Significance
Because posttraumatic stress disorder (PTSD) occurs in a subset of trauma-exposed persons, expression profiling in the context of an animal model that focuses on individual differences in stress response permits identification of the relevant signaling pathways that lead to sustained impairment or resilience. The inclusion of blood and brain samples from both sexes is important because it allows the detection of convergent susceptibility pathways and concomitant identification of blood-based biomarkers. The across tissue and sex involvement of glucocorticoid receptor signaling with exposure-related individual differences suggests that targeting this signaling pathway may lead to a promising therapeutic strategy in PTSD.
Keywords: predator stress, transcription regulation, NR3C1, preventive treatment, psychiatry
Abstract
Delineating the molecular basis of individual differences in the stress response is critical to understanding the pathophysiology and treatment of posttraumatic stress disorder (PTSD). In this study, 7 d after predator-scent-stress (PSS) exposure, male and female rats were classified into vulnerable (i.e., “PTSD-like”) and resilient (i.e., minimally affected) phenotypes on the basis of their performance on a variety of behavioral measures. Genome-wide expression profiling in blood and two limbic brain regions (amygdala and hippocampus), followed by quantitative PCR validation, was performed in these two groups of animals, as well as in an unexposed control group. Differentially expressed genes were identified in blood and brain associated with PSS-exposure and with distinct behavioral profiles postexposure. There was a small but significant between-tissue overlap (4–21%) for the genes associated with exposure-related individual differences, indicating convergent gene expression in both sexes. To uncover convergent signaling pathways across tissue and sex, upstream activated/deactivated transcription factors were first predicted for each tissue and then the respective pathways were identified. Glucocorticoid receptor (GR) signaling was the only convergent pathway associated with individual differences when using the most stringent statistical threshold. Corticosterone treatment 1 h after PSS-exposure prevented anxiety and hyperarousal 7 d later in both sexes, confirming the GR involvement in the PSS behavioral response. In conclusion, genes and pathways associated with extreme differences in the traumatic stress behavioral response can be distinguished from those associated with trauma exposure. Blood-based biomarkers can predict aspects of brain signaling. GR signaling is a convergent signaling pathway, associated with trauma-related individual differences in both sexes.
Posttraumatic stress disorder (PTSD) develops in only some persons who are exposed to extremely traumatic life events (1). Animal models that focus on identifying different patterns and adaptations in the behavioral response to trauma are of particular clinical relevance (2). Development of animal models can be accomplished by studying animals at the extremes of the behavioral response distribution (vulnerable vs. resilient). Along these lines, Cohen and Zohar developed an animal model of PTSD in which adult outbred rats are exposed briefly to predator-scent-stress (PSS), an ecologically valid stressor that mimics a life-threatening situation for a rodent (3). This exposure resulted in animals displaying a wide-range of behavioral and physiological responses to later provocations (3). Statistically validated cut-off behavioral criteria (CBC) were used to classify exposed rats according to their performance on behavioral tests; for example, anxiety behavior in an elevated plus-maze (EPM) and arousal assessed as the acoustic-startle response (ASR): 25% of Sprague–Dawley rats display an extreme behavioral response (EBR, “PTSD-like” vulnerability), 25% a minimal behavioral response (MBR, resilience), and 50% a partial behavioral response (PBR) (3).
This approach has been used to validate candidate biological markers identified in PTSD, such as the blunted glucocorticoid response to stress (4, 5). The emergence of system- and genome-wide approaches permits the opportunity for unbiased identification of novel pathways. Because PTSD is more prevalent in women than men (1), and sex is a potential source of response variation to trauma in both animals (6) and humans (7), it is also critical to include both sexes in such studies.
In the present study, PSS-exposed male and female rats were behaviorally tested in EPM and ASR tests a week after PSS and divided in EBR and MBR groups [at this point, the behavioral response of the rats is stable in terms of prevalence of EBRs vs. MBRs (3)]. Genome-wide expression was evaluated 24 h after the behavioral testing in the amygdala, hippocampus, and blood, and the control (CON) group consisted of stress-naïve but behaviorally tested rats (Fig. S1A). Data were analyzed with the aim at identifying differentially expressed genes (DEG) together with the respective transcription factors and signaling pathways in the brain, and their blood correlates. A candidate signaling pathway for PTSD, the glucocorticoid receptor (GR) signaling pathway (8), was detected to be associated with exposure-related individual differences in this animal model, and preventive treatment targeting this pathway was then evaluated in both sexes (Fig. S1B).
Results
According to previously defined CBC (3), 10 male (26.3%) and 12 female (28.6%) rats fulfilled criteria for EBR, and 10 males (26.3%) and 10 females (23.8%) were classified as MBR (Fig. S1 C and D). A χ2 analysis indicated that sex did not affect the prevalence of extremes in the behavioral response to PSS (χ21, 80 = 0.087, not significant). Expression profiling in amygdala, hippocampus, and blood of EBRs, MBRs and CONs detected a comparable number of probes in both sexes (Fig. S2A). The differential gene expression analysis identified multiple differentially expressed probes in all tissue and both sexes (Fig. S2B and Dataset S1 A–F), which were validated (Fig. S3 and Dataset S1G) by quantitative PCR (qPCR). There was a distinct distribution of differentially expressed probes for each tissue and sex.
The inclusion of unexposed-to-PSS rats together with exposed groups with extreme differences in phenotype permitted the identification of gene expression associated with stress-exposure and with exposure-related individual differences. The total number of DEG linked to individual differences differed in various tissues and in males and females (from 86 genes to 334 genes), representing 36.5–98.9% of the total DEG (Fig. S4A). Furthermore, in general, vulnerability was associated with more DEG than resilience (Fig. S4 C–H). The overlap of vulnerability-related and resilience-related genes was universally very small (0.0–2.4%), indicating that the vulnerability or resilience behavioral constructs were distinct at the trascriptome level.
Because of their limited overlap, the vulnerability- and resilience-related DEG were pooled in subsequent analyses because together they reflected exposure-related individual differences. An analysis comparing those genes between tissue in each sex separately revealed a small between-tissue overlap in both sexes [1.1–20.9% for males, (Fig. 1A, Center); 0.6–16.4% for females (Fig. 1B, Center)]. Only CD93 was present in all three tissues (in females). In both sexes, the overlap between amygdala and hippocampus DEG was significant (P < 0.001). Additionally, the overlap between amygdala and blood DEG was significant in males (P = 0.009 for the amygdala gene pool, P = 0.016 for the blood gene pool), and between hippocampus and blood in females (P = 0.005 for the hippocampus gene pool, P = 0.031 for the blood gene pool).
Fig. 1.
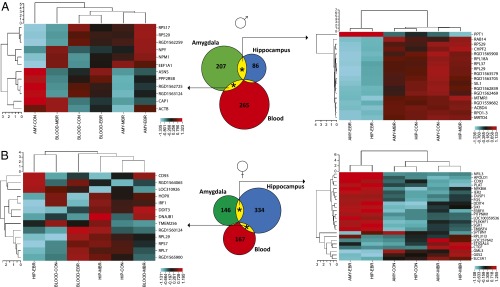
Numbers of DEG associated with individual differences in the behavioral response to PSS in amygdala (AMY), hippocampus (HIP), and blood in males (A) and females (B). The area-proportional Venn diagrams (Center) indicate a tissue-specific DEG distribution, but also a between-tissue overlap in both sexes. The four significant between-tissue overlaps are indicated by an asterisk (*) and the respective DEG were submitted to unsupervised hierarchical clustering. The vertical dendrograms of the four (two left and two right) heatmaps reflect gene clustering, and the horizontal dendrograms reflect group clustering (i.e., EBRs, MBRs, and CONs). The color scale of the heatmaps is a z-score of transformed gene expression values, with red corresponding to high expression and blue to low. Both (Right) heatmaps of the amygdala-hippocampus overlaps revealed good concordance among groups, whereas the (Left) heatmaps of the overlaps of blood with amygdala (A) or with hippocampus (B) did not reveal a similar concordance.
Unsupervised hierarchical clustering of DEG associated with exposure-related individual differences in both amygdala and hippocampus revealed that the same experimental groups clustered together in both males (Fig. 1A, Right) and females (Fig. 1B, Right). The values of the EBR group clustered together and away from the respective values of the MBRs and CONs, indicating that likely, up-regulation or down-regulation of those genes was vulnerability-related in both regions. No such clustering was observed for the shared DEG between amygdala and blood (Fig. 1A, Left) or hippocampus and blood (Fig. 1B, Left).
An upstream regulator analysis identified transcription factors in amygdala, hippocampus, and blood, respectively, for which activation/deactivation was associated with exposure-related individual differences in gene expression. The identified transcription factors were merged across sex, yielding 29 transcription factors in the amygdala (Dataset S2A), 31 in the hippocampus (Dataset S2B), and 48 in the blood (Dataset S2C). The same analysis using the stress-exposure–associated DEG sets predicted other and overlapping transcription factors in all tissues (Dataset S2 A–C). A small portion of the transcription factors predicted by individual differences associated DEG sets regulates stress-exposure–associated DEG in amygdala (13.8%) and hippocampus (12.9%), and in the blood this portion is large (52.1%).
Seventy-three unique transcription factors were predicted to regulate DEG associated with individual differences (Dataset S2D), and a between-tissue overlap (Fig. 2, Venn diagram) revealed that 47 were tissue-specific (11 in amygdala, 11 in hippocampus, and 25 in blood), 3 shared between amygdala and hippocampus, 6 shared between amygdala and blood, 8 shared between hippocampus and blood, and 9 convergent across tissue [CREB1, FOXO3, JUN, MYC, MYCN, NFE2L2, NFKBIA (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha), NR3C1 (nuclear receptor subfamily 3, group C, member 1), TP53]. From the latter, only GR (NR3C1), which is a ligand-dependent nuclear receptor, is a target of known pharmaceutical agents (Dataset S2D). The nine across-tissue transcription factors target 265 individual differences DEG (Dataset S2E). These genes were ranked (Dataset S2F) based on the number of transcription factors that target them: 3 were targeted by seven transcription factors [FN1, FOS (FBJ murine osteosarcoma viral oncogene homolog), SOD2], 1 by six transcription factors [SGK1 (serum/glucocorticoid regulated kinase 1)], 6 by five transcription factors [CTGF, DUSP1, IL1B (interleukin 1, beta), JUNB, NFKBIA, TXN], 14 by four transcrption factors, 22 by three transcription factors, 58 by two transcription factors, and 161 by only one transcription factor.
Fig. 2.
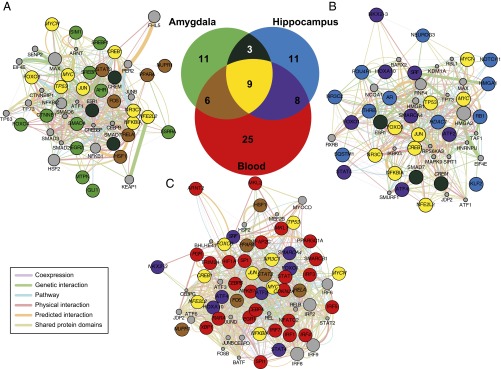
Gene network analysis of the amygdala (A), hippocampus (B), and blood (C) transcription factors (Dataset S2 A–C) regulating DEG associated with individual differences in the behavioral response to PSS. The area proportional Venn-diagram (Center) represents the overlap between the amygdala (A), hippocampus (B), and blood (C) transcription factors. The three weighted gene networks were built using the GeneMANIA database (9). The coloring of the network nodes is the same as in the Venn diagram to represent the 47 tissue-specific transcription factors (11 light green nodes in amygdala, 11 light blue in hippocampus, and 25 red in blood), three shared between amygdala and hippocampus (dark turquoise), six shared between amygdala and blood (brown), eight shared between hippocampus and blood (purple), and nine convergent across tissue (yellow). The nodes with their name italicized represent transcription factors that regulate also stress-exposure–associated DEG in the same tissue (Dataset S2 A–C). The gray nodes are predicted interactors and their diameter denotes the prediction score (Dataset S3 A–C). The between-nodes edges represent relationships, the color of the edges represent the type of the relationship (coexpression, genetic interactions, physical interactions, common pathway, shared protein domains), and the thickness of the edges denotes weight (i.e., strength of the pairwise relationship) (Dataset S3 A–C).
The independent submission of the amygdala, hippocampus, and blood transcription factors lists to GeneMania (9), a large functional association database, permitted the building of self-organizing gene networks in each tissue separately (for amygdala in Fig. 2A and Dataset S3A; for hippocampus in Fig. 2B and Dataset S3B; for blood in Fig. 2C and Dataset S3C), where all of the factors were interlinked at multiple levels (coexpression, genetic and physical interactions, common pathway, shared protein domains).
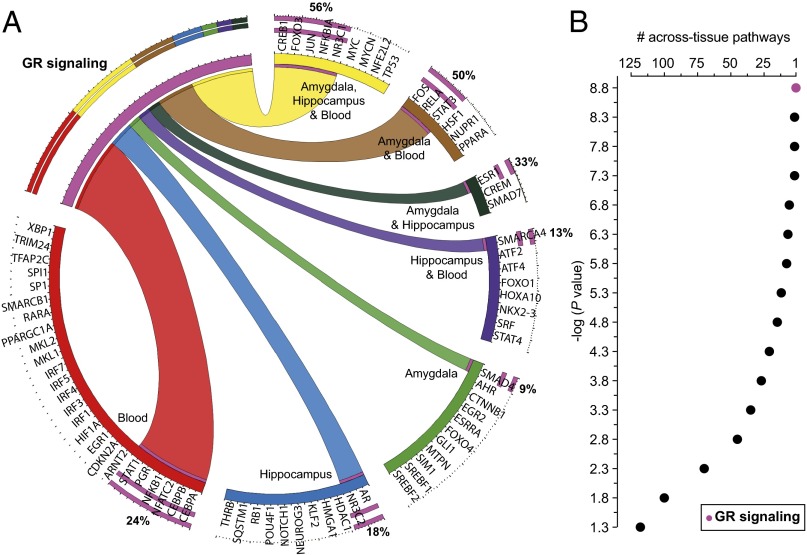
The canonical pathway annotation (Ingenuity Pathway Analysis; IPA) of the 73 identified transcription factors (Dataset S2D) revealed that the blood-specific transcription factors belong in more canonical pathways (average 25) than the amygdala-specific (average 6) or the hippocampus-specific (average 5) factors, and that the GR signaling is the pathway including the highest portion of factors (19 of 73) (Fig. 3A and Dataset S4A). A canonical pathway enrichment analysis (IPA), using the amygdala, hippocampus, and blood transcription factor lists separately, uncovered multiple tissue-specific and across-tissue enriched canonical pathways. The amygdala transcription factor list predicted the enrichment of 178 signaling pathways with a significant Fischer’s exact test P value and 171 pathways after a Benjamini–Holzberg multiple testing correction (top 10 in Fig. S5A and Dataset S4B). The respective numbers for the hippocampus list were 150 and 136 (top 10 in Fig. S5B and Dataset S4C), and for the blood list 199 and 198 (top 10 in Fig. S5C and Dataset S4D). To uncover the convergent across tissue and sex pathways, the amygdala, hippocampus, and blood-enriched pathways were compared using different statistical thresholds. At the least-strict statistical threshold, 118 pathways were shared by all tissue, whereas at the most-stringent threshold, only one pathway (GR signaling) remained common (Fig. 3B).
Fig. 3.
Circos diagram (A) depicting which of the 73 transcription factors predicted to regulate the DEG associated with exposure-related individual differences are also part of the GR signaling (19 of 73 = 26%) according to the IPA. The diagram was prepared using the Circos Table Viewer v0.63–9 (http://mkweb.bcgsc.ca/tableviewer/). Number of across-tissue enriched canonical pathways (B) in association with exposure-related individual differences at different statistical thresholds. The canonical pathways, significantly enriched (IPA) in the amygdala, hippocampus, and blood transcription-factor lists associated with exposure-related individual differences were compared. At the least-strict statistical threshold, -log(P value) = 1.3, 118 pathways were enriched in all three tissue types, whereas at the most stringent threshold possible, -log(P value) = 8.8, only GR signaling was still enriched (pink/purple datapoint).
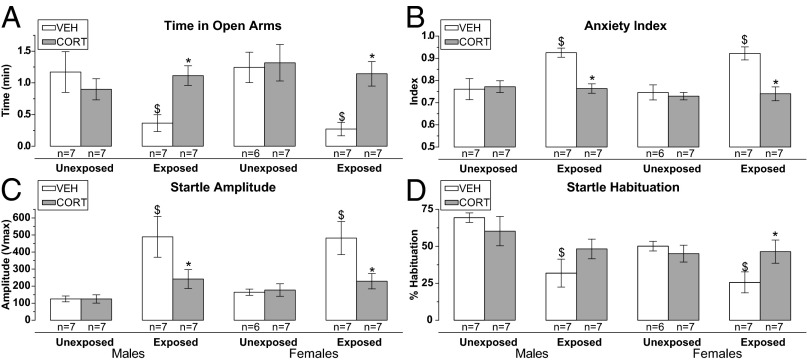
GR and GR-related pathways involvement in the PSS behavioral response in both sexes was confirmed in a follow-up study, where corticosterone (CORT) was intraperitoneally injected 1 h after PSS and behavior was tested 7 d later. A significant interaction effect between PSS-exposure and CORT-treatment (F1, 24 = 6.2, P = 0.020) on time spent in the open arms of the EPM (Fig. 4A) was observed in males. CORT-treated exposed spent more time in the open arms than vehicle (VEH) -treated exposed rats (P = 0.017). The results were appreciably the same when the anxiety index was used as outcome measure (Fig. 4B). In females, there were main effects of PSS-exposure (F1, 23 = 7.4, P = 0.006), CORT-treatment (F1, 23 = 4.9, P = 0.003), and their interaction at a trend level (F1, 23 = 3.3, P = 0.081) on time spent on the open arms (Fig. 4A). CORT-treated exposed spent more time in the open arms than VEH-treated exposed rats (P = 0.002). The results were similar when the anxiety index measure was used (Fig. 4B). Thus, in both sexes, CORT preventive treatment was associated with lower anxiety behavior in PSS-exposed rats.
Fig. 4.
Effects of CORT preventive treatment on the behavioral response to PSS. A high CORT dose (25.0 mg/kg), 1 h after PSS, increased rats’ time in open arms (A) and decreased anxiety (B) in the EPM compared with VEH treated in both sexes. CORT also reduced the ASR amplitude (C) of PSS-exposed rats in both sexes, but reversed the PSS-induced ASR habituation (D) deficit only in females. $ vs. corresponding value of unexposed; * vs. corresponding value of the VEH-treated. Data are presented as mean ± SEM. Significance was set at P < 0.05.
The reversal of the PSS-induced phenotype by CORT-treatment was also apparent in the ASR-test. In males, there were main effects of PSS-exposure (F1, 24 = 12.6, P = 0.002), and trend level effects of CORT-treatment (F1, 24 = 3.3, P = 0.080) and their interaction (F1, 24 = 3.3, P = 0.080) on startle amplitude (Fig. 4C). CORT-treated exposed startled less than VEH-treated exposed rats (P = 0.016). In females, the same effects were observed (PSS-exposure: F1, 23 = 9.9, P = 0.005, CORT-treatment: F1, 23 = 4.1, P = 0.052 and their interaction: F1, 23 = 5.1, P = 0.033). CORT-treated exposed startled less than VEH-treated exposed rats (P = 0.005) (Fig. 4C). CORT-treatment did not affect startle habituation in males (there was only a main effect of PSS-exposure: F1, 24 = 10.2, P = 0.004). However, it affected startle habituation in females (PSS-exposure: F1, 23 = 3.4, P = 0.077, interaction effect between PSS-exposure and CORT-treatment: F1, 23 = 3.3, P = 0.051) (Fig. 4D). CORT-treated exposed displayed greater habituation than VEH-treated exposed rats (P = 0.026).
Discussion
Examination of EBR, MBR, and CON groups permitted the identification of the gene expression signatures (genes, transcription factors, and signaling pathways) associated with vulnerable or resilient behaviors in blood and two stress-regulatory brain regions in both sexes. This approach has previously been used in the social-defeat stress paradigm, where DEG were identified in the nucleus accumbens and ventral tegmental area in association with exposure-related individual differences (10). In both studies, the small overlap between vulnerability- and resilience-related differential gene expression indicates the specificity of the identified DEG in their association with either extreme behavioral response. These findings may have a diagnostic value, particularly if one were to use gene-expression differences from CONs as a benchmark for judging direction in the diagnosis of phenotype.
Different numbers of genes were associated with vulnerability or resilience in males and females, whereas the prevalence of vulnerable and resilient phenotypes was similar in both sexes. The presence of sex differences in gene expression is consistent with numerous reports (11, 12), but few studies compared males and females under the same provocation. In PTSD, sex differences have been noted in both prevalence (7) and gene expression profiles (13). Pooling data across-sex allowed the identification of convergent upstream regulation and the respective signaling pathways associated with exposure-related individual differences and potentially blood-based biomarkers for PTSD risk and resilience that operate in both sexes.
The small, but significant, between-tissue overlap of DEG associated with individual differences revealed the existence of both tissue-specific and convergent across-tissue gene regulation and signaling. The GR signaling pathway was identified as one of the most convergent pathways associated with extreme differences in the behavioral response to PSS-exposure. It is noteworthy that, using an agnostic approach, this genome-wide expression study identified what careful hypothesis-driven research in human and animal studies also demonstrated (8, 14, 15). GR signaling is hypothesized to be low before (16) or immediately after (5) trauma exposure in PTSD compromising stress-induced adaptation and making that time-window a potential target for prevention with glucocorticoids. A high dose of a natural or synthetic GR ligand shortly before or after stress exposure prevented PTSD-like phenotypes in the PSS model (4, 17, 18) or in another model of single stress exposure (19). The behavioral rescue was accompanied by a recovery of stress-induced changes in the dendritic tree morphology in the dentate gyrus of hippocampus (18) and basolateral amygdala (19). In this study, the effectiveness of secondary prevention with CORT in both sexes supported further the involvement of GR signaling in the development of the PSS behavioral response. High-dose CORT-treatment at a critical poststress time-window may recalibrate GR responsiveness and GR signaling (15) but also affect other GR-dependent interrelated pathways. Consistent with this idea, previous data in men and women reflect that NFκB-signaling is up-regulated in PTSD (13), and this could be prevented by glucocorticoids or other more specific inhibitors (20).
Response variation in the PSS-model might be an outcome of a priori differences (in the genetic make-up or early-life experiences) or an interaction of a priori differences with the stress exposure. Because we used outbred rats, genetic differences are possible. Assaying gene expression and genetic variation simultaneously on a genome-wide basis could detect the more genetically driven expression quantitative trait loci associated with individual differences. Interestingly, inbred rats and mice, although genetically identical, show individual differences in the response to PSS (4, 21). However, some of the inbred strains tested with the PSS protocol showed high prevalence of EBRs (e.g., 50% in Lewis, 55% in C57BL/6), and others (e.g., Fisher 344, DBA/2) showed lower rates (4, 21). It is worth noticing that C57BL/6 mice displayed a similar prevalence of extreme response to PSS (21) and to chronic social-defeat stress (10), supporting the existence of a priori nongenetic differences that contribute to the behavioral response to stress independent of the genetic background or the nature/severity of the exposures. Early adversity can alter the epigenetic status of the GR or other stress regulatory genes and predict individual differences in the adult physiologic and behavioral stress response (22), and this type of experiences might be a source of the response variation in the PSS-model as well. We have recently reported alterations in the methylation of a GR gene promoter in relation to PTSD (23) and its risk (24).
Longitudinal assessment of gene expression and behavior in relation to the development of poststress phenotypes is necessary to delineate the predictive nature of molecular biomarkers. Apart from blood sampling or biopsy-collection issues, an additional difficulty in such an undertaking is that gene expression has to be assessed in the prestress samples of the whole population of rats, and not only in the animals that eventually display extreme differences in phenotype, to really demonstrate the predictive value of the identified biomarkers. This approach has been fruitful in PTSD animal models (25), and also in human studies of PTSD where, for example, predeployment peripheral blood GR levels (16) or expression of immune genes (26) could predict postdeployment PTSD-risk and resilience.
The present data should be regarded in the context of recent studies with other animal models of PTSD or stress-related disorders that also studied expression patterns at long-term stress recovery and not only immediately after stress exposure. Two studies investigated differential gene expression in the amygdala during fear-conditioning based on previous experience of a homotypic (27) or heterotypic stressor (28). Gene expression was also investigated in blood and brain tissue of mice exposed to an anxiogenic vs. an anxiolytic drug using the convergent functional genomics approach (29). Another study examined within-brain correlations of expression of mitochondrial and mitochondria-related nuclear genes of rats exposed to inescapable stress (restraint stress with tail-shock) compared with unexposed CONs (30). Finally, expression of blood and brain core modules (31) and hippocampal DEG (32) were associated with short- and long-term recovery to chronic social defeat and restraint stress, respectively. There are genes (e.g., FOS, IL1B, ITGB1, NFKBIA, SGK1) and signaling pathways (e.g., GR, cancer, inflammation, immune) that are common between these studies and ours, but the differences also reflect the different focus of each model (fear mechanisms, stress exposure, or individual differences).
In conclusion, blood and brain expression profiling using an animal model that captures individual differences of the traumatic-stress response can distinguish genes and pathways associated with exposure-related individual differences from those associated with stress exposure per se. The data also illustrate that identification of blood-based biomarkers is possible using valid animal models and genomic tools. This report is an initial step in a longer process to fully unpack these important issues using replication, parallel genome-wide genetic, epigenetic, and expression analyses, and computational integrative methods. The GR signaling pathway in blood and brain is involved in exposure-related individual differences in both sexes and glucocorticoid-based therapeutics immediately when trauma occurs or after PTSD occurs are both expected to be efficacious (15).
Materials and Methods
Details are described in SI Material and Methods. The study was approved by the James J. Peters Veterans Affairs Medical Center Research & Development Committee.
Animals.
All animal experiments were approved by the Ben-Gurion University of the Negev Institutional Animal Care and Use Committee, and procedures were carried out under strict compliance with ethical principles and guidelines of the NIH Guide for the Care and Use of Laboratory Animals. Adult male and female (50 and 53, respectively) Sprague–Dawley rats (175–225 g) were habituated to housing conditions for at least 10 d and handled once daily. Four same-sex rats were housed together in the vivarium with food and water ad libitum under stable temperature and a reversed 12-h light/dark cycle (lights off at 0800 hours). PSS-exposed rats were placed for 10 min on well-soiled cat litter (used by a cat for 2 d, sifted for stools) in a plastic cage (stress exposure), and CONs were exposed to unused litter for 10-min (sham exposure).
Experimental Design.
Exp. I (Fig. S1A).
After an inescapable PSS or sham exposure (on day 0), rats were assessed behaviorally in the EPM and ASR tests on day 7. Animals were decapitated with a guillotine in a separate room from the one used for behavioral tests, 24 h after the last behavioral tests (between 1400 and 1430 hours) on day 8. Care was taken to minimize situational stress; the area was thoroughly cleaned between each decapitation and bodies removed. One-way ANOVA was used for the analysis of the behavioral data with PSS-exposure as the between subjects factor.
Exp. II (Fig. S1B).
Rats were intraperitoneally injected with VEH (saline NaCl: 0.9%) or 25.0 mg/kg CORT (Sigma) 1 h after an inescapable PSS or sham exposure (day 0) and assessed behaviorally on day 7. The VEH or CORT solutions were freshly prepared in a volume of 1 mL/kg body weight. Two-way ANOVA was used for the analysis of the behavioral data with the PSS-exposure and CORT-treatment as the between-subjects factors.
Behavioral Assessment.
EPM and ASR tests were performed as previously described (4, 17) and CBC were used to classify PSS-exposed rats according to their behavior response on day 7 (3, 17). Male and female behavioral data were analyzed separately, but with the same CBC. Within each sex, exposed rats were divided in three groups: EBRs, MBRs, and PBRs. The graphic representation of the behavioral data depicts the same type of behavioral clustering in males and females (Fig. S1 C and D, respectively).
DNA Microarray Analysis.
Blood, amygdala, and hippocampus RNA of EBR, MBR, and CON rats was used for microarray analysis with the Rat Ref-12 Expression BeadChip (Illumina). RNA samples from males and females were run at the same time (blood: 8 arrays per group per sex, 48 arrays; brain: 5 arrays per group per sex per region, 60 arrays). Quality control was performed using the lumi R (www.R-project.org) package (33). Because our primary interest was large expression changes, we removed poorly expressed genes. Detected probes were called for males and females separately (Fig. S2A). Fewer probes were detected in blood (approximately 6,000) than in the brain (approximately 10,000), but there were similar numbers of probes in both sexes.
Differential Gene Expression.
Data were log2-transformed and normalized using robust spline normalization and differentially expressed probes were identified using the Significance Analysis of Microarray (median false-discovery rate < 5%) considering the three pairwise group comparisons (EBR vs. MBR, EBR vs. CON, MBR vs. CON). The lists of the differentially expressed probes were further trimmed according to the log2 ratios of group geometric means; only probes with ratios more than 0.3 (absolute value) were retained. The lists of differentially expressed probes were annotated by IPA (Ingenuity Systems, 2013 Summer Release) and can be found in Dataset S1 (males in A–C, females in D–F), and were unevenly distributed across tissue in both sexes (Fig. S2B). qPCR was performed to validate differential gene expression results using four biological replicates. See Table S1 for qPCR primers' target sequences and associated Universal Probe Library probes. Data analysis for qPCR was performed using qBase v2.5 (Biogazelle NV). The correlations between microarray data and qPCR data were high (R = 0.77–0.84, P < 0.001), when including DEG above the log2 ratio cut-off, DEG below the log2 ratio cut-off and non-DEG, and similarly high (R = 0.79–0.85, P < 0.001) when including only DEG above the cut-off (Fig. S3 A–C and Dataset S1G).
Because the study aimed to describe gene expression profiles associated with individual differences in the behavioral response to PSS, the differentially expressed probes were divided into probes associated with stress exposure (i.e., probes for which both EBRs and MBRs were different from CONs but not from each other, either both up-regulated or both down-regulated compared with CONs) and probes associated with exposure-related individual differences (Fig. S4A). The latter were subdivided into probes associated with either vulnerability to a rat PTSD-like syndrome or resilience. For the vulnerability-related probes the following condition applied: “EBRs were different than MBRs and CONs”; for the resilience-related probes this condition applied: “MBRs were different than EBRs and CONs.” The Venn-diagram template in Fig. S4B depicts the intersection and union of the vulnerability and resilience probe-sets. For the intersection (I-set), the following conditions applied: “EBRs were different from MBRs, and both EBRs and MBRs were different from CONs”; the union (U-set), as a whole, was related to exposure-related individual differences. The vulnerability- and resilience-related DEG were compared in each sex separately (amygdala in Fig. S4 C and D, hippocampus in Fig. S4 E and F, and blood in Fig. S4 G and H). Finally, the U-set sets were compared between-tissue and unsupervised hierarchical clustering analysis was performed for the DEG that belonged in the statistically significant overlaps determined by permutation tests in R.
Transcription Factor, Network, and Canonical Pathway Analyses.
Transcription factor prediction was carried out with the Upstream Regulator Analysis (IPA) for the male and female DEG associated with stress exposure and exposure-related individual differences. An across-sex transcription factor signature was constructed for amygdala, hippocampus, and blood by pulling together respective male and female predicted transcription factors (Dataset S3 A–C). The transcription-factor lists associated with individual differences were then compared between-tissue, and submitted to Gene Network Analysis (9) and Canonical Pathway Analysis (IPA).
Supplementary Material
Acknowledgments
We thank Dr. Li Shen for advice on the bioinformatic analyses. This work was supported in part by Department of Defense Grant W81XWH-08-2-0021 (to R.Y.) and United States Army Medical Research and Materiel Command W81XWH-13-1-0071 (to R.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession nos. GSE60280, GSE60302, GSE60303, and GSE60304).
See Commentary on page 13253.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401660111/-/DCSupplemental.
References
- 1.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52(12):1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 2.Yehuda R, LeDoux J. Response variation following trauma: A translational neuroscience approach to understanding PTSD. Neuron. 2007;56(1):19–32. doi: 10.1016/j.neuron.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Cohen H, Zohar J. An animal model of posttraumatic stress disorder: The use of cut-off behavioral criteria. Ann N Y Acad Sci. 2004;1032:167–178. doi: 10.1196/annals.1314.014. [DOI] [PubMed] [Google Scholar]
- 4.Cohen H, et al. Blunted HPA axis response to stress influences susceptibility to posttraumatic stress response in rats. Biol Psychiatry. 2006;59(12):1208–1218. doi: 10.1016/j.biopsych.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Yehuda R, McFarlane AC, Shalev AY. Predicting the development of posttraumatic stress disorder from the acute response to a traumatic event. Biol Psychiatry. 1998;44(12):1305–1313. doi: 10.1016/s0006-3223(98)00276-5. [DOI] [PubMed] [Google Scholar]
- 6.Cohen H, Yehuda R. Gender differences in animal models of posttraumatic stress disorder. Dis Markers. 2011;30(2-3):141–150. doi: 10.3233/DMA-2011-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslau N, Davis GC, Andreski P, Peterson EL, Schultz LR. Sex differences in posttraumatic stress disorder. Arch Gen Psychiatry. 1997;54(11):1044–1048. doi: 10.1001/archpsyc.1997.01830230082012. [DOI] [PubMed] [Google Scholar]
- 8.Yehuda R. Post-traumatic stress disorder. N Engl J Med. 2002;346(2):108–114. doi: 10.1056/NEJMra012941. [DOI] [PubMed] [Google Scholar]
- 9.Warde-Farley D, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan V, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131(2):391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Jazin E, Cahill L. Sex differences in molecular neuroscience: From fruit flies to humans. Nat Rev Neurosci. 2010;11(1):9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 12.McEwen BS. Stress, sex, and neural adaptation to a changing environment: Mechanisms of neuronal remodeling. Ann N Y Acad Sci. 2010;1204(Suppl):E38–E59. doi: 10.1111/j.1749-6632.2010.05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Donovan A, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30(2-3):123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daskalakis NP, Yehuda R, Diamond DM. Animal models in translational studies of PTSD. Psychoneuroendocrinology. 2013;38(9):1895–1911. doi: 10.1016/j.psyneuen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Daskalakis NP, Lehrner A, Yehuda R. Endocrine aspects of post-traumatic stress disorder and implications for diagnosis and treatment. Endocrinol Metab Clin North Am. 2013;42(3):503–513. doi: 10.1016/j.ecl.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 16.van Zuiden M, et al. Pre-existing high glucocorticoid receptor number predicting development of posttraumatic stress symptoms after military deployment. Am J Psychiatry. 2011;168(1):89–96. doi: 10.1176/appi.ajp.2010.10050706. [DOI] [PubMed] [Google Scholar]
- 17.Cohen H, Matar MA, Buskila D, Kaplan Z, Zohar J. Early post-stressor intervention with high-dose corticosterone attenuates posttraumatic stress response in an animal model of posttraumatic stress disorder. Biol Psychiatry. 2008;64(8):708–717. doi: 10.1016/j.biopsych.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Zohar J, et al. High dose hydrocortisone immediately after trauma may alter the trajectory of PTSD: Interplay between clinical and animal studies. Eur Neuropsychopharmacol. 2011;21(11):796–809. doi: 10.1016/j.euroneuro.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Rao RP, Anilkumar S, McEwen BS, Chattarji S. Glucocorticoids protect against the delayed behavioral and cellular effects of acute stress on the amygdala. Biol Psychiatry. 2012;72(6):466–475. doi: 10.1016/j.biopsych.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen H, Kozlovsky N, Matar MA, Zohar J, Kaplan Z. The characteristic long-term upregulation of hippocampal NF-κB complex in PTSD-like behavioral stress response is normalized by high-dose corticosterone and pyrrolidine dithiocarbamate administered immediately after exposure. Neuropsychopharmacology. 2011;36(11):2286–2302. doi: 10.1038/npp.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen H, Geva AB, Matar MA, Zohar J, Kaplan Z. Post-traumatic stress behavioural responses in inbred mouse strains: Can genetic predisposition explain phenotypic vulnerability? Int J Neuropsychopharmacol. 2008;11(3):331–349. doi: 10.1017/S1461145707007912. [DOI] [PubMed] [Google Scholar]
- 22.Caldji C, Hellstrom IC, Zhang TY, Diorio J, Meaney MJ. Environmental regulation of the neural epigenome. FEBS Lett. 2011;585(13):2049–2058. doi: 10.1016/j.febslet.2011.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Yehuda R, et al. Lower methylation of glucocorticoid receptor gene Ppromoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Yehuda R, et al. Influences of maternal and paternal PTSD on epigenetic regulation of the glucocorticoid receptor gene in Holocaust survivor offspring. Am J Psychiatry. 2014;171(8):872–880. doi: 10.1176/appi.ajp.2014.13121571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegmund A, Kaltwasser SF, Holsboer F, Czisch M, Wotjak CT. Hippocampal N-acetylaspartate levels before trauma predict the development of long-lasting posttraumatic stress disorder-like symptoms in mice. Biol Psychiatry. 2009;65(3):258–262. doi: 10.1016/j.biopsych.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Glatt SJ, et al. Marine Resiliency Study Investigators Blood-based gene-expression predictors of PTSD risk and resilience among deployed marines: A pilot study. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(4):313–326. doi: 10.1002/ajmg.b.32167. [DOI] [PubMed] [Google Scholar]
- 27.Ponomarev I, Rau V, Eger EI, Harris RA, Fanselow MS. Amygdala transcriptome and cellular mechanisms underlying stress-enhanced fear learning in a rat model of posttraumatic stress disorder. Neuropsychopharmacology. 2010;35(6):1402–1411. doi: 10.1038/npp.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andero R, et al. Amygdala-dependent fear is regulated by Oprl1 in mice and humans with PTSD. Sci Transl Med. 2013;5(188):188ra173. doi: 10.1126/scitranslmed.3005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le-Niculescu H, et al. Convergent functional genomics of anxiety disorders: Translational identification of genes, biomarkers, pathways and mechanisms. Transl Psychiatr. 2011;1:e9. doi: 10.1038/tp.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia M, et al. Biomarkers in an animal model for revealing neural, hematologic, and behavioral correlates of PTSD. J Vis Exp. 2012;(68):e3361. doi: 10.3791/3361. doi:10.3791/3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang R, et al. Core modular blood and brain biomarkers in social defeat mouse model for post traumatic stress disorder. BMC Syst Biol. 2013;7:80. doi: 10.1186/1752-0509-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.