Significance
The innate immune system is a highly conserved mode of defense that induces gene expression programs to restrict microbial infections. However, much remains unknown about how the target genes are poised for their rapid induction. Using a Drosophila model, we demonstrate that Nup98 plays an essential antiviral role in insects against human insect-borne viruses. Although Nup98 is known for its role in nuclear-cytoplasmic transport, our data suggest that this antiviral function is not at the nuclear pore, rather at promoters controlling expression of a subset of virus-induced genes. Our findings suggest that the Nup98 primes virus-stimulated genes by regulating the occupancy of active RNA polymerase at these promoters poising them for rapid induction, thereby coordinating a robust and complex antiviral response.
Keywords: innate immunity, nuclear pore, nucleoporin, pol II regulation, P-TEFb
Abstract
In response to infection, the innate immune system rapidly activates an elaborate and tightly orchestrated gene expression program to induce critical antimicrobial genes. While many key players in this program have been identified in disparate biological systems, it is clear that there are additional uncharacterized mechanisms at play. Our previous studies revealed that a rapidly-induced antiviral gene expression program is active against disparate human arthropod-borne viruses in Drosophila. Moreover, one-half of this program is regulated at the level of transcriptional pausing. Here we found that Nup98, a virus-induced gene, was antiviral against a panel of viruses both in cells and adult flies since its depletion significantly enhanced viral infection. Mechanistically, we found that Nup98 promotes antiviral gene expression in Drosophila at the level of transcription. Expression profiling revealed that the virus-induced activation of 36 genes was abrogated upon loss of Nup98; and we found that a subset of these Nup98-dependent genes were antiviral. These Nup98-dependent virus-induced genes are Cdk9-dependent and translation-independent suggesting that these are rapidly induced primary response genes. Biochemically, we demonstrate that Nup98 is directly bound to the promoters of virus-induced genes, and that it promotes occupancy of the initiating form of RNA polymerase II at these promoters, which are rapidly induced on viral infection to restrict human arboviruses in insects.
Innate immunity is an evolutionarily conserved mode of defense against invading pathogens. A major facet of innate immunity involves the recognition of pathogen-associated molecular patterns by pattern recognition receptors to initiate signaling pathways to induce antimicrobial gene expression (1–3). This system is robust and is the sole mode of protection against invading pathogens in insects and plants. The gene expression programs activated on pathogen detection are tightly orchestrated to regulate downstream immune responses. The best-characterized example is the lipopolysaccharide (LPS)-dependent gene expression program (2, 4). This response is divided into two stages; within minutes, a rapid primary response independent of new protein synthesis is initiated, which instructs the downstream translation-dependent secondary response (2, 5). Many primary response genes have active chromatin marks and features of transcriptional pausing, including high occupancy of the initiating form of RNA polymerase II (RNAPII), S5 phosphorylated (S5P) (2, 4, 6), along with negative elongation factor complex (NELF) and DRB Sensitivity-Inducing Factor complex (DSIF), which prevent transcriptional elongation (4, 6–10). Paused RNAPII can be activated by the positive transcription elongation factor b (P-TEFb) in a stimulus-dependent manner, which phosphorylates NELF, DSIF, and RNAPII to release the pause and promote transcriptional elongation and thus the production of mature mRNAs (9, 11–13). Indeed, a large number of LPS-induced primary response genes are controlled at the level of pausing including the classical gene TNF-α (4). Furthermore, this is conserved in Drosophila as the LPS-inducible homolog of TNF-α (Eiger) is also regulated by pausing (6). Furthermore, depletion of the pausing factor NELF reduced RNAPII occupancy on the promoters of LPS-stimulated genes in Drosophila (6).
Although many signaling pathways that regulate antibacterial and antifungal gene expression programs have been well characterized in insects, our understanding of antiviral gene expression programs is less clear (14, 15). We recently found that viral infection can lead to a rapid antiviral gene expression program, and that one-half of these virus-inducible genes are regulated at the level of transcriptional pausing (14). We also found that NELF is required for RNAPII occupancy at these pausing-regulated genes (14). These data suggest a conserved role for this mode of gene regulation in the control of antiviral gene expression; however, whether there are specific factors required to promote high RNAPII occupancy at these promoters or to promote the future activation at particular loci remains unclear.
Nucleoporins (Nups), first identified for their role in nuclear-cytoplasmic transport, have recently been found to have roles outside of the nuclear pore. Initially, a subset of Nups was found to be mobile, able to move off and on the pore (16). The intranuclear accumulation of one such Nup, Nup98, is linked to ongoing nuclear transcription and chemical inhibition of RNA polymerase II was shown to abrogate its intranuclear mobility (17, 18). Moreover, Nup98 was subsequently found to directly control gene expression of a subset of developmentally regulated genes (19–21). Nup98 is recruited to these loci during developmental transcriptional activation, and this association is required for the expression of such genes, particularly for the rapid induction of hormone-activated developmental gene targets (19). It was recently shown that Nup98 is similarly involved in the transcriptional regulation of IFN-γ–induced gene expression (22), suggesting that the transcriptional roles of Nups may be involved in immunity.
Based on these findings, and given the fact that many developmental and immune genes are regulated by transcriptional pausing (4, 6, 8, 14, 23, 24), we hypothesized that Nup98 also may regulate virus-induced antiviral genes. We found that Nup98 plays a broadly antiviral role in insects; cells or adult flies depleted of Nup98 are more susceptible to infection against a panel of disparate RNA viruses. This includes human arboviruses from diverse families of viruses: Sindbis virus (SINV), an alphavirus; vesicular stomatitis virus (VSV), a rhabdovirus; and West Nile virus (Kunjin strain), a flavivirus. Interestingly, we found that in our experimental system, transient depletion of Nup98 did not affect general nuclear pore function; nuclear import of NFkB and nuclear export of mRNAs and proteins were intact. This led us to discover a role for Nup98 in promoting antiviral gene expression in Drosophila. Through transcriptional profiling, we found that 36 genes were virus-induced and Nup98-dependent. Importantly, we found that a subset of Nup98-dependent virus-induced genes were antiviral themselves, suggesting that Nup98 directly regulates expression of these antiviral genes. Moreover, single-molecule RNA fluorescent in situ hybridization (FISH) revealed reduced levels of virus-induced mRNAs, but not their localization, again supporting a role in the direct regulation of gene expression at the level of transcription. These Nup98-dependent virus-induced genes are translation-independent and regulated by the pausing-release factor P-TEFb. Mechanistically, we found that Nup98 binds to the promoter of these antiviral genes and positively regulates the levels of active RNAPII at these loci at the basal state. Taken together, our data suggest that Nup98 binds to these loci and facilitates the engagement of RNAPII; subsequent virus challenge signals P-TEFb to activate transcription at these loci, inducing antiviral gene expression. These findings demonstrate a previously unidentified requirement for Nup98 in antiviral defense via direct transcriptional initiation of antiviral genes and its coordination with transcriptional pausing to restrict viral infection.
Results
Nup98 Restricts Sindbis Virus Infection.
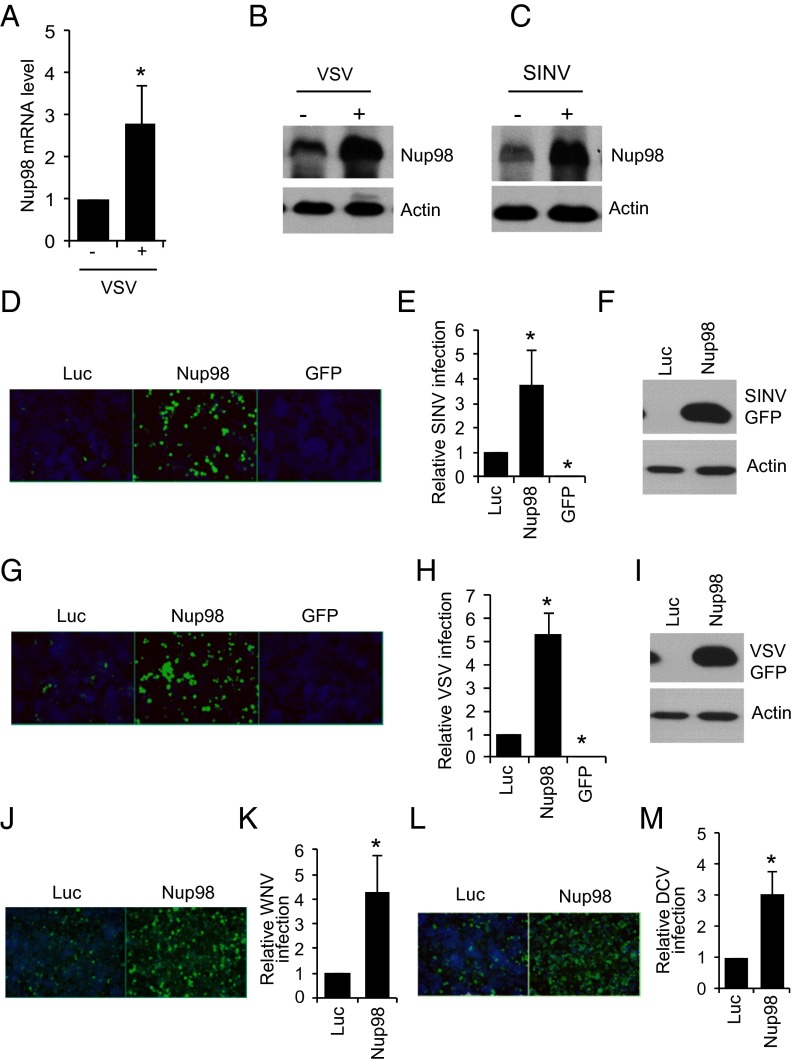
We recently reported that Nup98 mRNA is induced on VSV infection in Drosophila (14). Because many virus-induced genes are antiviral, we explored the role of Nup98 in viral infection. We first verified that Nup98 is induced at the mRNA level and protein level on infection with VSV (Fig. 1 A and B). We then tested whether Nup98 was also induced by SINV infection by immunoblot analysis and found increased levels (Fig. 1C). Finally, we depleted Nup98 using RNAi in Drosophila cells and infected these cells with SINV that expresses a GFP reporter (25). Viral infection was monitored by fluorescence microscopy and quantitative image analysis. We found that Nup98-depleted cells were threefold to fourfold more susceptible to infection with SINV (Fig. 1 D and E). We also measured viral replication by immunoblot analysis and found significantly increased viral gene expression on depletion of Nup98 (Fig. 1F). We further validated our results using several nonoverlapping RNAi reagents and found that the protein levels of Nup98 were reduced by multiple reagents (Fig. S1A), and that this resulted in enhanced SINV infection (Fig. S1 B and C).
Fig. 1.
Nup98 is antiviral in vitro. (A) Drosophila DL1 cells were infected with VSV (10 MOI) and the level of Nup98 mRNA was examined by RT-qPCR. (B-C) DL1 cells were infected with VSV (B) or SINV (C) for 2 h, and Nup98 protein expression was examined by immunoblot. (D–F) DL1 cells were treated with dsRNA against control (Luc and GFP) or Nup98 and 72 h later infected with SINV-GFP (10 MOI). (D) At 42 hpi, cells were fixed and processed for microscopy and image analysis. Representative image is shown. (E) Quantification from three independent experiments. (F) At 42 hpi, cells were harvested and GFP expression was examined by immunoblot. Actin expression was examined as a loading control. (G–I) Drosophila DL1 cells were treated with dsRNA against control or Nup98 and 72 h later infected with VSV-GFP (5 MOI) for 40 h for microscopy. A representative image is shown in G, and quantification is presented in H. (I) VSV-GFP expression was examined by immunoblot analysis at 40 hpi; actin expression served as a loading control. (J and K) DL1 cells were treated with the indicated dsRNAs for 72 h and then infected with WNV (Kunjin) (10 MOI) for 48 h. Cells were fixed and processed for quantitative imaging. Representative images are shown in J, and percent infection is quantified in K. (L and M) DL1 cells depleted of Nup98 were infected with DCV for 20 h. Cells were processed for quantitative imaging. A representative image is shown in L, and percent infection quantified from three independent experiments is shown in M. The data represent at least three independent experiments and are presented as mean ± SD. *P < 0.05.
Nup98 Is Antiviral Against Disparate Viruses.
We next examined the role of Nup98 in viral infections against additional RNA viruses. First, we infected Nup98-depleted cells with VSV expressing GFP (26) and observed a threefold to fourfold increase in infection as measured by microscopy and immunoblot analyses (Fig. 1 G–I and Fig. S1 D and E). Second, we challenged Nup98-depleted cells with West Nile virus (strain Kunjin virus) and observed increased infection (Fig. 1 J and K). Because these viruses do not naturally infect Drosophila, but rather infect mosquitoes, we examined the role of Nup98 in the infection of a natural Drosophila pathogen, Drosophila C virus (DCV). We found that cells depleted for Nup98 were also more susceptible to DCV infection (Fig. 1 L and M).
To investigate whether all nuclear pore proteins have similar antiviral roles, we tested two additional Nups, Megator (Mtor) and GP210, and verified depletion with available antibodies (Fig. S1 I and M). We nex infected the Mtor- or GP210-depleted cells with SINV or VSV. We found that unlike Nup98 depletion, Mtor or GP210 depletion did not enhance SINV or VSV infection (Fig. S1 F–H and J–L). These data suggest that Nup98 plays a specific antiviral role and likely is not related to its function at the nuclear pore.
Nup98 Restricts Virus Infection in Adult Flies.
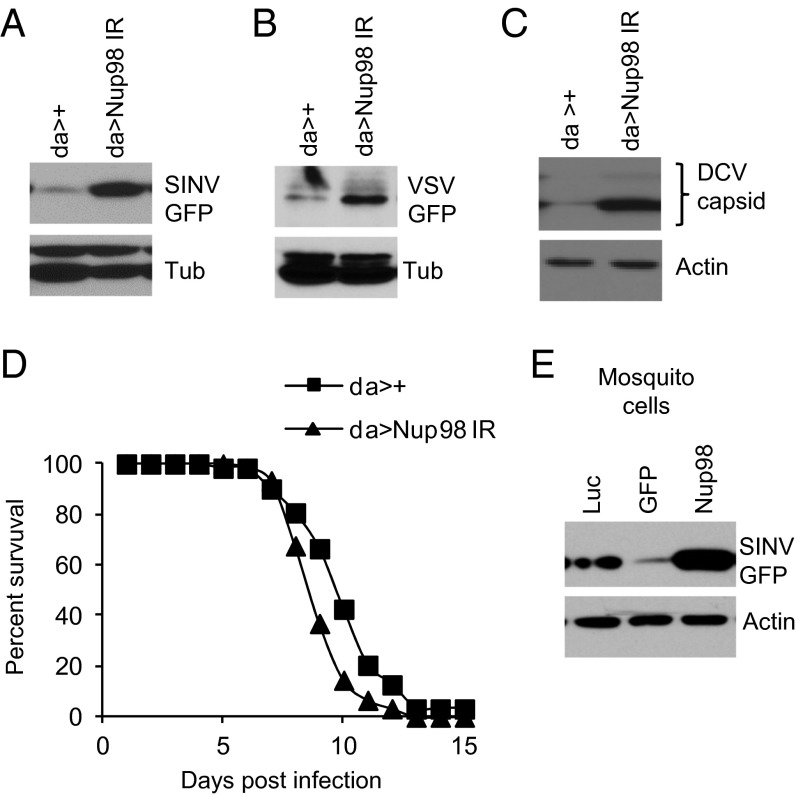
To determine whether Nup98 plays an antiviral role at the organismal level, we generated flies mutant for Nup98 using in vivo RNAi with a previously validated transgenic against Nup98 driven by a ubiquitous but low-level promoter [daughterless (da)] (19). We challenged the control flies (da >+) as well as Nup98-depleted flies (da > Nup98 IR) with SINV and monitored virus replication by immunoblot analysis. We observed increased levels of viral replication in Nup98-depleted flies (Fig. 2A). We next examined the role for Nup98 against VSV and DCV, two additional viruses that we found to be restricted in cell culture. Nup98 depletion led to increased VSV and DCV replication in flies (Fig. 2 B and C). Nup98-depleted flies not only demonstrated increased viral replication, but also succumbed to DCV infection earlier than control flies (P < 0.05) (Fig. 2D). Taken together, these results suggest a broad antiviral role for Nup98 in adult flies.
Fig. 2.
Nup98 is antiviral in adult flies. (A–C) Flies of the indicated genotypes were infected with SINV-GFP for 6 d (A), VSV-GFP for 6 d (B), or DCV for 4 d (C). The flies were then processed for immunoblot analysis. (D) Flies of the indicated genotypes were infected with DCV, and percent survival was examined (P < 0.01, log-rank test). (E) Mosquito Aag2 cells were treated with dsRNA against AENup98 (AAEL007586) for 72 h and then infected with SINV-GFP for 18 h. GFP expression was examined by immunoblot analysis.
Because mosquitoes, particularly Aedes aegypti, are the relevant vectors for numerous arboviruses, we examined whether Nup98 can restrict viral infection in mosquito cells. We used RNAi to deplete Nup98 in A. aegypti Aag2 cells and found significantly higher levels of SINV infection compared with control cells as determined by immunoblot analysis (Fig. 2E). This result suggests that Nup98 plays a conserved antiviral role in insects.
Nucleo-Cytoplasmic Transport Might Not Be Affected on Nup98 Depletion.
The nuclear pore complex, which is composed of ∼30 Nups, controls nucleo-cytoplasmic transport of diverse macromolecules. In a recent study using a genome-wide RNAi screen, Farny et al. (27) tested the requirements for each gene in poly(A) RNA transport and found that depletion of only three Nups (including Nup98) affected transport. In contrast, Sabri et al. (28) tested the role for Nups using GFP reporters tagged with either a nuclear localization signal or a nuclear export signal (NES) and found that depletion of only one Nup affected export, whereas depletion of three affected import (none of these include Nup98). Thus, at least during transient depletion, nuclear pore function seems to be maintained on depletion of many Nups, including Nup98.
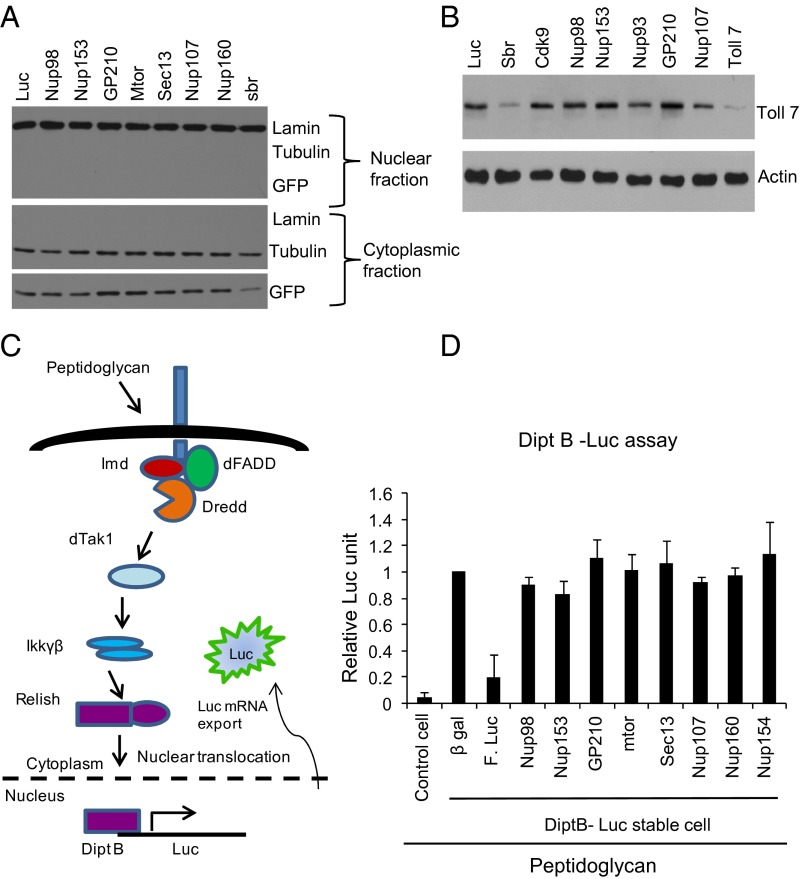
Nevertheless, we tested whether Nup98 depletion affects general nuclear pore-dependent transport. We first generated a Drosophila DL1 cell line stably expressing the NES-GFP reporter used previously in S2 cells and examined whether depletion of Nup98 or other Nups affected nuclear export (28). As a control, we depleted sbr (dNXF1), which is an essential export factor in metazoans (29, 30). Indeed, depletion of sbr led to decreased export of NES-GFP (Fig. 3A). As expected, we found that depletion of Nup98 and several other Nups did not alter the nuclear export of GFP (Fig. 3A). We complemented these studies by examining the expression of Toll-7, a membrane protein required for antiviral defense against VSV (31). Again we found that whereas depletion of the major nuclear export factor sbr resulted in reduced Toll-7 protein levels, depletion of Nup98 did not affect Toll-7 levels (Fig. 3B), suggesting that Toll-7 mRNA export might not be affected.
Fig. 3.
Transient depletion of individual Nups does not globally affect nuclear-cytoplasmic transport. (A) DL1 cells stably expressing a GFP-NES reporter were treated with the indicated dsRNAs and the nuclear and cytoplasmic distribution of the indicated proteins was examined by immunoblot analysis. (B) DL1 cells were treated with indicated dsRNAs for 72 h, and then endogenous Toll-7 expression was examined by immunoblot analysis. (C) Schematic representation of PGN-induced antimicrobial peptide expression reporter assay. (D) S2 cells stably expressing DiptB-Luc were treated with indicated dsRNA for 3 d and then treated with PGN for 6 h. Luciferase expression was measured. The data are mean ± SD from three independent experiments.
We next took advantage of a luciferase reporter assay that monitors both the import of a protein and export of an mRNA. On stimulation with peptidoglycan (PGN), NFκB (Rel) is activated and translocates through the nuclear pore into the nucleus, where it binds to the κB sites in the diptericin B (DiptB) promoter inducing the luciferase reporter (32, 33). This mRNA is then translocated into the cytoplasm through the nuclear pore, where it is translated and luciferase activity is monitored (Fig. 3C) (33). Using this system, we first validated that treatment with PGN induces luciferase and that knockdown of luciferase mRNA decreases the induced signal (Fig. 3D).
We next tested whether depletion of Nup98 or several additional Nups affects luciferase production. We found that depletion of these genes did not significantly reduce the induced luciferase activity, demonstrating that nuclear import of NFκB and export of the induced reporter mRNA are not affected by transient depletion of Nup98 (Fig. 3D). This finding is particularly important because it shows that other innate immune gene expression programs can be induced during infection in the context of Nup98 depletion.
Nup98 Occupancy at the Nuclear Pore Is Reduced on Virus Infection.
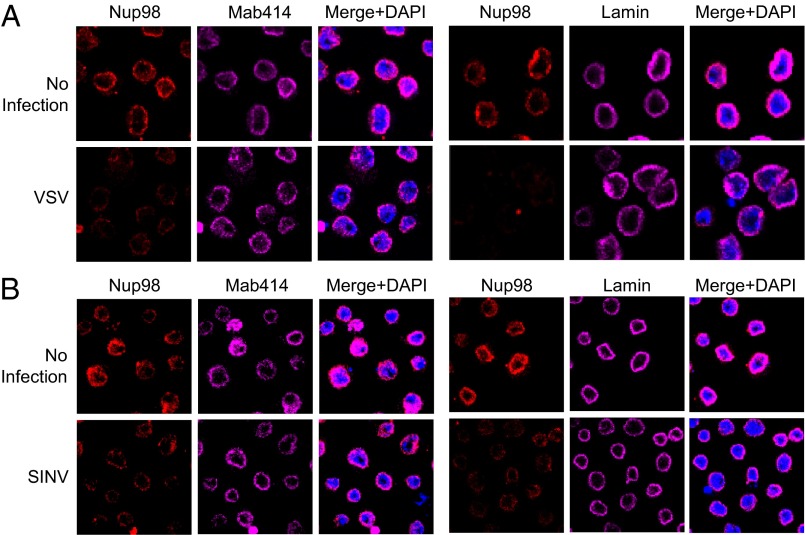
Studies have shown that Nup98 localizes predominantly to the nuclear pore complex, but has an additional population in the nucleoplasm and can exhibit altered nuclear distribution in response to some stimuli, such as developmental signaling or IFN-γ (19, 20, 22, 34). In our system, we found that Nup98 levels are increased on viral infection (Fig. 1 A–C); thus, we examined whether Nup98 localization is altered by viral infection. As expected, in control cells, endogenous Nup98 is localized predominantly to the nuclear periphery and colocalizes with Mab 414, a classical marker of the nuclear pore and recognizes several phenylalanine-glycine repeat Nups, as well as with lamin, which stains the nuclear envelope (Fig. 4A). However, within 4 h after VSV infection, the Nup98 signal at the nuclear envelope was reduced, whereas the patterns of Mab414 and lamin remained unchanged (Fig. 4A). We observed similar changes in Nup98 on infection with SINV (Fig. 4B). The increased total Nup98 protein levels at this time point suggests a change in localization.
Fig. 4.
Viral infection alters Nup98 localization. (A) DL1 cells were infected with VSV (20 MOI) and at 4 hpi, cells were fixed and processed for microscopy and probed with the indicated antibodies. (B) DL1 cells were infected with SINV (10 MOI) and at 4 hpi, probed with the indicated antibodies by microscopy. Representative images from three independent experiments are shown.
We next investigated the intracellular distribution of Nup98 by nucleo-cytoplasmic fractionation. We found that on infection, most of the Nup98 remained nuclear (Fig. S2), suggesting that although Nup98 intensity is reduced at the nuclear envelope, it is induced and present within the nucleus on infection. This may further suggest that on infection, the primary antiviral activity of Nup98 is executed independent of its nuclear pore localization.
Nup98 Is Required for Antiviral Gene Expression.
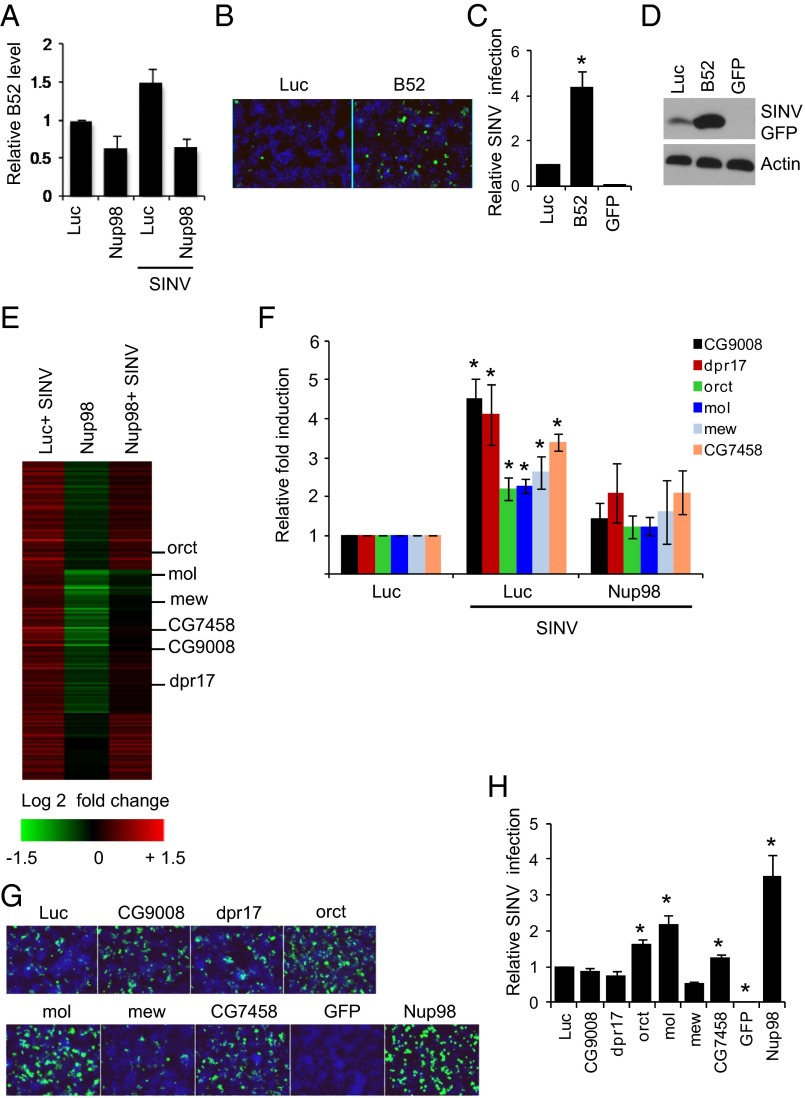
Recent studies have revealed a direct role for Nup98 in gene expression, particularly in the transcriptional regulation of developmental and immune genes (19, 20, 22, 35). Thus, we hypothesized that Nup98 may similarly regulate the expression of antiviral genes independent of its role in nucleo-cytoplasmic transport. As a first test of this hypothesis, we determined whether any of the 37 genes that we identified as antiviral against SINV in a genome-wide RNAi screen (36) were bound by Nup98, as measured by chromatin immunoprecipitation microarray (ChIP-chip) analysis in Drosophila cells (19). Only one gene, B52, was identified as both antiviral and bound by Nup98; thus, we hypothesized that B52 is transcriptionally regulated by Nup98. Indeed, we found that B52 was modestly induced by viral infection (Fig. 5A). Furthermore, both virus-induced and basal levels of B52 were Nup98-dependent, and Nup98 depletion led to reduced B52 mRNA levels (Fig. 5A). Although we had identified B52 as antiviral in our genome-wide RNAi screen, we had not validated this gene outside of the screening format. Therefore, we depleted B52 using independent RNAi reagents (Fig. S3A) and found enhanced SINV replication, as measured by microscopy (Fig. 5 B and C) and immunoblot analyses (Fig. 5D). We also found that in addition to SINV, B52-depleted Drosophila cells are more susceptible to infection with VSV and DCV (Fig. S3 B and C).
Fig. 5.
Nup98 is required for antiviral gene expression. (A) DL1 cells were depleted of Nup98 and then mock-infected or infected with SINV (MOI 20) for 2 h. B52 expression was examined by RT-qPCR. (B and C) DL1 cells were treated with dsRNA against B52 and 72 h later infected with SINV-GFP (MOI 5) for 42 h. Representative images of SINV-GFP–infected cells are shown in B, and data (mean ± SD) from three independent experiments are shown in C. *P < 0.05. (D) DL1 cells depleted of B52 were infected with SINV-GFP (MOI 5) for 40 h, and GFP expression was examined by immunoblot analysis. (E) Microarray analysis of SINV-induced Nup98-dependent genes. Column 1, comparison of uninfected and SINV-infected control cells; column 2, comparison of uninfected control cells and uninfected Nup98-depleted cells; column 3, comparison of Nup-98-depleted or control cells with SINV infection. Key shown below for log twofold change in expression. (F) RT-qPCR validation of genes identified in the microarray analysis. Data are mean ± SEM of four independent experiments. *P < 0.05. (G and H) DL1 cells were treated with indicated dsRNA and then infected with SINV for 44 h. Representative images are shown in G, and quantification of three independent experiments (mean ± SD) is shown in H. *P < 0.05.
We next examined whether B52 is antiviral in adult flies. We again used the Gal4/UAS system to perform in vivo RNAi against B52. We found that depletion of B52 led to increased SINV infection as measured by immunoblot analysis at two time points postinfection (Fig. S3D). Moreover, DCV infection in adult flies was increased on the loss of B52, concomitant with increased mortality (P < 0.05) (Fig. S3 E and F). These results suggest that B52 is broadly antiviral in Drosophila.
Because depletion of Nup98 had a stronger phenotype than loss of B52, we hypothesized that Nup98 may be controlling a larger set of antiviral genes during infection. Thus, we performed global gene expression profiling of control or Nup98-depleted Drosophila cells in the presence or absence of SINV infection at 2 h postinfection (hpi). We identified 169 genes that were significantly induced on viral infection, and found that 36 of these genes are Nup98-dependent for their expression on viral infection (Fig. 5E). When we analyzed the Gene Ontology (GO) molecular functions of these virally induced genes, we found significantly enriched transcription factor activity and kinase regulator activity (Fig. S4A). Furthermore, analysis of the GO cellular localization category revealed that these genes are enriched for plasma membrane localization and are integral membrane proteins (Fig. S4B). We next independently tested six virus-induced, Nup98-dependent genes from the microarray and verified that all six genes were virus-induced and Nup98-dependent for their induction (Fig. 5F).
None of these genes had any previously known role in viral infection. However, because many antiviral genes are transcriptionally induced by viral infection, we examined whether these genes are antiviral against SINV in cell culture. We first validated the depletion of these genes by RT-qPCR (Fig. S4 C–H). We then infected Drosophila cells depleted of these genes individually, and found that three of the six genes were modestly antiviral for SINV, but not for VSV (Fig. 5 G and H and Fig. S4I). Again, because Nup98 depletion had a stronger phenotype compared with the depletion of any one downstream target, these data suggest that Nup98 regulates the expression of a panel of antiviral genes that together restricts infection.
Nup98 Regulates Expression, but Not Localization, of Virus-Induced Genes.
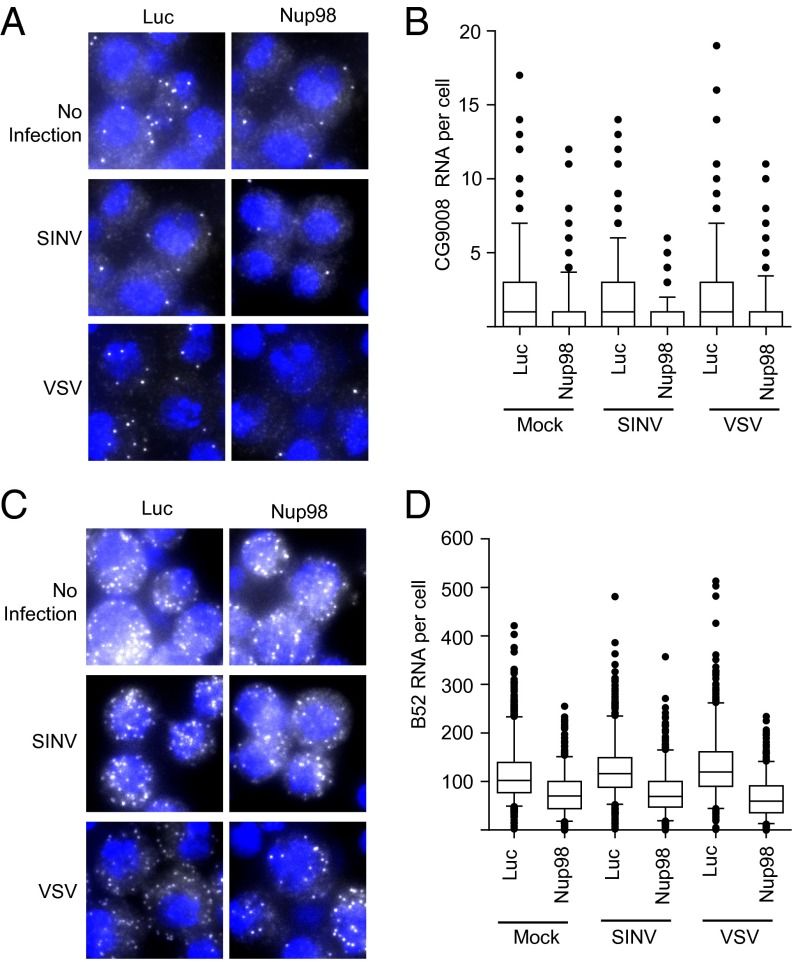
We further examined whether Nup98 affects the subcellular localization (mRNA export) or generation (transcription) of virus-induced Nup98-dependent mRNAs using single-molecule mRNA FISH. This technique can measure both the level and location of individual mRNAs. Using specific probes, we analyzed CG9008 and B52 mRNAs in control and Nup98-depleted cells in the presence or absence of infection, and found a reduced level of CG9008 mRNA on Nup98 knockdown (Fig. 6 A and B). Although the number of CG9008 mRNAs was low, we observed that the largely cytoplasmic distribution of CG9008 mRNA was unchanged on Nup98 knockdown (Fig. 6A). We also monitored B52 mRNA and found decreased levels on Nup98 depletion (Fig. 6 C and D). We also observed a modest increase in B52 RNA levels on VSV and SINV infection consistent with our RT-qPCR results. There was no apparent change in the localization of B52 mRNAs; the mRNAs were predominantly cytoplasmic both with and without Nup98 depletion. These data reveal that virus-induced Nup98-dependent genes are not dependent on Nup98 for their export to the cytoplasm, but that their levels depend on Nup98.
Fig. 6.
Nup98 regulates expression but not the localization of virally induced mRNAs. Control cells or Nup98-depleted cells were mock-infected or infected with SINV or VSV for 2 h and then processed for RNA FISH. (A and C) Localization of CG9008 (A) and B52 (C) mRNA. (B and D) Quantification of CG9008 (B) and B52 (D) mRNA. Data are shown as boxplots of >300 cells from duplicate experiments.
Nup98 Localizes to the Promoters of Virus-Induced Genes.
Nup98-dependent control of gene expression has been linked to its occupancy at target promoters (19, 20). Indeed, we chose B52 because Nup98 was found at the B52 promoter in ChIP-chip studies performed in Drosophila S2 cells (19). Thus, we examined whether Nup98 is directly bound to the promoters of two virus-induced genes, B52 and CG9008, using ChIP-qPCR analysis. As predicted, we found that Nup98 binds to the promoters of B52 and CG9008 (Fig. 7 A and B). This binding was increased, albeit modestly, on SINV infection. This binding signal was specific to Nup98, as demonstrated by the fact that depletion of Nup98 reduced the occupancy to a similar extent (approximately twofold) as the knockdown.
Fig. 7.
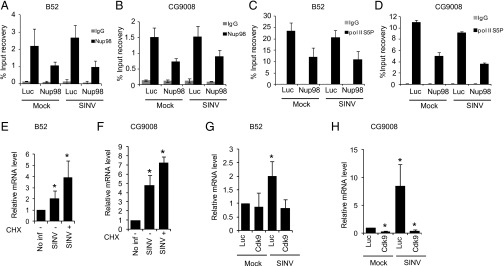
Nup98 binds to the promoter and promotes RNAPII occupancy of virus-induced Nup98-dependent genes. (A–D) Control cells or Nup98-depleted cells were either uninfected or infected with SINV for 2 h and then ChIP was performed using anti-Nup98 (A and B) or anti-RNAPII S5P (C and D) antibodies. The recovery of promoters relative to the input is shown for B52 (A and C) and for CG9008 (B and D). Data represent mean ± SD of four independent samples from two independent experiments. (E and F) Control or CHX-treated DL1 cells were infected with SINV, and gene expression was analyzed by RT-qPCR. Relative gene expression compared with uninfected control cells is shown. Data represent mean ± SD from three independent experiments. *P < 0.05. (G and H) Control DL1 cells or Cdk9-depleted cells were either mock-infected or infected with SINV. Gene expression was examined by RT-qPCR. Relative gene expression compared with control uninfected cells from three independent experiments is shown; data are mean ± SD. *P < 0.05.
These results suggest that Nup98 positively regulates gene activation at these promoters and thus may recruit or regulate RNAP II at these promoters. Therefore, we examined whether occupancy of the initiating form of RNAPII (S5P) was affected by the depletion of Nup98. We performed ChIP using the RNAP II S5P antibody (CTD4H8) and found that depletion of Nup98 led to a decrease in the RNAPII S5P present at the promoters of B52 and CG9008 (Fig. 7 C and D). These results suggest that Nup98 is required to recruit or maintain active RNAPII at the promoters of virus-induced Nup98-dependent genes, providing direct evidence for Nup98 in transcriptional activation of antiviral genes.
Many rapidly inducible genes are regulated in a translation-independent manner and thus are termed primary response genes (4). To determine whether the induction of B52 or CG9008 requires new protein synthesis, we examined their mRNA levels on viral infection in the presence or absence of the translation inhibitor cycloheximide (CHX). We found that virus-induced activation of both B52 and CG9008 was independent of new protein synthesis (Fig. 7 E and F). We expanded this analysis to the five additional virus-induced Nup98-dependent genes that we validated, and found that they were all translation-independent (Fig. S5 A–E). We next examined whether Nup98 controls this response and found that Nup98-dependent regulation of virus-induced gene activation is independent of translation (Fig. S5 F and G).
In a previous study, we observed that half of the virus-induced genes are regulated by P-TEFb (14), and that a subset of the primary response genes have RNAPII S5P occupancy at their promoters (4). Here we observed Nup98 occupancy and RNAPII S5P at the promoters before gene induction, indicative of a transcriptionally paused locus. Thus, we hypothesized that Nup98-dependent genes are pausing-regulated. To examine this, we depleted cyclin-dependent kinase 9 (Cdk9), the catalytic subunit of P-TEFb, and determined the expression levels of these seven virus-induced Nup98-dependent genes. We found that expression of all seven genes was dependent on Cdk9, because Cdk9 depletion attenuated the virus-induced expression of these genes (Fig. 7 G and H and Fig. S5 H–L). Taken together, these results provide a link between Nup98-dependent gene regulation and P-TEFb-dependent gene induction in the regulation of antiviral genes. Our findings suggest that Nup98 and transcriptional pausing machinery concertedly regulate transcriptional activation of antiviral gene expression and host defense.
Discussion
Here we describe a previously unknown role for Nup98 in regulating antiviral gene expression. Using Drosophila as a model system, we found that Nup98 is required for antiviral defense against disparate viruses including human arboviruses. We also found that Nup98 is required for the regulation of a subset of virus-induced antiviral genes. Mechanistically, we found that Nup98 binds to the promoters of these genes and promotes RNAPII S5P occupancy at these promoters, poising them for P-TEFb–dependent activation on infection.
The classically described role for Nups is within the nuclear pore complex, with specific functions in the transport of macromolecules in and out of the nucleus (37–39); however, recent studies have found both on-pore and off-pore roles for a subset of Nups (19, 20). Using two independent assays, we found no defect in nuclear export or import on depletion of Nup98 or several other nuclear pore proteins. This finding is consistent with two recent studies that found no role for Nup98 in nuclear transport when Nup98 was compromised (22, 28). In addition, using single-molecule RNA FISH, we detected no defect in nuclear export of B52 or CG9008 mRNA on depletion of Nup98. Nup98 may regulate the transport of additional antiviral genes, but nevertheless our results suggest that a subset of antiviral genes is regulated by Nup98 at the transcriptional level. Altogether, this suggests an off-pore transport-independent role for Nup98 in antiviral defense.
Viral infection leads to the rapid induction of an antiviral transcriptional response (14, 15, 40, 41). In metazoans, RNAPII recruitment and activation are known to regulate signal-dependent gene transcription (9, 42, 43); however, the factors involved in recruiting and stabilizing RNAPII at the promoter in a context- and gene-specific manner in response to diverse stimuli, including viral infection, are unclear. We demonstrate that Nup98 depletion reduces the level of RNAPII S5P at the promoters of virally induced genes and, consequently, the level of transcripts. This suggests that Nup98 either promotes recruitment of RNAPII or maintains the initiating form of RNAPII at the promoters to facilitate transcription. Along with identifying this Nup98-dependent pathway in antiviral defense, our findings also shed light on the mechanistic involvement of Nups in transcription. Although several studies have identified a functional role for Nups in transcriptional activation (19–21, 35, 44), the detailed mechanism behind this role has not been fully deciphered. Our results suggest a specific step in the transcriptional process, RNAPII S5 activity at the target gene promoter, which is regulated by Nup98.
We recently showed that transcriptional pausing regulates one-half of the virus-induced genes in Drosophila, which is significantly enriched compared with the genome as a whole (14). Transcriptionally paused genes have high occupancy of RNAPII S5P, are primary response genes, and are dependent on P-TEFb for their induction (4). In this study, we found that the seven virus-induced Nup98-dependent genes that we validated are primary response genes; there are high levels of RNAPII S5P at the promoters basally, and they are dependent on P-TEFb for their induction. This suggests that Nup98 maintains active RNAPII at these genes, keeping them poised for future activation by P-TEFb. In the absence of Nup98, along with the loss of RNAPII S5P, these antiviral genes are no longer efficiently induced, resulting in elevated levels of viral infection. This model is consistent with the recent results demonstrating recruitment of Nup98 to the promoter of developmentally regulated genes independent of transcription elongation, because Nup98 recruitment is insensitive to flavopiridol treatment (19), and that many developmental genes are known to be regulated at the level of pausing (8, 37). Consistent with the hypothesis that Nup98 is a specific transcription activator, it also has been demonstrated that Nup98 interacts with histone-modifying enzymes CBP/p300 and histone deacetylases (45, 46).
Given that Nup98 regulates gene expression of developmental genes in mammals and Drosophila (19, 20, 35) and we observed regulation of antiviral genes in Drosophila, we suggest that Nup98 may have functions in regulating cell intrinsic antiviral gene expression in mammalian systems. Indeed, it has long been recognized that Nup98 is involved in antiviral defense (34, 47–49). Nup98 depletion reduces the nuclear export of specific immune regulated genes, including IFN-stimulated genes (47). Furthermore, viral infection was enhanced in Nup98-deficient MEFs (47). These antiviral phenotypes were ascribed to transport defects; however, it has not been explored whether Nup98 also directly regulates the induction of antiviral primary response genes. Interestingly, a recent study found that Nup98 impacts the regulation of IFN-γ–responsive genes (22). In this study, Nup98 was required to set the reactivation state of IFN-γ–responsive genes without affecting the initial activation (22). Mechanistically, they found that Nup98 binds the promoter of IFN-γ–responsive genes and is required for the maintenance of histone H3K4 dimethylation during transcriptional memory (22). Whether Nup98 affects the transcriptional memory of immune-regulated genes in Drosophila remains to be seen.
The signal-dependent antiviral response is tightly regulated to induce the appropriate immune response. Our results shed light on how virus-specific gene regulation is controlled. This role for Nup98 in the regulation of a subset of antiviral genes unravels one additional layer in this complex control of innate immune gene expression programs. Given our finding that Nup98 is induced, it is possible that the newly synthesized Nup98 regulates a robust secondary response or alters the memory of these loci, as has been observed for IFN-γ–responsive genes. Future work examining the role of Nup98 in other immune contexts in insects and in viral infection in mammals will lead to better understanding of the transcriptional regulation of immune system and may help develop better therapeutic interventions against viral diseases.
Materials and Methods
Cells, Viruses, and Reagents.
DL1, SL2, and Aag2 cells were maintained as described previously (50). SINV-GFP was propagated in C6/36 cells (25), VSV-GFP was propagated in BHK-21 cells (26), WNV (Kunjin) was propagated in BHK21 cells (51), and DCV was propagated in DL2 cells (52). Viral titers for multiplicity of infection (MOI) calculations were determined in BHK-21 cells. The following antibodies were used: anti-DCV capsid (52), anti-WNV Ns1 (51), anti-Nup98 for immunofluorescence and ChIP (19), anti-Nup98 for immunoblot analysis (a gift from Cordula Schulz, University of Georgia, Athens, GA) (53), anti-RNAPII S5P (CTD4H8; Milipore), control IgGs (Santa Cruz Biotechnology), anti-GFP (Santa Cruz Biotechnology), anti-actin (Santa Cruz Biotechnology); anti-tubulin (Sigma-Aldrich); anti-lamin (Developmental Studies Hybridoma Bank); HRP-conjugated secondary antibodies (Amersham), and Alexa Fluor-conjugated secondary antibodies (Life Technologies). The following reagents were used: Firefly luciferase britelite (Perkin-Elmer); protein G magnetic beads (Life Technologies); and all other chemicals were obtained from Sigma-Aldrich.
RNAi and Virus Infection.
RNAi infection was performed as described previously (50). Drosophila DL1 cells in 96- or 384-well plates were infected with SINV at an MOI of 20 for 42 h. In 12-well plates, DL1 cells were infected with SINV at an MOI of 5 for 24–36 h. DL1 cells were infected with VSV at an MOI of 5 for 24–26 h. Aag2 cells were infected with SINV at an MOI of 3 for 16 h. DL1 cells were infected with DCV to achieve a 10–20% infection. DL1 cells were infected with WNV (Kunjin) at 1 MOI for 48 h.
Immunoblot Analysis.
DL1 cells were harvested in RIPA buffer and processed for immunoblot analysis. Total protein was determined by the Bradford protein assay, and equal amounts were separated on a SDS/PAGE gel. Representative experiments from at least three replicates are shown in the figures.
Adult Fly Infections.
Nup98 RNAi flies were reported previously (19). B52 RNAi transgenics were from the Vienna Drosophila Resource Center. These were crossed to daGAL4, and the indicated genotypes of the adult progeny (4–7 d old) were used for infections as described previously (52).
Fluorescence Microscopy.
For high-magnification studies, DL1 cells were infected with VSV-GFP or SINV-GFP for 4 h. Cells were replated on glass coverslips and then fixed with 4% formaldehyde, permeabilized for 10 min with 0.1% Triton X-100, and stained with the indicated antibodies for 16 h at 4 °C. Images were captured with a Leica DMI 5000 confocal microscope using a 63× objective. Representative images from at least three independent experiments are shown. Percent infection analysis was done by automated imaging and image analysis as described previously, using ImageXpress Micro and Metaxpress software (36).
DiptB Luciferase Assay.
S2 cells stably expressing a firefly luciferase reporter downstream of the DiptB promoter (33) were transfected with dsRNA using calcium phosphate and then incubated for 24 h. These cells were treated with ecdysone (10 μg/uL) for 24 h, then with PGN for 6 h, and monitored for luciferase activity.
Cellular Fractionation.
DL1 cells were resuspended in cytoplasmic lysis buffer (30 mM Hepes, 2 mM magnesium acetate, 0.1% Nonidet P-40, 5 mM DTT, protease inhibitors, and PMSF). Cells were lysed by passage through a 30-G needle and centrifuged at 1,000 × g for 5 min to obtain the cytoplasmic supernatant. The nuclear pellet was washed twice with wash buffer (30 mM Hepes, 2 mM magnesium acetate, 0.1% Nonidet P-40, 5 mM DTT, and PMSF), followed by lysing in RIPA buffer, centrifugation at 15,000 × g for 15 min, and collection of nuclear supernatant.
Microarray Analysis.
For expression profiling on Affymetrix Drosophila GeneChip microarrays (Affymetrix), Drosophila cells were treated with Luc or Nup98 dsRNA and then either uninfected or infected with SINV (MOI of 20) for 2 h, in two independent biological replicates. Total RNA was isolated by TRIzol, and microarray experiments were performed at the University of Pennsylvania Microarray Facility following the manufacturer’s protocol (Affymetrix). Arrays were analyzed using the Affy (54) and limma packages (55) for R (R Foundation for Statistical Computing) and Microsoft Excel. Fold changes relative to uninfected controls were calculated, and genes with a false discovery rate (FDR) of <0.01 were considered significant. For Nup98 dependence, fold changes relative to control samples were calculated, and genes with 1.5-fold down-regulation and an FDR of <0.1 were considered significant. MeV software was used for hierarchical clustering and visualization.
Bioinformatics Analysis.
GO analysis was performed using the DAVID tool (http://david.abcc.ncifcrf.gov/).
RT-qPCR.
RNA was extracted using TRIzol and reverse-transcribed using MMLV RT. For qPCR, cDNA was subjected to PCR using SYBR Green and analyzed by the ΔΔCT method, with normalization to Rp49. Data are presented as relative mRNA expression compared with the control samples and are displayed as mean ± SD values for at least three independent experiments.
RNA FISH.
Single-molecule RNA FISH was performed following the Stellaris RNA FISH protocol (56, 57). We labeled oligonucleotide pools (Biosearch Technologies) of 26 oligonucleotides against B52 with Alexa Fluor 594 and 29 oligonucleotides against CG9008 with Atto 647N. The oligonucleotides were desgined using the online Stellaris probe design software (56). All images were acquired using a Nikon Ti-E widefield microscope with a 100× 1.4 NA objective and a Pixis 1024BR CCD camera. mRNA in each cell was quantified using custom image processing scripts written in MATLAB (56).
ChIP.
ChIP experiments were carried out as described here with some minor modifications (http://www.epigenesys.eu/en/protocols/chromatin-immunoprecipitation-chip Prot 48). In brief, DL1 cultures were cross-linked with 1% formaldehyde for 10 min. Nuclei were isolated with swelling buffer (25 mM Hepes, 1.5 mM MgCl2, 10 mM, KCl, 0.2% Igepal, and protease inhibitor mixture) using a Dounce homogenizer with a tight pestle (10–15 strokes). The nuclei were resuspended in sonication buffer (50 mM Hepes pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and 0.1% SDS), and the chromatin was fragmented into <500 bp using a bioruptor (Diagenode). For each sample, 500 μg of chromatin was immunoprecipitated overnight at 4 °C using rabbit anti-Nup98 antibody (15 μL), rabbit anti-IgG (40 μL), mouse anti-RNAPII Ser5P (10 μL), or mouse anti-IgG (40 μL) as a control and 40 μL of protein G magnetic beads. Beads were washed for 5 min at 4 °C with 1 mL of each of the following wash buffers: four times with sonication buffer, once with wash buffer B (20 mM Tris pH 8.0, 250 mM LiCl, 0.5% Igepal, 0.5% Na-deoxycholate, and 1 mM EDTA), and once with TE buffer (10 mM Tris pH 8.0 and 1 mM EDTA). The DNA was eluted; cross-linking was reversed, and DNA was recovered by phenol-chloroform extraction and ethanol precipitation. RT-qPCR analysis of the recovered DNA was performed, and the percentage of the DNA recovery from duplicate samples from two independent experiments after ChIP was plotted and compared with the amount of input material.
Statistical Analysis.
The Student t test was performed in each individual experiment. Experiments were performed at least three times. A P value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank C. Schulz for Nup98 antibody, N. Silverman for the DiptB stable cell line, N. Sabri for GFP-NES plasmid, and the Bloomington Stock Center and Vienna Drosophila Resource Center for fly stocks. We thank members of the S.C. laboratory for helpful discussions and advice. This work was supported by National Institutes of Health Grants R01AI074951, U54AI057168, R21AI103441, and R01AI095500 (to S.C.). S.C. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410087111/-/DCSupplemental.
References
- 1.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Cherry S. Viruses and antiviral immunity in Drosophila. Dev Comp Immunol. 2014;42(1):67–84. doi: 10.1016/j.dci.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138(1):129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140(6):833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci USA. 2009;106(43):18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: Promoter-proximal pausing and beyond. Biochim Biophys Acta. 2013;1829(1):98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Gaertner B, Zeitlinger J. RNA polymerase II pausing during development. Development. 2014;141(6):1179–1183. doi: 10.1242/dev.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13(10):720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133(4):581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 11.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Lenasi T, Barboric M. P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 2010;7(2):145–150. doi: 10.4161/rna.7.2.11057. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, et al. Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe. 2012;12(4):531–543. doi: 10.1016/j.chom.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemp C, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. 2013;190(2):650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6(11):1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 17.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13(4):1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15(4):1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capelson M, et al. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140(3):372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140(3):360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Vaquerizas JM, et al. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6(2):e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Light WH, et al. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11(3):e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145(4):502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Reports. 2013;4(5):959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367(1):212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Ramsburg E, et al. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J Virol. 2005;79(24):15043–15053. doi: 10.1128/JVI.79.24.15043-15053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farny NG, Hurt JA, Silver PA. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22(1):66–78. doi: 10.1101/gad.1616008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabri N, et al. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol. 2007;178(4):557–565. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7(12):1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 30.Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276(23):20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 31.Nakamoto M, et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36(4):658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann JA, Reichhart JM. Drosophila innate immunity: An evolutionary perspective. Nat Immunol. 2002;3(2):121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 33.Flatt T, et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol. 2008;211(Pt 16):2712–2724. doi: 10.1242/jeb.014878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enninga J, Levy DE, Blobel G, Fontoura BM. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295(5559):1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- 35.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9(2):e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panda D, et al. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Reports. 2013;5(6):1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raices M, D’Angelo MA. Nuclear pore complex composition: A new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13(11):687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 38.Ptak C, Aitchison JD, Wozniak RW. The multifunctional nuclear pore complex: A platform for controlling gene expression. Curr Opin Cell Biol. 2014;28:46–53. doi: 10.1016/j.ceb.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Nucleoporin Nup98: A gatekeeper in the eukaryotic kingdoms. Genes Cells. 2010;15(7):661–669. doi: 10.1111/j.1365-2443.2010.01415.x. [DOI] [PubMed] [Google Scholar]
- 40.Dostert C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat Immunol. 2005;6(9):946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 41.Mudiganti U, Hernandez R, Brown DT. Insect response to alphavirus infection—establishment of alphavirus persistence in insect cells involves inhibition of viral polyprotein cleavage. Virus Res. 2010;150(1-2):73–84. doi: 10.1016/j.virusres.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev. 2011;21(2):231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taddei A, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441(7094):774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 45.Kasper LH, et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19(1):764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai XT, et al. Trans-repressive effect of NUP98-PMX1 on PMX1-regulated c-FOS gene through recruitment of histone deacetylase 1 by FG repeats. Cancer Res. 2006;66(9):4584–4590. doi: 10.1158/0008-5472.CAN-05-3101. [DOI] [PubMed] [Google Scholar]
- 47.Satterly N, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci USA. 2007;104(6):1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Huang S, Chen Z. Human cellular protein nucleoporin hNup98 interacts with influenza A virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J Gen Virol. 2010;91(Pt 10):2474–2484. doi: 10.1099/vir.0.022681-0. [DOI] [PubMed] [Google Scholar]
- 49.von Kobbe C, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6(5):1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 50.Rose PP, et al. Natural resistance-associated macrophage protein is a cellular receptor for Sindbis virus in both insect and mammalian hosts. Cell Host Microbe. 2011;10(2):97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanna SL, et al. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79(21):13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5(1):81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parrott BB, et al. Nucleoporin98-96 function is required for transit amplification divisions in the germ line of Drosophila melanogaster. PLoS ONE. 2011;6(9):e25087. doi: 10.1371/journal.pone.0025087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 56.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27(11):1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.