et al. Phospholipase C epsilon plays a suppressive role in incidence of colorectal cancer. Med Oncol. 2012;29(2):1051–1058. doi: 10.1007/s12032-011-9981-1. [DOI] [PubMed] [Google Scholar]
- 12.Buck E, et al. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther. 2007;6(2):532–541. doi: 10.1158/1535-7163.MCT-06-0462. [DOI] [PubMed] [Google Scholar]
- 13.Shitashige M, et al. Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology. 2008;134(7):1961–1971. doi: 10.1053/j.gastro.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Satow R, et al. β-catenin inhibits promyelocytic leukemia protein tumor suppressor function in colorectal cancer cells. Gastroenterology. 2012;142(3):572–581. doi: 10.1053/j.gastro.2011.11.041. [DOI] [PubMed] [Google Scholar]
- 15.Conacci-Sorrell M, et al. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: The roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163(4):847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugli A, et al. Prognostic significance of the wnt signalling pathway molecules APC, beta-catenin and E-cadherin in colorectal cancer: A tissue microarray-based analysis. Histopathology. 2007;50(4):453–464. doi: 10.1111/j.1365-2559.2007.02620.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci USA. 2011;108(29):11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau MT, Klausen C, Leung PC. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via β-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30(24):2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 19.Shin SI, Freedman VH, Risser R, Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci USA. 1975;72(11):4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laprise P, Langlois MJ, Boucher MJ, Jobin C, Rivard N. Down-regulation of MEK/ERK signaling by E-cadherin-dependent PI3K/Akt pathway in differentiating intestinal epithelial cells. J Cell Physiol. 2004;199(1):32–39. doi: 10.1002/jcp.10432. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, et al. Loss of E-cadherin promotes the growth, invasion and drug resistance of colorectal cancer cells and is associated with liver metastasis. Mol Biol Rep. 2012;39(6):6707–6714. doi: 10.1007/s11033-012-1494-2. [DOI] [PubMed] [Google Scholar]
- 22.Mohri Y. Prognostic significance of E-cadherin expression in human colorectal cancer tissue. Surg Today. 1997;27(7):606–612. doi: 10.1007/BF02388215. [DOI] [PubMed] [Google Scholar]
- 23.Roger L, Jullien L, Gire V, Roux P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J Cell Sci. 2010;123(Pt 8):1295–1305. doi: 10.1242/jcs.061002. [DOI] [PubMed] [Google Scholar]
- 24.Makrodouli E, et al. BRAF and RAS oncogenes regulate Rho GTPase pathways to mediate migration and invasion properties in human colon cancer cells: A comparative study. Mol Cancer. 2011;10:118. doi: 10.1186/1476-4598-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onder TT, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura Y, et al. Phospholipase Cdelta1 is required for skin stem cell lineage commitment. EMBO J. 2003;22(12):2981–2991. doi: 10.1093/emboj/cdg302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarty S, Radjendirane V, Appelman H, Varani J. Extracellular calcium and calcium sensing receptor function in human colon carcinomas: Promotion of E-cadherin expression and suppression of beta-catenin/TCF activation. Cancer Res. 2003;63(1):67–71. [PubMed] [Google Scholar]
- 28.Ilyas M, Straub J, Tomlinson IP, Bodmer WF. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35(14):1986–2002. doi: 10.1016/s0959-8049(99)00298-1. [DOI] [PubMed] [Google Scholar]
- 29.Kanemaru K, et al. Epidermal phospholipase Cδ1 regulates granulocyte counts and systemic interleukin-17 levels in mice. Nat Commun. 2012;3:963. doi: 10.1038/ncomms1960. [DOI] [PubMed] [Google Scholar]
- 30.Kouchi Z, Fujiwara Y, Yamaguchi H, Nakamura Y, Fukami K. Phosphatidylinositol 5-phosphate 4-kinase type II beta is required for vitamin D receptor-dependent E-cadherin expression in SW480 cells. Biochem Biophys Res Commun. 2011;408(4):523–529. doi: 10.1016/j.bbrc.2011.04.045. [DOI] [PubMed] [Google Scholar]
- 31.Hirano T, et al. Identification of novel small compounds that restore E-cadherin expression and inhibit tumor cell motility and invasiveness. Biochem Pharmacol. 2013;86(10):1419–1429. doi: 10.1016/j.bcp.2013.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.