Significance
The 2003 severe acute respiratory syndrome (SARS) epidemic and recent emergence of Middle East respiratory syndrome highlight the potential lethality of zoonotic coronavirus infections in humans. No specific antiviral treatment options are available. Coronaviruses possess the largest known RNA virus genomes and encode a complex replication machinery consisting of 16 viral nonstructural proteins (nsps). Our study reveals that the SARS-coronavirus RNA polymerase (nsp12) needs to associate with nsp7 and nsp8 to activate its capability to replicate long RNA. Moreover, this complex associates with nsp14, the proofreading subunit required to safeguard coronavirus replication fidelity. Our study thus defines the core of an RNA-synthesizing machinery that is unique in the RNA virus world and includes several key targets for antiviral drug development.
Keywords: replicative complex reconstitution, processivity factor
Abstract
In addition to members causing milder human infections, the Coronaviridae family includes potentially lethal zoonotic agents causing severe acute respiratory syndrome (SARS) and the recently emerged Middle East respiratory syndrome. The ∼30-kb positive-stranded RNA genome of coronaviruses encodes a replication/transcription machinery that is unusually complex and composed of 16 nonstructural proteins (nsps). SARS-CoV nsp12, the canonical RNA-dependent RNA polymerase (RdRp), exhibits poorly processive RNA synthesis in vitro, at odds with the efficient replication of a very large RNA genome in vivo. Here, we report that SARS-CoV nsp7 and nsp8 activate and confer processivity to the RNA-synthesizing activity of nsp12. Using biochemical assays and reverse genetics, the importance of conserved nsp7 and nsp8 residues was probed. Whereas several nsp7 mutations affected virus replication to a limited extent, the replacement of two nsp8 residues (P183 and R190) essential for interaction with nsp12 and a third (K58) critical for the interaction of the polymerase complex with RNA were all lethal to the virus. Without a loss of processivity, the nsp7/nsp8/nsp12 complex can associate with nsp14, a bifunctional enzyme bearing 3′-5′ exoribonuclease and RNA cap N7-guanine methyltransferase activities involved in replication fidelity and 5′-RNA capping, respectively. The identification of this tripartite polymerase complex that in turn associates with the nsp14 proofreading enzyme sheds light on how coronaviruses assemble an RNA-synthesizing machinery to replicate the largest known RNA genomes. This protein complex is a fascinating example of the functional integration of RNA polymerase, capping, and proofreading activities.
A virus-encoded RNA-dependent RNA polymerase (RdRp) is the central enzyme in the replicative cycle of RNA viruses (1). In the case of mammalian positive-strand RNA (+RNA) viruses, the enzyme is generated by the translation of the incoming viral genome, which yields a polyprotein precursor from which the RdRp-containing subunit is released by proteolytic cleavage. Subsequently, the RdRp is integrated into a membrane-associated viral enzyme complex that drives the production of negative-strand RNA (−RNA), new genome molecules, and in many virus groups also subgenomic (sg) messenger RNAs (mRNAs) (2–4). Compared with the replication of either viral or cellular DNA sequences, RNA virus genomes are copied with relatively low fidelity, because the products of replication are believed to be neither proofread nor edited (5). This property is a major factor in the evolution, adaptation, and epidemiology of RNA viruses.
Among +RNA viruses, coronaviruses (CoVs) (order Nidovirales) stand out for having the largest single-stranded RNA genomes known to date (6, 7). Research into the molecular and structural biology of CoVs was boosted significantly by the emergence, in 2003, of a previously undiscovered CoV that caused the severe acute respiratory syndrome (SARS) epidemic (6). Whereas established human CoVs are commonly associated with mild upper respiratory tract infections, SARS-CoV caused a severe form of pneumonia with a case fatality rate of about 10%. Two years after the outbreak, bats were identified as the likely reservoir from which SARS-CoV had jumped to humans, probably using other animals as intermediate hosts (8, 9). Since April 2012, history seems to repeat itself with the emergence of another zoonotic CoV, Middle East respiratory syndrome coronavirus (MERS-CoV) (10, 11), which has thus far infected over 800 people with a case fatality rate of about 35%. MERS-CoV is most closely related to several bat CoVs (12) and to a newly discovered coronavirus (EriCoV) found in hedgehogs (13). This new episode emphasizes the ability of CoVs to cross species barriers and threaten public health worldwide.
The CoV RNA genome can be up to 32 kb long (14), carries a 5′-terminal cap structure, and is polyadenylated at its 3′ end (15). The genome is polycistronic with the 5′-proximal two-thirds being occupied by two large overlapping open reading frames (ORFs) (ORFs 1a and 1b) (16). These are translated to yield replicase polyproteins pp1a and pp1ab, with synthesis of the latter requiring −1 ribosomal frameshifting near the 3′ end of ORF1a. Subsequently, the two polyproteins are cleaved by two or three ORF1a-encoded proteases to release a total of 16 nonstructural proteins (nsp1 to nsp16). Next, these nsps assemble into a replication and transcription complex (RTC) that is associated with modified intracellular membranes (17) and directs a complex mechanism of viral RNA synthesis. This not only entails the synthesis of new genomes from a full-length negative-stranded template, but also a process of discontinuous RNA synthesis to produce subgenome-length negative-stranded RNAs (18, 19). The latter serve as template to produce a nested set of sg mRNAs required to express the viral structural and accessory proteins from genes not accessible to ribosomes translating the viral genome RNA.
The CoV replicase harbors a wide variety of enzymatic activities (7, 16, 20). Several nsps have already been characterized as multifunctional subunits. Whereas the RdRp and helicase enzymes form the C-terminal domains of nsp12 and nsp13, respectively, and are typically found in +RNA viruses (21, 22), less common or even unique RNA-processing activities are associated with other nsps. For example, the N-terminal domain of the bifunctional nsp14 includes a 3′ to 5′ exoribonuclease (ExoN) (23) that has been implicated in a unique but poorly characterized form of RNA viral proofreading (24–26) and nsp15 carries an endoribonuclease activity (NendoU) of unknown function (27, 28). In addition, SARS-CoV nsp8 was cocrystallized with another viral protein [nsp7 (29)] as a unique hexadecameric ring structure, which was reported to exhibit a “noncanonical,” de novo-initiating RdRp activity (30–32). Moreover, evidence from genetics studies previously implicated nsp8 in a specific interaction with RNA structures near the 3′ end of the CoV genome (33). The nsp7/nsp8 protein complex was therefore proposed to act as a cofactor for the canonical nsp12 RdRp. Indeed, bioinformatics analyses of the nsp12 RdRp domain identified the so-called “G motif,” which has been implicated in the recognition of the primers predicted to be required for the initiation of RNA synthesis by nsp12 (34).
Insights into how the RTC is assembled and operates are critical for the advancement of our basic understanding of CoV replication and for the development of efficient means to control CoV infections. Over the past 10 years, the SARS-CoV RTC has been studied by using different approaches including bioinformatics, (reverse) genetics, as well as biochemical and structural characterization of recombinant nsps. Clearly, there must be a network of interacting replication proteins that controls the CoV replication cycle. During the past years, we and others have elucidated how the in vitro activity of various CoV enzymes can be modulated by interactions with other viral nsps (24, 35). Moreover, the SARS-CoV RTC was partially purified from infected cells and shown to produce the full spectrum of viral mRNAs (36). Exogenous RNA templates are, however, not replicated by this large membrane-bound complex, whose subunit composition was not fully resolved.
Several attempts have been made to characterize nsp12 RdRp activity in vitro. A weak polymerase activity was described for recombinant SARS-CoV nsp12 expressed in fusion with a GST-tag. The uncharacterized N-terminal domain of nsp12 was concluded to be required for activity (37). Further enzymatic studies showed that a full-length, C-terminally His6-tagged nsp12 could initiate homopolymeric RNA synthesis in a primer-dependent manner (21). However, the polymerase activity described in these studies appeared generally weak and nonprocessive. The processivity of RNA synthesis, defined as the number of nucleotides polymerized during a single encounter of the RNA polymerase with its template, bears important biological significance: a highly processive RNA polymerase may synthesize a full-length copy of a viral RNA genome without falling off the template.
Here, we report that the processivity of the SARS-CoV nsp12 polymerase in primer extension mode can be dramatically increased upon formation of a complex with nsp7 and nsp8. Thus, this tripartite complex forms—uniquely among RNA viruses—a holoenzyme, which is also capable of initiating RNA synthesis de novo, i.e., in the absence of an exogenous RNA primer. We further established that nsp14, which harbors a N7-guanine cap methyltransferase activity (38) downstream of the ExoN domain (23), can join the nsp7/nsp8/nsp12 complex, without significantly affecting either its polymerase activity or its processivity. The formation of this multifunctional nsp7/nsp8/nsp12/nsp14 complex demonstrates the physical association of subunits possessing RNA polymerization, proofreading, and capping activities needed to replicate the unusually long CoV RNA genome.
Results
Nsp7 and Nsp8 Cooperate in Activating the Primer-Dependent Activity of the Nsp12 RdRp.
The nsp12 subunit is the essential RdRp of the coronavirus replicative machinery. Purified recombinant nsp12 is able to extend a homopolymeric primer-template substrate by a few dozen nucleotides in vitro, but in a nonprocessive (distributive) manner (21). Moreover, nsp12 was previously reported to interact with nsp8 in GST pull-down experiments (39), which displayed a primase-like RdRp activity (30–32). Because one of the two described structures of the nsp7/nsp8 complex resembles that of DNA processivity factors such as proliferating cell nuclear antigen (PCNA), it was hypothesized also that nsp7/nsp8 may modulate nsp12 RdRp activity (29, 40).
Recombinant SARS-CoV proteins carrying affinity tags were expressed in Escherichia coli using a T7 RNA polymerase-free system and were purified by affinity chromatography (Fig. S1A). Pull-down experiments confirmed the interaction between nsp12 and nsp8 (Fig. S1B), of which the latter in turn interacts with nsp7 (Fig. S1C), suggesting that a tripartite complex may be formed. However, such a direct interaction between these three proteins remained undetected (Fig. S1D).
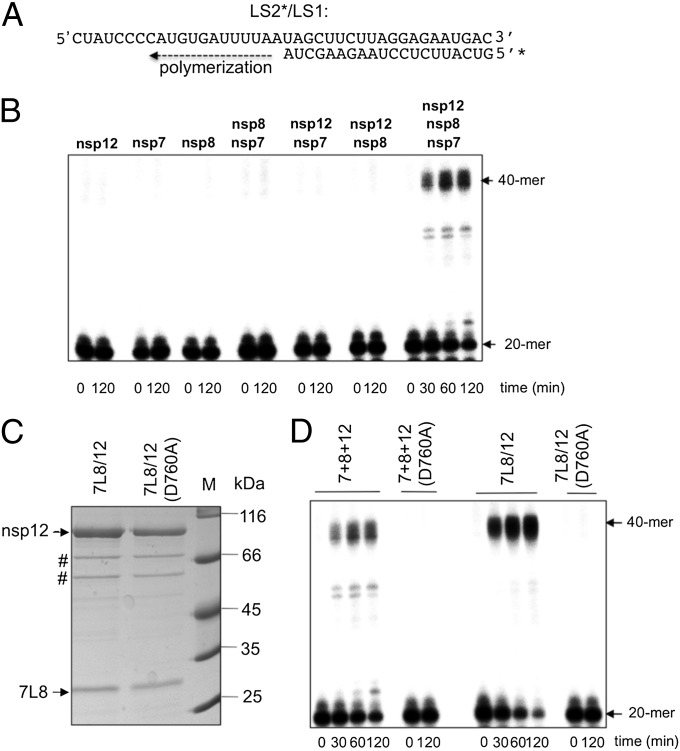
Nsp12 alone was unable to extend the primer strand of a heteropolymeric primer/template (P/T), of which the template mimicked the 3′-terminal 40 nt of the SARS-CoV genome [excluding the poly(A) tail], hereafter called LS2/LS1 (Fig. 1 A and B). In the same manner, nsp8 alone was inactive on the LS2/LS1 RNA primer/template, as previously described (30). Pairwise combinations of nsp7, nsp8, and nsp12 displayed no significant polymerase activity either (Fig. 1B). However, RNA synthesis activity was found when nsp7, nsp8, and nsp12 were simultaneously present in the reaction mixture (Fig. 1B). To evaluate the requirements for RdRp activity in more detail, we preincubated all possible pairwise combinations of nsp7, nsp8, and nsp12 for various periods of time, before adding the third protein and assaying RNA synthesis. The highest RdRp activity was obtained upon addition of an equimolar amount of nsp12 to a preincubated 1:1 mixture of nsp7 and nsp8, suggesting that the level of association of nsp7 and nsp8 may constitute a limiting factor for obtaining a high RNA polymerase activity.
Fig. 1.
SARS-CoV nsp12 polymerase activity is activated by nsp7 and nsp8. (A) Sequence of the RNA primer/template used in this study; the 20-nt primer LS2 was 5′-radiolabeled (marked by *) and annealed to the 40-nt template LS1. (B) Primer extension polymerase assays were performed using LS2*/LS1 as substrate and different combinations of separately purified nsp12, nsp8, and nsp7. RNA products were separated by denaturing gel electrophoresis (20% polyacrylamide/7 M urea) and analyzed by autoradiography. The positions of the primer (20-mer) and the full-length extension product (40-mer) are indicated. (C) WT or mutant (D760A) nsp12 were coexpressed in E. coli with the nsp7-L-nsp8 fusion protein (7L8). After purification of the 7L8/nsp12 complex on a Strep-Tactin column, analysis by 12% SDS/PAGE and Coomassie blue staining of the proteins constituting the complex (nsp12 WT or D760A mutant) was done. # indicates the position of E. coli protein contaminants (defined by MALDI-TOF analysis). (D) Comparison of primer extension polymerase activities of WT and mutant (D760A) nsp12 in the presence of nsp7 and nsp8. Nsp7, nsp8, and nsp12 were either purified and added separately (lanes labeled 7+8+12) or copurified from E. coli as described above (lanes labeled 7L8/12). The reactions were performed on RNA template LS2*/LS1 (see B). Primer conversion rates (at 60 min): 40% for 7+8+12; 67% for 7L8/12; and 0% for 7+8+12(D760A) and 7L8/12(D760A).
To overcome this limitation, we designed a set of gene constructs to express fusion proteins in which nsp7 was either directly fused to nsp8 (imitating the uncleaved nsp7–nsp8, designated 7–8) or through a peptide linker of different lengths (7L8). Upon coexpression with nsp12 in E. coli, all five engineered fusion proteins bound to and copurified with nsp12 (Fig. S2A). Interestingly, the protein complex consisting of nsp12 and a fusion protein, in which the nsp7 C terminus is linked to the nsp8 N terminus by an engineered peptide of 6 (or 12) amino acids residues, is able to efficiently polymerize RNA (Fig. S2B). In contrast, when nsp12 is complexed with an uncleaved nsp7–nsp8 polyprotein (either 7–8/12 or 7–8HC/12), these protein complexes fail to show polymerase activity (Fig. S2B). Combined, these results indicate that inserting a linker between nsp7 and nsp8 may be sufficient to imitate the effect of the nsp7–nsp8 cleavage and associated structural rearrangements on the functioning of nsp7 and nsp8 in the activation of nsp12 RNA polymerase activity. These data are also in agreement with those of Deming et al. (41), who showed that murine hepatitis virus could not be recovered when the cleavage between nsp7–nsp8 was prevented. Being the most efficient in terms of its RNA primer conversion rate, the 7L8/12 complex, with the linker L consisting of six histidines, was selected for further experiments. This affinity-purified complex (Fig. 1C) is at least threefold more active than a preincubated mixture of the three separately purified subunits (7+8+12; Fig. 1D), confirming that, in our experimental conditions, the nsp7/nsp8 association is a limiting factor to obtain a highly active RNA polymerase complex. The C terminus of nsp7 and the N terminus of nsp8 are far apart in the published SARS-CoV nsp7–nsp8 structure, indicating that the 7L8 fusion protein is highly unlikely to assemble into a similar hexadecameric structure, due to topological constraints. Another assembly type than the SARS-CoV hexadecamer was reported for feline coronavirus, in which the nsp7/nsp8 complex forms a heterotrimer (32), clearly pointing to the plasticity of this CoV protein complex that could be relevant for 7L8.
To investigate whether the observed primer extension was indeed catalyzed by the nsp12-RdRp domain, we mutated the conserved motif C of the RdRp (SDD to SAD, or D760A). This nsp12 (D760A) mutant is still able to bind to 7L8 (Fig. 1C) but is incapable of RNA synthesis in the presence of either separately purified nsp7 and nsp8 or copurified 7L8 (Fig. 1D).
The 7L8/12 Polymerase Complex Catalyzes de Novo RNA Synthesis.
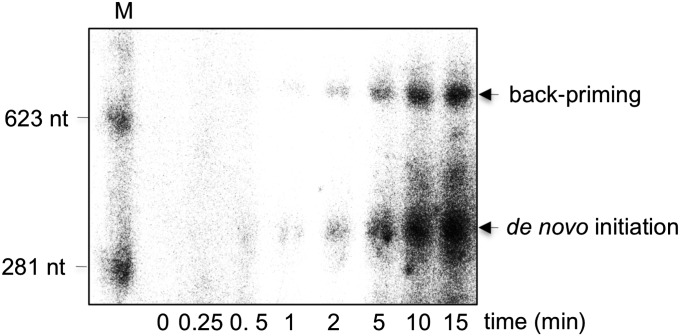
An assay to investigate de novo initiation of RNA synthesis by 7L8/12 was designed using a 339-nt-long heteropolymeric RNA template, corresponding to the 3′-nontranslated region (3′-NTR) of the SARS-CoV genome (named 3R). The protein complex and template 3R were used for a time course experiment in the presence of [α-32P]ATP and reaction products were analyzed using denaturing agarose gel electrophoresis and autoradiography. Two radiolabeled products are detected (Fig. 2), one with a length similar to that of the RNA template and the other approximately twice as long. The size of the shorter product suggests it to correspond to the complementary strand of 3R, implying that de novo initiation of RNA synthesis has occurred. The longer product is interpreted as the result of “backpriming,” i.e., the formation of a hairpin structure at the 3′ end of the template, which can serve as primer to be extended into a product of about twice the template length (42, 43). Noteworthy, no de novo polymerase activity is detected using a mixture of the three proteins purified independently from E. coli. Previously, nsp8 was proposed to be an RNA primase, possibly mediating the synthesis of small primers (30–32) that could be elongated by nsp12. To revisit this hypothesis, we performed de novo RNA synthesis assays using template 3R and a protein complex consisting of 7L8 copurified with the active site knockout mutant of nsp12 (D760A). Under these conditions, nsp8-mediated synthesis of short primers is not detected (Fig. S3).
Fig. 2.
The 7L8/12 polymerase complex catalyzes de novo RNA synthesis. De novo RNA polymerase assays were performed using 500 nM 7L8/12 with 500 nM RNA template (3R), representing the 3′-terminal 339 nt of the SARS-CoV genome. Reaction products, collected at the indicated time points, were analyzed by denaturing agarose gel electrophoresis and autoradiography. Reaction products are identified on the Right of the gel, and radiolabeled RNA size marker (M) is shown to the Left of the panel.
To exclude that the products detected in the de novo assay were due to 3′-terminal nucleotide transferase activity of 7L8/12, the 3′ end of template 3R was blocked using periodate oxidation. This chemical modification results in cleavage of the 2′C—3′C bond in the ribose of the 3′-terminal nucleotide, thus preventing any further nucleotide addition to the 3′ end of the RNA template. When using this modified RNA template, the putative backpriming product is no longer observed, whereas a full-length product complementary to the template is still produced, albeit with lower efficiency (Fig. S4A, lane 1). This reduction in RNA synthesis likely reflects reduced binding of the polymerase complex to the periodate-treated 3′ end of the template, as previously described for dengue virus RdRp (44).
To better mimic the in vivo context for de novo initiation, we also tested a variant of the 3R template carrying a 3′-terminal 20-residue poly(A) tail (named 3RA). The efficiency of 7L8/12-mediated RNA synthesis is unchanged on this template compared with that of the template without poly(A) tail (Fig. S4B). Taken together, these results show that the 7L8/12 polymerase complex is capable of de novo initiation of RNA synthesis on a template with the same sequence as the 3′-proximal domain of the viral RNA genome, and of subsequent elongation of RNA products to >300 nt.
The Nsp7/Nsp8 Complex Confers Processivity to Nsp12 RdRp.
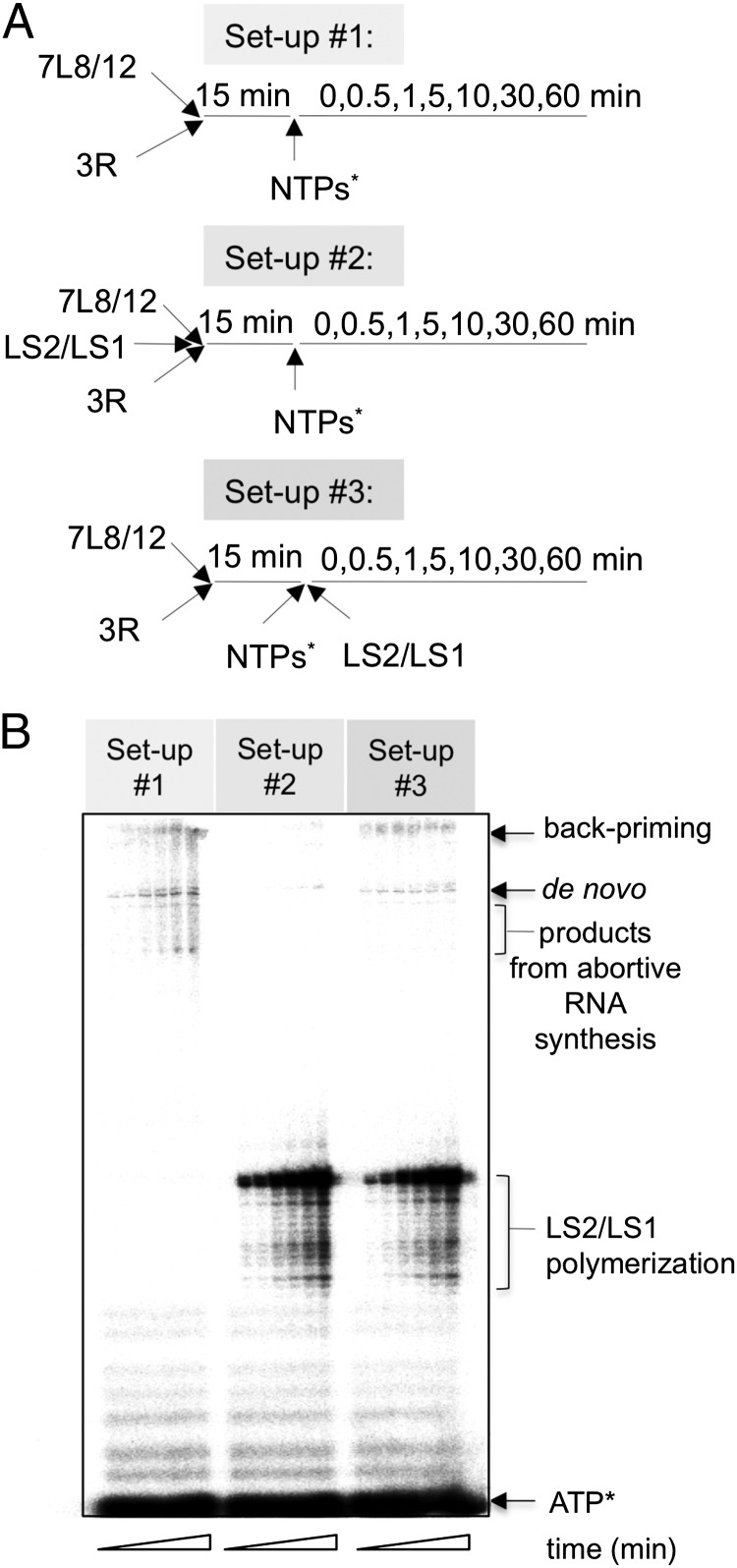
The activation of nsp12 RdRp activity in the presence of nsp7 and nsp8 suggests that the nsp7/nsp8 complex enhances processivity of nsp12-mediated RNA synthesis. We next analyzed the processivity of 7L8/12 on the 3R template. To avoid recycling of the polymerase once it had terminated a series of nucleotide additions, we made use of a 200-fold excess of an exogenous RNA substrate, the previously used LS2/LS1 primer/template (Fig. 1A) that served as a “polymerase trap.” The latter was added either at the same time as the 3R template, or after 7L8/12/3R template complex formation had been allowed to occur (Fig. 3A, setups #2 and #3, respectively). In the latter setup, exogenous LS2/LS1 should trap all free polymerase complexes as soon as they would fall off the 3R template. Thus, this last condition would allow visualization of RNA products resulting from a single de novo (or backpriming) initiation event. All polymerase assays were started by the addition of the NTP mixture, including [α-32P]ATP (NTP*). As a control (setup #1), a reaction was performed without the LS2/LS1 trap. Fig. 3B (left lanes of the gel) shows that it yields some smaller “abortive products,” presumably deriving from polymerase complexes falling off the 3R template, in addition to the two expected full products resulting from de novo initiation and backpriming. The products from abortive RNA synthesis are only detected after longer incubation times (>5 min), in line with the polymerase recycling hypothesis. When the 3R template and LS2/LS1 trap were introduced simultaneously (setup #2), the synthesis of long products derived from template 3R is barely detectable (Fig. 3B, compare #1 and #2) indicating that LS2/LS1 outcompetes 3R. However, when the same excess of LS2/LS1 was added after 7L8/12/3R complex formation had occurred (setup #3), the two long products (from de novo and backpriming initiation modes) are the only products >40 nt detected (Fig. 3B, setup #3). The absence of any shorter product resulting from abortive RNA synthesis shows that RNA synthesis proceeds with a very low dissociation rate once a productive polymerase/3R complex is formed.
Fig. 3.
The 7L8/12 polymerase complex processively replicates the 3R RNA template. (A) Schematic representation of the three experimental setups used, all including 500 nM of 7L8/12 polymerase complex and 100 nM of the 339-nt template 3R; the LS2/LS1 template (Fig. 1A) was used as a trap at 20 µM. NTPs* represents a mix of the four NTPs (500 µM GTP, UTP, CTP, and 50 μM ATP) and 0.17 µM [α-32P]ATP (0.5 µCi/µL). Nucleotide incorporation was followed in time as indicated in the three flow charts. (B) Reactions were started by the addition of NTPs*. RNA products were separated by denaturing acrylamide/urea gels and visualized by autoradiography. RNA products and residual radiolabeled ATP* are indicated on the Right of the autoradiograph.
To gain insight into the association kinetics of RNA template and 7L8/12, we performed trapping experiments similarly to those presented in Fig. 3. The obtained koff value (Fig. S5; 7.9·10−3 s−1) indeed reflects a very low dissociation rate. Together, these results clearly demonstrate that the 7L8 cofactor activates and confers processivity to the SARS-CoV nsp12 RdRp.
Nsp7 and Nsp8 Confer RNA Binding Capacity to Nsp12.
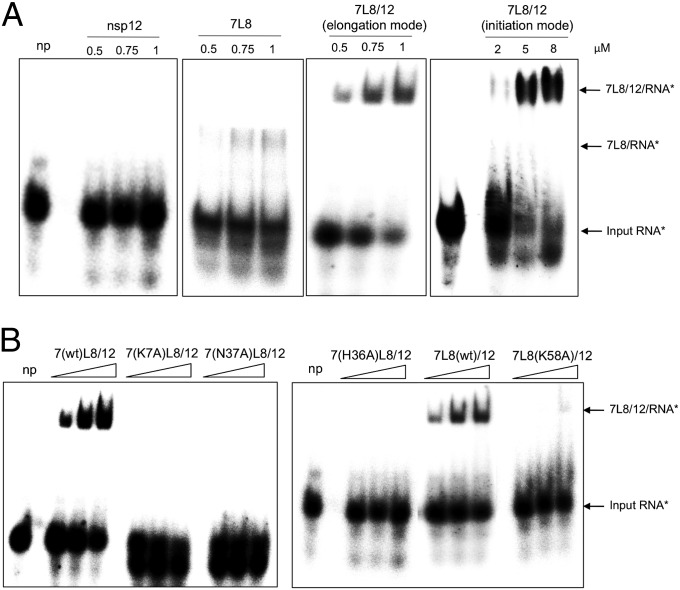
How does the nsp7/nsp8 complex stimulate nsp12 RdRp activity? We first compared the RNA binding properties of nsp12, 7L8, and the 7L8/12 complex. Electrophoretic mobility shift assays (EMSAs) indicate that nsp12 itself does not bind significantly to RNA (LS2/LS1), that 7L8 interacts weakly with it, whereas 7L8/12 strongly binds to RNA (Fig. 4A). Moreover, the 7L8/12 RNA binding affinity is higher in elongation than in initiation mode, based on protein complex quantities used to observe an RNA shift (Fig. 4A). Altogether, these data suggest that 7L8 mediates affinity of the complex for RNA.
Fig. 4.
Nsp7 and nsp8 confer RNA-binding capacity to the nsp12 RdRp. EMSAs were performed with 100 nM radiolabeled RNA template (LS2*/LS1) and increasing concentrations of the indicated proteins. With the exception of 7L8/12 in initiation mode, all of the other reactions were done with NTPs (500 μM CTP, UTP, GTP and 50 μM 3′-dATP) to facilitate formation of a stalled RNA/enzyme complex, and with 3′-dATP preventing synthesis of full-length RNA product. Reactions were incubated for 60 min at 30 °C and products were analyzed by native 6% PAGE. (A) No protein (np) control and increasing concentrations of nsp12, 7L8, and 7L8/12, as indicated above each panels. (B) No protein (np) control and increasing concentrations (0.5, 0.75, and 1 µM) of 7L8/12 polymerase complexes containing different nsp7 and nsp8 mutants, as indicated above each panel. 7L8/12 in elongation mode gave 42.5% ± 7.5% of shifted probe; 7L8, 9.0% ± 3.5%; 7L8(K58A)/12, 10.0% ± 6.5%, whereas nsp12 alone and the other mutants presented here did not show any ability to bind the probe (0%). These values are averages from quantification of three independent experiments.
Next, we sought to identify nsp7 and nsp8 residues that may confer RNA template-binding ability to the nsp7/nsp8/nsp12 complex. Based on sequence conservation across Coronaviridae, their exposure on the surface of the published nsp7/nsp8 crystal structures (29, 32), and amino acid properties, 5 nsp7 residues and 16 nsp8 residues were selected for substitution with alanine (Fig. S6). Primer extension assays were first performed using the five nsp7 mutants in combination with separately purified WT nsp8 and WT nsp12. Nsp7 residues K7 and N37 are found to be essential for RdRp activity, whereas replacement of H36 confers a moderate decrease in primer-dependent polymerase activity (38% residual activity; Table 1). Next, to address the impact on de novo polymerase activity, 7L8 proteins carrying these three mutations were expressed in E. coli and copurified with nsp12. Complexes containing the nsp7 H36A and N37A mutants show strongly reduced de novo polymerase activity (less than 7% of the WT control), whereas the complex containing K7A exhibits 33% of the activity of the WT control. Then, EMSAs were performed with complexes containing these same nsp7 mutants (K7A, H36A, and N37A). Replacement of each of these nsp7 residues severely reduces the RNA binding capacity of the nsp7/nsp8/nsp12 complex during elongation, indicating that nsp7 plays a crucial role in RNA binding (Fig. 4B and Table 1).
Table 1.
Biochemical properties of SARS-CoV nsp7 mutants
 |
Summary of quantification for nsp7 mutants in primer extension and de novo polymerase activities, as well as RNA binding. Quantifications of polymerization activities were done after 60 min of incubation and by using Image Gauge software. ND, not determined. WT absolute values corresponding to 100% activity are as follows: 42% ± 4% of primer conversion (at 60 min) for primer-dependent experiments; 42.5% ± 7.2% of shifted probe for EMSAs. For the de novo assays, polymerase activities of the different mutants were always analyzed on denaturating gel in parallel with the WT polymerase protein complex, whose band intensities were set at 100%. SDs are calculated from three independent experiments.
Nsp7 WT plus nsp8 WT plus nsp12 WT primer extension activity was set at 100%.
7L8/12 WT complex for de novo polymerase activity was set at 100%.
RNA binding of 7L8/12 WT complex was set at 100%. Mutants highlighted in dark gray indicate important residues for in vitro polymerase activities.
The same approach was used to analyze mutants in which one of 16 conserved nsp8 residues had been mutated. Polymerase activities of these nsp8 mutants were assayed either in the context of separately purified (nsp7 plus nsp8 plus nsp12) for primer extension assays or as part of the copurified 7L8/12 polymerase complex for de novo polymerase assays. Of note, primer extension activities of nsp8 WT and a set of nsp8 mutants were also assayed both in the context of separately purified nsp7, nsp8, and nsp12 and in the copurified 7L8/12 polymerase complex. Comparable relative values of primer extension activities are observed in both setups, supporting the biological relevance of data obtained with the 7L8 fusion protein (Table S1). Five nsp8 mutations (K58A, D99A, P116A, P183A, and R190A) are associated with a strong decrease of both primer-dependent and de novo polymerase activities (Table 2). The 7L8/12 complex containing the nsp8 K58A mutant shows strongly reduced RNA binding (Fig. 4B), consistent with the observed loss of polymerase activity, but without providing an explanation for the difference in activity observed between primer extension and de novo initiation assays (70% versus 99% loss of activity, respectively; Table 2).
Table 2.
Biochemical properties of SARS-CoV nsp8 mutants
 |
Summary of quantification for nsp8 mutants in primer extension and de novo polymerase activities, as well as in nsp8/nsp12 interaction assays. Quantifications of polymerization activities were done after 60 min of incubation and by using Image Gauge software. ND, not determined. WT absolute values corresponding to 100% activity are as follows: 42% ± 4% of primer conversion (at 60 min) for primer-dependent experiments and 42% ± 8.2% of 7L8 band intensity (compared with nsp12 band intensity) for nsp8/nsp12 interaction assays. For the de novo assays, polymerase activities of the different mutants were always analyzed on denaturating gel in parallel with the WT polymerase protein complex, whose band intensities were set at 100%. SDs are calculated from three independent experiments.
Nsp7 WT plus nsp8 WT plus nsp12 WT primer extension activity was set at 100%.
7L8/12 WT complex for de novo polymerase activity was set at 100%.
Level of copurification of 7L8(WT) relative to nsp12 was taken to represent 100% of nsp12/nsp8 protein interaction. Mutants highlighted in dark gray indicate important residues for in vitro polymerase activities.
In conclusion, our study identifies three nsp7 residues (K7, H36, and N37) and one nsp8 residue (K58) that appear to be important for the interaction of the polymerase complex with RNA, thus also providing the first evidence (to our knowledge) for a role of nsp7 in CoV RNA synthesis.
The Nsp8/Nsp12 Interaction Is Critical for SARS-CoV RdRp Activity.
To further unravel the involvement of nsp8 residues in nsp12 RdRp stimulation, we characterized the four other nsp8 mutants exhibiting a decrease in both primer extension and de novo polymerase activities (D99A, P116A, P183A, R190A; Table 2). These mutants, in the context of 7L8, were analyzed for their ability to interact with nsp12 in a protein–protein interaction study also including the D50A, K82A, and S85A nsp8 mutants (displaying unchanged primer-dependent polymerase activities) and nsp8 mutant K58A, which was associated with a critical decrease of the interaction between polymerase complex and RNA. Using the E. coli coexpression system, WT or mutant 7L8 proteins were copurified with nsp12. Proteins were separated by SDS/PAGE, and peaks corresponding to nsp12 and 7L8 were quantified. Table 2 presents the level of copurification between nsp12 and 7L8(mutants) relative to 7L8(WT), which was taken to define 100% of nsp12/nsp8 protein interaction. Nsp8 mutants D99A, P116A, P183A, and R190A lose their ability to interact with nsp12, likely explaining the lack of polymerase activity, whereas the interaction between nsp12 and nsp8 D50A, K58A, K82A, and S85A mutants is unaffected (Table 2).
Our study thus identifies four nsp8 residues (D99, P116, P183, and R190) that are critical for the interaction of nsp8 with nsp12 in vitro, and thereby essential for processive RNA polymerization to occur.
Reverse Genetics Confirms the Critical Role of Nsp7 and Nsp8 in SARS-CoV Replication.
The importance of the coronavirus nsp7 and nsp8 subunits for virus replication in vivo has barely been addressed thus far. Based on our in vitro studies, mutations of nsp7 and nsp8 residues that are important for polymerase activity (i.e., nsp7 K7, H36, and N37; nsp8 K58, D99, P116, P183, and R190; Tables 1 and 2) were reverse-engineered into the SARS-CoV genome using a BAC-based full-length cDNA clone (45). Also, nsp8 residues S11 (with constrained Ser/Thr variability in coronaviruses) and N43, one of the few invariant nsp8 residues among CoVs (Fig. S6), were targeted in this reverse-genetics study, as well as three residues previously identified as important for in vitro nsp8 function (i.e., D52, K82, and S85) (30, 31), which have phenotypes comparable to that of the WT control in our biochemical studies (Table 2).
Mutant viruses were launched by electroporation of in vitro-generated RNA transcripts into BHK-Tet-SARS-N cells, which express the viral nucleocapsid (N) protein (46). These cells can be transfected efficiently and release progeny virus, but do not support further viral spread because they lack the ACE2 receptor used by SARS-CoV. For this reason, transfected cells were mixed with (susceptible) Vero-E6 cells and the replication of mutant viruses was characterized at different time points using immunofluorescence microscopy and progeny virus titration (plaque assays). Each mutant was tested twice, by transfection of RNA derived from two independently generated cDNA clones. For mutants producing infectious progeny, the 42-h posttransfection (p.t.) harvest was used to infect fresh VeroE6, from which RNA was isolated to amplify the nsp7-, nsp8-, and nsp12-coding regions by RT-PCR. This material was used to confirm the presence of the engineered mutation(s) and investigate the presence of possible second-site reversions. The properties of the various mutants are summarized in Table S2.
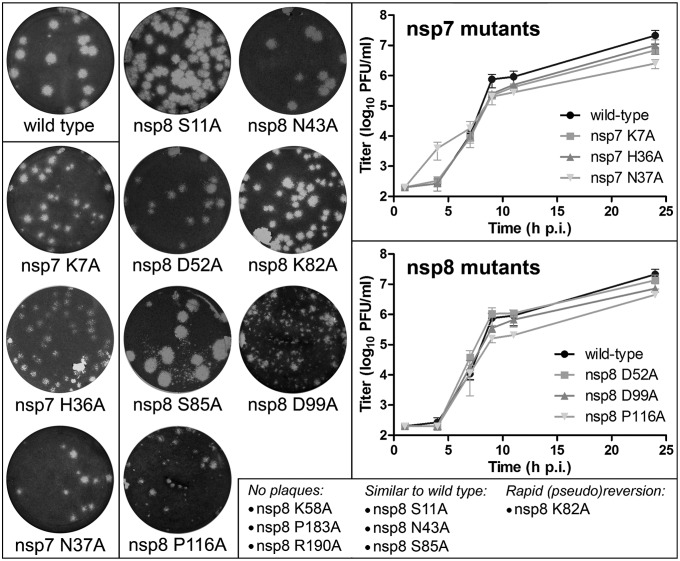
Substitution of the invariant nsp7 residues H36 and N37 by alanine supports the importance of nsp7 for virus replication in vivo. Although both mutants were viable, their replication was impaired compared with that of the WT virus as was initially evident from their reduced plaque size (Fig. 5). Similar observations were made for mutant nsp7 K7A. The phenotype of these three nsp7 mutants was further compared by determining their growth-curves, for which fresh Vero-E6 cells were infected [multiplicity of infection (M.O.I.) of 5] with supernatant harvested at 42 h p.t. from cells transfected with mutant RNA (Fig. 5). This more detailed comparison confirms that all three nsp7 mutants are (about equally) crippled compared with the WT control, reaching final progeny titers [24 h postinfection (p.i.)] that are about 0.5–1 log reduced (Fig. 5).
Fig. 5.
Reverse-genetics analysis of the impact of selected SARS-CoV nsp7 and nsp8 mutations. Plaque phenotypes of viable but crippled mutants with engineered nsp7 and nsp8 mutations are illustrated on the Left. Plaque assays were performed with early progeny virus, harvested from cells at 18 h post full-length RNA transfection (Materials and Methods). On the Right, growth curves are shown for crippled nsp7 and nsp8 mutants and the WT parental virus. For these experiments, mutant virus was harvested from transfected cells at 42 h p.t. After titration of these virus stocks, fresh Vero-E6 cells were infected (M.O.I., 5), and virus production at the given time points was measured by plaque assay. Graphs display mean titers and SDs derived from three independent experiments. Nonviable mutants and mutants with a phenotype similar to WT virus are listed at the Bottom.
The 10 nsp8 mutants tested displayed a variety of phenotypes (Table S2), ranging from nonviable (K58A, P183A, and R190A), via crippled (D52A, K82A, D99A, and P116A), to quite similar to the WT control (S11A, N43A, and S85A). For the mutants in the first and third groups, the in vivo phenotypes are in general agreement with the results from the biochemical assays (Table 2). This is less clearcut for the second group, which for example contains nsp8 mutants (D99A, P116A) with major defects in the biochemical assays but a viable, although crippled in vivo phenotype, as reflected in their reduced plaque size and somewhat lower progeny titers (Fig. 5). Conversely, two mutants (D52A, K82A) not showing major defects in biochemical assays were impaired when tested by reverse genetics. Mutant D52A displayed a reduced plaque size, although its growth kinetics was quite similar to what was found for the WT virus (Fig. 5). For K82A, no evidence of replication was observed at the earliest time point after transfection analyzed (18 h p.t.; Table S2), but this changed later on. This transition coincided with the discovery of second-site mutations, specifically a D78N substitution in one experiment and a mix of E77T and D78N in the second experiment. The results obtained for these two mutants once again highlight how the use of diverse assays to analyze the effect of mutations in viral proteins is a critical part of their functional characterization. Interestingly, with regard to the only three invariant residues identified upon the comparative analysis of CoV nsp8 sequences (N43, K58, and P183; Fig. S6), replacements of K58 and P183 impair polymerase activity and resulted in a nonviable phenotype in reverse genetics, whereas mutant N43A surprisingly displayed a WT-like phenotype both in vitro and in vivo.
Although the lethal effect of an nsp8 full-length deletion previously suggested the overall importance of nsp8 (41), this study is the first (to our knowledge) to establish the direct functional importance of specific nsp8 residues (K58, P183, and R190) for virus replication.
The Nsp7/Nsp8/Nsp12 Polymerase Complex Is Able to Associate with an Active Bifunctional Nsp14.
In agreement with the proposed role of the nsp14 ExoN domain in coronavirus proofreading during replication (24, 25, 47), the nsp14 replicase subunit binds to nsp12 (Fig. S1B and ref. 39). We therefore addressed the question whether nsp14 is still able to bind to nsp12 in the context of the 7L8/12 complex and if such an interaction could potentially affect any of the three-enzymatic activities (RdRp, 3′-5′ ExoN, and N7-MTase).
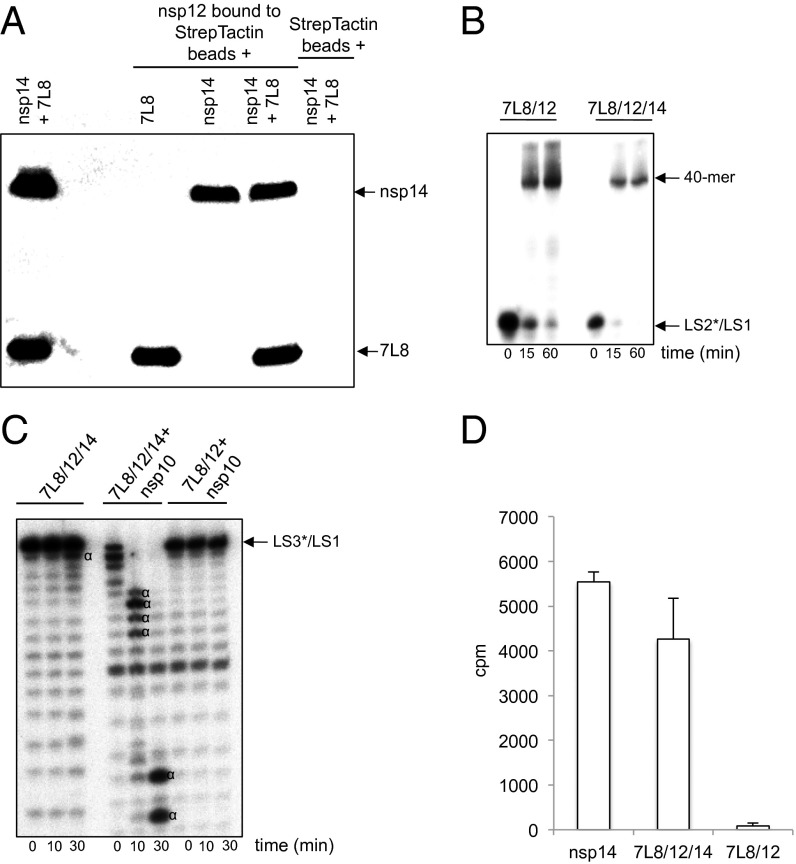
Pull-down experiments were performed as described in the legend of Fig. S1. Interaction of 7L8, nsp12, and nsp14 is readily detected (Fig. 6A), and the protein complex (hereafter 7L8/12/14) can be purified (Fig. S7). The polymerase activities of the 7L8/12 and 7L8/12/14 complexes are comparable (Fig. 6B). In these polymerase assays, RNA degradation is not detected in the presence of nsp14, indicating that nsp14 ExoN activity is either very weak due to the stimulating-factor absence (24) or suppressed when the 7L8/12/14 complex is in “polymerization mode.” The 3′-5′ ExoN activity of the 7L8/12/14 complex was then assayed using the optimal experimental conditions previously described for ExoN (24). Under these conditions, ExoN activity is barely detected (Fig. 6C). When nsp10 is added to the 7L8/12/14 complex, the RNA exonuclease activity is significantly enhanced (Fig. 6C), indicating that nsp10 is able to either enhance nsp14-ExoN activity in situ as previously reported (24), or to partially displace nsp14 from 7L8/12/14 and activate nsp14-ExoN activity separately. Moreover, nsp14 N7-MTase activity is also found associated with the 7L8/12/14 complex (Fig. 6D). We therefore conclude that the nsp7/nsp8/nsp12/nsp14 complex represents a unique coronavirus nsp assembly that incorporates RdRp, exoribonuclease, and N7-MTase activities.
Fig. 6.
The SARS-CoV 7L8/12/14 complex possesses RdRp, ExoN, and N7-MTase activities. (A) Strep-tagged SARS-CoV nsp12 was bound to Strep-Tactin beads and incubated with 7L8, nsp14, or both simultaneously. After SDS/PAGE and Western blotting, his-tagged proteins (7L8 and nsp14) were revealed using an anti-His5-HRP antibody. (B) Time course primer extension polymerase assays were performed using either the 7L8/12 (500 nM) or the 7L8/12/14 (500 nM) complexes with LS2*/LS1 as primer*/template where LS2 was 5′-radiolabeled (marked by *). RNA products were separated in a denaturing polyacrylamide/urea gel and visualized by autoradiography. (C) Time course exoribonuclease assays were performed using the 7L8/12/14 (500 nM) complex in the absence or presence of 100 nM nsp10, and as control with 7L8/12 (500 nM) plus nsp10 (100 nM). The RNA substrate was a 40-nt RNA (LS1) annealed with 5′-radiolabeled LS3 primer carrying one noncomplementary base at its 3′ end (LS3*) and named LS3*/LS1. Digestion products were separated by denaturing polyacrylamide/urea gel electrophoresis and visualized by autoradiography (Fuji). The “α” symbol indicates RNA cleavage products. (D) AdoMet-dependent N7-MTase activity of the 7L8/12/14 complex. The different purified proteins or protein complexes (nsp14, 300 nM; 7L8/12/14, 300 nM; and 7L8/12, 300 nM) were incubated with substrate GpppAC4 RNA oligonucleotide in the presence of [3H]AdoMet. The methyl transfer to the capped RNA substrate was determined by using a filter-binding assay (as described in ref. 73). All experiments were done in triplicate (SDs are presented).
Discussion
In the infected cell, viral RNA replication is challenged at several levels. RNA synthesis must be regulated to minimize exposure to the host cell’s defense mechanisms. Moreover, compared with DNA-based organisms, RNA viruses are generally believed to replicate their genetic material with low accuracy, as they are believed to lack proofreading and correction mechanisms (48, 49). Hence, RNA replication has evolved toward an equilibrium at which a heterogeneous population of viral RNAs, commonly referred to as a quasispecies, is reproduced with high efficiency (50, 51). The viral RdRp, fine-tuned during evolution, is the central enzyme in this process. It is thought to possess the optimum combination of RNA synthesis efficiency and nucleotide incorporation fidelity (52, 53). At the enzyme level, speed and accuracy can be measured and evaluated using a number of parameters. Among them, the processivity of the RNA polymerase, reflected in the number of nucleotides polymerized during a single encounter with its template, is considered critical for efficient virus replication (54–58).
Coronaviruses carry RNA genomes at least twice larger (∼30 kb) than that of the majority of animal RNA viruses (∼3 to ∼15 kb). Our present results suggest that to ensure processivity of their nsp12-polymerase, coronaviruses have evolved a mechanism of RNA synthesis that is unique among RNA viruses. Based on in vitro assays, SARS-CoV nsp12 RdRp was previously reported to be a nonprocessive primer-dependent RNA polymerase (21, 37). We now demonstrate that the simultaneous addition of nsp7 and nsp8 activates and confers processivity to the nsp12 RdRp. By displaying a very low dissociation rate from RNA, the nsp7/nsp8/nsp12 complex can synthesize an RNA of, at least, 359 nt when synthesis is started without a primer (de novo). Clearly, association with other nsps may be required to copy the 30-kb full-length genome.
The three amino acids in nsp7 (K7, H36, and N37) that confer RNA-binding properties to the polymerase complex (Fig. 4B) are also important for virus replication in vivo (Fig. 5). Indeed, in reverse-genetics studies, these mutations delayed virus growth compared with the WT control, although their impact is less severe than one might have anticipated given their phenotype in the in vitro assays. The lack of correlation for some mutants between in vitro and in vivo studies suggests that the in vitro biochemical assays reflect only a part of the complexity of coronavirus RNA synthesis in vivo. It is conceivable that the effects of single-point mutations on properties like RNA binding are more dramatic in the context of the trimeric complex in vitro than in the context of a 16-nsp complex in vivo, where, e.g., the presence of other replicase subunits may provide some form of compensation. Nonetheless, these data constitute the first evidence (to our knowledge) for a role of nsp7 in coronavirus replication. Moreover, our biochemical (Tables 1 and 2) and reverse-genetics (Fig. 5 and Table S2) data strengthen the case for a role for nsp8 in viral RNA synthesis, with three nsp8 residues (K58, P183, and R190) being identified as critical for SARS-CoV genome replication. Our biochemical assays further indicate that nsp8 residues P183 and R190 are involved in nsp8/nsp12 interaction, whereas residue K58 modulates the interaction of the polymerase complex with RNA.
Nsp8 was previously suggested to be a second, noncanonical coronavirus RdRp, which would be able to synthesize small oligomers (<6 nt) alone (30) or in complex with nsp7 (31, 32). In an effort to understand how de novo RNA synthesis is achieved and possibly demonstrate the role of nsp8 as primase in this complex, we tested nsp12 catalytic mutant in de novo assays, in the presence of the nsp7/nsp8 complex. Full-length products were absent as expected. However, we were also unable to detect the nsp8-driven formation of short RNAs (Fig. S3), even in the presence of Mn2+, an element described as essential for nsp8-RdRp activity (30). One possible explanation of this discrepancy may be that nsp8 in complex with nsp12 and nsp7 adopts a structural conformation that prevents the synthesis of short RNAs. The ability of the nsp7/nsp8 complex to adapt different conformations (refs. 29 and 32; see below) may reflect its involvement in distinct functions, including primer synthesis. It is thus plausible that a particular function of coronavirus nsp7 and nsp8 may in fact be triggered by the nature of their partnership and/or quaternary structure.
Nsp7 and nsp8 are shown to interact with each other, and nsp7/nsp8 complexes were crystallized and solved for different coronaviruses. The presence of nsp7 seems to be required to crystallize nsp8. The structure of the SARS-CoV nsp7/nsp8 complex (29) revealed a hexadecameric complex with a positively charged central tunnel supposedly accommodating RNA. This structure is reminiscent of processivity-promoting factors such as, e.g., the eukaryotic PCNA sliding clamp (59–61), or the β-subunit bound to E. coli DNA polymerase III (62). In contrast, feline coronavirus nsp7 and nsp8 were shown to form a heterotrimer by association of two nsp7 molecules with one molecule of nsp8 without the formation of a hollow structure (32). It is thus likely that coronavirus nsp7 and nsp8 may in fact be endowed with several functions triggered by the nature of their partnership and/or quaternary structure. They would thus constitute additional examples of RNA virus proteins that have evolved a remarkable plasticity to accommodate multiple functions. Another recent example of this kind of functional flexibility is the Ebola virus matrix protein VP40, which was shown to be subject to structural rearrangements giving rise to at least three distinct structures, each executing a distinct function during the viral replication cycle (63).
Polymerase processivity factors have been identified for DNA viruses such as the gp45 protein of bacteriophage T4 (64), the UL42 protein of herpes simplex virus 1 (65, 66), or E. coli thioredoxin for bacteriophage T7 (67, 68). To our knowledge, coronaviruses now are the first RNA viruses known to use an RdRp processivity factor, to expedite replication of their ∼30-kb RNA genome. Remarkably, the function of the coronavirus RNA polymerase processivity factor appears to be compatible with the capability of the nsp7/nsp8/nsp12 complex to associate with an active nsp14-exonuclease that is able to remove terminal mismatches from an RNA duplex (24). The nsp7/nsp8/nsp12 complex binds to the bifunctional exoribonuclease/N7-guanine cap methyltransferase nsp14, with no apparent modulation of polymerase, ExoN, or N7-MTase activities. The reconstitution of an nsp7/nsp8/nsp12/nsp14 complex makes it now possible to dissect in vitro the coronavirus proofreading mechanism discovered through prior research (25, 26, 47).
In any case, this complex provides a fascinating example of the integration of RNA polymerization, proofreading, and cap-modifying activities into a multifunctional protein assembly. This macromolecular assembly, possibly in combination with the helicase/RNA triphosphatase nsp13 and the 2′O-MTase nsp16 may constitute the core of the coronavirus replication/transcription machinery, which is able to engage in coordinated RNA synthesis and processing activities.
Materials and Methods
Cloning, Expression, and Purification of SARS-CoV Proteins.
All SARS-CoV proteins used in this study were expressed in a bacteriophage T7 RNA polymerase-free system, with the exception of nsp10 and nsp14. All plasmids were expressed in E. coli C2523 cells (NE Biolabs), whereas E. coli C41 (DE3; pLysS) (Novagen) was used for nsp10 and nsp14 expressions. SARS-CoV nsp7 fused to a C-terminal hexahistidine tag (named nsp7) was cloned into the vector pASK3-Kana, a modified pASK3 vector (IBA) in which the ampicillin resistance cassette is replaced by that for kanamycin. Nsp7 expression was done overnight at 17 °C after induction with 200 μg/L anydrotetracyclin. Protein purification was performed as described previously for nsp9 (69). SARS-CoV nsp8 with a C-terminal hexahistidine tag (named nsp8) was purified as described previously (31). The fusion protein 7L8 was generated by inserting a hexahistidine tag linker between nsp7- and nsp8-coding sequences. This gene was cloned into the pASK3-Kana backbone and protein purification was performed as described for nsp7. SARS-CoV nsp12 was expressed from a synthetic gene optimized for expression in E. coli and equipped with a C-terminal Strep-Tag, which was cloned into plasmid pJ404 (purchased from DNA2.0). Nsp12 expression was induced by addition of 500 μM isopropyl β-d-1-thiogalactopyranoside when the OD600nm value of the culture reached 0.5, and was allowed to proceed for 16 h at 17 °C. Lysis buffer [50 mM Hepes (pH 7.5); 150 mM KCl; 5 mM MgCl2; 5% (vol/vol) glycerol] supplemented with 10 µg/mL DNase I, 1 mM PMSF, and 0.25 mg/mL lysozyme were used to resuspend pellets. A Strep-Tactin (IBA) purification step was performed and nsp12 was recovered using lysis buffer supplemented with 2.5 mM d-desthiobiotin. A second chromatography step was then performed. The salt concentration of nsp12 protein solution was lowered to 25 mM KCl, and nsp12 was then incubated during 1 h with heparin beads and finally eluted with a linear gradient of KCl (25–500 mM).
The 7L8/12 complex was obtained after cotransformation of plasmids encoding 7L8 and nsp12, and was purified as described above for nsp12, with an additional size exclusion chromatography step performed on a KW804 column (Shodex).
Site-directed mutagenesis to generate plasmids expressing mutant proteins was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. For relevant nsp7 (K7A, H36A, and N37A) and nsp8 (K58A, D99A, P116A, P183A, and R190A) mutants as well as for nsp7 WT and nsp8 WT, correct folding was confirmed by circular dichroism (CD) analysis. The CD spectra were recorded on a Jasco 810 dichrograph using 1-mm-thick quartz cells.
RNA Templates.
Synthetic RNAs (LS1, LS2, and LS3) were purchased from Biomers (HPLC grade). Templates 3RA and 3R RNA templates, corresponding to the last 339 nt of the SARS-CoV (strain Frankfurt; GenBank accession no. AY291315) genomic 3′-NTR with or without 20 adenines, respectively, were produced using an in vitro T7 transcription kit, and purified as described by the manufacturer (Ambion). RNA marker (Promega; catalog no. G3191) was radiolabeled with the T4 polynucleotide kinase (PNK) (NEB) and [γ-32P]ATP.
Pull-Down, Western Blot, and Nsp8/Nsp12 Interaction Assays.
Nsp12 was bound to Strep-Tactin beads and incubated with 1 mg/mL BSA for 15 min at room temperature. Then, the Strep-Tactin beads with or without bound nsp12 were incubated for 2 h in the presence of 15 µg of his-tagged partner protein to be tested. Next, unbound proteins were extensively washed away with washing buffer (50 mM Tris, pH 8; 150 mM NaCl; 0.05% Triton). The beads were diluted in 2× SDS/PAGE loading buffer [100 mM Tris, pH 6.8; 200 mM DTT; 4% (vol/vol) SDS; 0.2% bromophenol blue; 20% (vol/vol) glycerol] and loaded on an SDS/PAGE gel. A Western blot was then performed using a single HRP-conjugate antibody (anti-His5 HRP conjugate; Qiagen), and bands were visualized with a Kodak Image Station 400 MM Pro (CareStream Health).
To study the interaction between nsp8 and nsp12, WT or mutant 7L8 were coexpressed with nsp12 in E. coli. After purification on a Strep-Tactin column, the eluted proteins were analyzed using SDS/PAGE, and the peak intensities corresponding to the 7L8 and nsp12 proteins were quantified.
Polymerase Assays.
All radioactive reagents were purchased from Perkin-Elmer. Polymerase assays were performed in polymerase assay buffer (20 mM Tris, pH 8; 10 mM KCl; 1 mM DTT; 2 mM MgCl2) with separately purified nsp12 (500 nM), nsp7 (1.5 µM), and nsp8 (1.5 µM) or 500 nM 7L8/12 complex and 250 nM RNA template, unless specified otherwise. Primers (LS2 or LS3) were radiolabeled at their 5′ ends using [γ-32P]ATP and PNK, and termed LS2* and LS3*, respectively. LS2* or LS3* were then annealed to the complementary template LS1 by heating at 70 °C for 10 min and then cooling down to room temperature (with a primer/template ratio of 1.2:1). Primer extension assays were always performed with LS2*/LS1 as template, and reactions were started by adding 500 μM NTP mix. De novo assays, using either 3R or 3RA as template, were started by adding 500 μM GTP, UTP, CTP, 50 μM ATP, and 0.17 μM [α-32P]ATP (0.5 μCi/μL). After incubation at 30 °C, reactions were quenched by the addition of an equal volume of loading buffer (formamide with 10 mM EDTA). RNA polymerization products of primer extension assays were analyzed in 20% (wt/vol) polyacrylamide/7 M urea gels, whereas products from de novo assays were analyzed in 1% agarose-formaldehyde gels, unless specified otherwise. RNA products were visualized using photostimulated plates and a phosphorimager (Fuji).
EMSAs.
Serial dilutions of SARS-CoV proteins in polymerase assay buffer were preincubated with 100 nM primer*/template (LS2*/LS1) for 30 min at 30 °C. Unless otherwise stated, 500 μM CTP, UTP, GTP, and 50 μM 3′-dATP were added to facilitate RNA–enzyme complex formation. The reaction mixture was incubated for 60 min at 30 °C, and samples were loaded directly onto 6% (vol/vol) acrylamide gels and were run in 0.5× TBE at 100 V for 60 min at 4 °C. Bands were visualized using a phosphorimager (Fuji).
Exonuclease Assay.
Nuclease reactions were performed as previously described (24) with 500 nM radiolabeled LS3*/LS1 substrate. Primer LS3 has the same sequence as LS2 with the addition of an adenosine at its 3′ end, causing an A:A mismatch when annealed with template LS1.
SARS-CoV Reverse Genetics.
Vero cells (ATCC; CCL-81) were cultured in Eagle’s MEM (Lonza) with 8% (vol/vol) FCS (PAA) and antibiotics. BHK-Tet-SARS-N cells (46) were cultured in the same medium containing 100 μg/mL G418. Mutations in the SARS-CoV nsp7- or nsp8-coding region were engineered in prSCV, a pBeloBac11 derivative containing a full-length cDNA copy of the SARS-CoV Frankfurt-1 sequence (45) as described in ref. 70. Full-length BAC clones were linearized with NotI, extracted with phenol-chloroform, and transcribed with T7 RNA Polymerase (mMessage-mMachine T7 kit; Ambion) using an input of 2 μg of BAC DNA per 20-μL reaction. RNA transcripts were precipitated with LiCl according to the manufacturer’s protocol. Subsequently, 6 μg of RNA was electroporated into 5 × 106 BHK-Tet-SARS-N cells, which express the SARS-CoV N protein after induction with 2 µM doxycyclin. Electroporations were done using the Amaxa Cell Line Nucleofector Kit T and program T-020 of an Amaxa Nucleofector (Lonza), according to the manufacturer’s instructions. BHK-Tet-SARS-N cells were mixed in a 1:1 ratio with Vero-E6 cells, and seeded on coverslips in six-well clusters for immunofluorescence microscopy analysis and analysis of virus production. Each mutant was launched twice from two independently generated BAC clones. All work with live SARS-CoV was performed inside biosafety cabinets in a biosafety level 3 facility at Leiden University Medical Center.
Sequence Analysis of the SARS-CoV Nsp7-, Nsp8-, and Nsp12-Coding Regions.
Fresh Vero-E6 cells were infected with harvests taken at 42 h p.t. and SARS-CoV RNA was isolated 18 h p.i. as described above and amplified by RT-PCR using random hexamers for the RT reaction, and primers 5′-CACCTTACAGTGTATCATGC-3′ and 5′-CTGGAACCACCTTGTAGGTTTG-3′ for PCR amplification of the nsp7–8 coding region. The nsp12-coding region was amplified in two overlapping fragments, by using the following primer sets: 5′-TTGCCTACTATAACAATTCG-3′ together with 5′-CAGAACTTCCTTCCTTAAAGAAACCTT-3′; 5′-GCAGCTTCTGGCAATTTATTGC-3′ together with 5′-GGAATGGTCTCCTAATACAGGC-3′. RT-PCR products were sequenced to verify the presence of the introduced mutations and possible second-site reversions.
Immunofluorescence Microscopy.
To monitor the presence and/or progression of SARS-CoV infection, transfected cells were seeded on coverslips and fixed at various time points. Immunofluorescence assays were done following a previously described protocol (71) using a rabbit antiserum against SARS-CoV nsp4 (36) and a mouse monoclonal antibody against the SARS-CoV N protein (72).
Virus Titration.
Vero-E6 cells seeded in six-well clusters were infected with serial 10-fold dilutions in PBS, 0.005% DEAE, and 2% (vol/vol) FCS of supernatants from transfected cells, incubated for 1 h at 37 °C, after which the inoculum was replaced by an overlay of 1.2% (wt/vol) Avicel (FMC BioPolymer) in DMEM supplemented with 50 mM Hepes, 2% (vol/vol) FCS, and incubated at 37 °C. After 3 d, the overlay was removed, and cells were fixed with 7.4% (vol/vol) formaldehyde in PBS and stained with crystal violet to visualize plaques. Titers were expressed in plaque-forming units per milliliter.
Supplementary Material
Acknowledgments
We thank Laure Gluais, François Ferron, Alice Blouet, Jérome Deval, Barbara Selisko, Aartjan te Velthuis, Alexander Kravchenko, Dmitry Samborskiy, Igor Sidorov, Isabelle Varlet, and Renaud Vincentelli for helpful discussions and/or technical assistance. We thank Christian Drosten and Susanne Pfefferle (University of Bonn) for providing the SARS-CoV reverse genetics system, and Ying Fang (Kansas State University) for providing the anti-N monoclonal antibody. We acknowledge Dr. Deborah Byrne for editorial assistance. This work was supported by the French National Research Agency under reference “ANR-08-MIEN-032” and “ANR-12-BSV3,” the Netherlands Organization for Scientific Research (TOP-GO Grant 700.10.352), and by the European Union Seventh Framework Programme (FP7/2007-2013) through the EUVIRNA project [European Training Network on (+)RNA Virus Replication and Antiviral Drug Development; Grant Agreement 264286], and the SILVER project (Small Inhibitor Leads Versus Emerging and Neglected RNA Viruses; Grant Agreement 260644), and finally by the Collaborative Agreement in Bioinformatics Between Leiden University Medical Center and Moscow State University (MoBiLe), and Leiden University Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323705111/-/DCSupplemental.
References
- 1.Cameron CE, Götte M. In: Viral Genome Replication. Raney K, editor. New York: Springer; 2009. [Google Scholar]
- 2.Kuhn RJ. Fields’ Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1001–1022. [Google Scholar]
- 3.Lai MMC, Perlman S, Anderson LJ. Fields’ Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1305–1335. [Google Scholar]
- 4.Lazarowitz SG. Fields’ Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 641–705. [Google Scholar]
- 5.Domingo E, et al. Basic concepts in RNA virus evolution. FASEB J. 1996;10(8):859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 6.Stadler K, et al. SARS—beginning to understand a new virus. Nat Rev Microbiol. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 9.Ge X-Y, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot RJ, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 12.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman VM, et al. Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J Virol. 2014;88(1):717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo PCY, et al. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J Virol. 2014;88(2):1318–1331. doi: 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Pe’ery T, Mathews MB. Fields’ Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 169–207. [Google Scholar]
- 16.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawicki SG, Sawicki DL. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Med Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 19.Pasternak AO, Spaan WJM, Snijder EJ. Nidovirus transcription: How to make sense...? J Gen Virol. 2006;87(Pt 6):1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 20.Nga PT, et al. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.te Velthuis AJW, Arnold JJ, Cameron CE, van den Worm SHE, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38(1):203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adedeji AO, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One. 2012;7(5):e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minskaia E, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouvet M, et al. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci USA. 2012;109(24):9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6(5):e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81(22):12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang H, et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol. 2007;81(24):13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricagno S, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA. 2006;103(32):11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai Y, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12(11):980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imbert I, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.te Velthuis AJW, van den Worm SHE, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Y, et al. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J Virol. 2012;86(8):4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Züst R, Miller TB, Goebel SJ, Thiel V, Masters PS. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J Virol. 2008;82(3):1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorbalenya AE, et al. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol. 2002;324(1):47–62. doi: 10.1016/S0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6(4):e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Hemert MJ, et al. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4(5):e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng A, et al. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology. 2005;335(2):165–176. doi: 10.1016/j.virol.2005.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106(9):3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imbert I, et al. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133(2):136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith EC, Denison MR. Coronaviruses as DNA wannabes: A new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9(12):e1003760. doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deming DJ, Graham RL, Denison MR, Baric RS. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol. 2007;81(19):10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomaguchi M, Ackermann M, Yon C, You S, Padmanabhan R. De novo synthesis of negative-strand RNA by dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J Virol. 2003;77(16):8831–8842. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85(Pt 5):1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 44.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 45.Pfefferle S, et al. Reverse genetic characterization of the natural genomic deletion in SARS-Coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in-vitro and in-vivo. Virol J. 2009;6:131. doi: 10.1186/1743-422X-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang G-H, et al. Establishment of the eukaryotic cell lines for inducible control of SARS-CoV nucleocapsid gene expression. Virol Sin. 2010;25(5):361–368. doi: 10.1007/s12250-010-3124-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith EC, Blanc H, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: Evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8):e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 49.Holmes EC. What does virus evolution tell us about virus origins? J Virol. 2011;85(11):5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holland JJ, De La Torre JC, Steinhauer DA. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 51.Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76(2):159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinhauer DA, Holland JJ. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 53.Holland J, et al. Rapid evolution of RNA genomes. Science. 1982;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 54.Werneburg BG, et al. DNA polymerase beta: Pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity. Biochemistry. 1996;35(22):7041–7050. doi: 10.1021/bi9527202. [DOI] [PubMed] [Google Scholar]
- 55.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 56.Mace DC, Alberts BM. T4 DNA polymerase. Rates and processivity on single-stranded DNA templates. J Mol Biol. 1984;177(2):295–311. doi: 10.1016/0022-2836(84)90458-3. [DOI] [PubMed] [Google Scholar]
- 57.Capson TL, et al. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31(45):10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 58.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26(25):8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 59.Takasaki Y, Deng JS, Tan EM. A nuclear antigen associated with cell proliferation and blast transformation. J Exp Med. 1981;154(6):1899–1909. doi: 10.1084/jem.154.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prelich G, et al. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326(6112):517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 61.McConnell M, et al. The mammalian DNA polymerase delta—proliferating cell nuclear antigen—template-primer complex: Molecular characterization by direct binding. Biochemistry. 1996;35(25):8268–8274. doi: 10.1021/bi9530649. [DOI] [PubMed] [Google Scholar]
- 62.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell. 1992;69(3):425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 63.Bornholdt ZA, et al. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154(4):763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reddy MK, Weitzel SE, von Hippel PH. Assembly of a functional replication complex without ATP hydrolysis: A direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci USA. 1993;90(8):3211–3215. doi: 10.1073/pnas.90.8.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallo ML, Dorsky DI, Crumpacker CS, Parris DS. The essential 65-kilodalton DNA-binding protein of herpes simplex virus stimulates the virus-encoded DNA polymerase. J Virol. 1989;63(12):5023–5029. doi: 10.1128/jvi.63.12.5023-5029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottlieb J, Marcy AI, Coen DM, Challberg MD. The herpes simplex virus type 1 UL42 gene product: A subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64(12):5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mark DF, Richardson CC. Escherichia coli thioredoxin: A subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1976;73(3):780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Modrich P, Richardson CC. Bacteriophage T7 deoxyribonucleic acid replication in vitro. Bacteriophage T7 DNA polymerase: An enzyme composed of phage- and host-specific subunits. J Biol Chem. 1975;250(14):5515–5522. [PubMed] [Google Scholar]
- 69.Campanacci V, et al. Structural genomics of the SARS coronavirus: Cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 9):1628–1631. doi: 10.1107/S0907444903016779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 71.van der Meer Y, et al. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73(9):7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fang Y, Pekosz A, Haynes L, Nelson EA, Rowland RRR. Production and characterization of monoclonal antibodies against the nucleocapsid protein of SARS-CoV. Adv Exp Med Biol. 2006;581:153–156. doi: 10.1007/978-0-387-33012-9_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Decroly E, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol. 2008;82(16):8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.