Significance
Microbial specific pathogen-associated molecular patterns (PAMPs) constitute a key feature by which a host organism detects the presence of microbes and mounts specific immune responses. Here, we report the discovery of two proteins (bjALP1 and 2) that interact with muramyl dipeptide, a pan-bacterial specific PAMP via a novel pattern recognition domain ApeC. Our studies have revealed that bjALP1 is a secreted immune effector, whereas bjALP2 functions as an intracellular pattern recognition receptor (PRR), both having an important role in protecting the host from microbial pathogens. Specifically, bjAPL1 functions in the extracellular space to reduce the harmful effect of pathogenic microbes, whereas bjALP2 functions as a PRR that serves as a sentinel for intracellular bacterial invasion.
Keywords: pattern recognition receptor, immune response, NF-κB signaling pathway
Abstract
Animals exploit different germ-line-encoded proteins with various domain structures to detect the signature molecules of pathogenic microbes. These molecules are known as pathogen-associated molecular patterns (PAMPs), and the host proteins that react with PAMPs are called pattern recognition proteins (PRPs). Here, we present a novel type of protein domain structure capable of binding to bacterial peptidoglycan (PGN) and the minimal PGN motif muramyl dipeptide (MDP). This domain is designated as apextrin C-terminal domain (ApeC), and its presence was confirmed in several invertebrate phyla and subphyla. Two apextrin-like proteins (ALP1 and ALP2) were identified in a basal chordate, the Japanese amphioxus Branchiostoma japonicum (bj). bjALP1 is a mucosal effector secreted into the gut lumen to agglutinate the Gram-positive bacterium Staphylococcus aureus via PGN binding. Neutralization of secreted bjALP1 by anti-bjALP1 monoclonal antibodies caused serious damage to the gut epithelium and rapid death of the animals after bacterial infection. bjALP2 is an intracellular PGN sensor that binds to TNF receptor-associated factor 6 (TRAF6) and prevents TRAF6 from self-ubiquitination and hence from NF-κB activation. MDP was found to compete with TRAF6 for bjALP2, which released TRAF6 to activate the NF-κB pathway. BjALP1 and bjALP2 therefore play distinct and complementary functions in amphioxus gut mucosal immunity. In conclusion, discovery of the ApeC domain and the functional analyses of amphioxus ALP1 and ALP2 allowed us to define a previously undocumented type of PRP that is represented across different animal phyla.
The innate immune system plays an essential role in the first line of host defense against pathogenic microbes. Microbes express signature molecules known as pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS) in Gram-negative bacteria, peptidoglycan (PGN) and lipoteichoic acid (LTA) in Gram-positive bacteria, mannan in yeast, double-stranded RNA in viruses, and unmethylated CpG motifs in bacteria, which are essential for survival and hence difficult for microbes to change via adaptive evolution (1, 2). The innate immune system relies on germ-line-encoded pattern recognition proteins (PRPs) to detect and react with PAMPs. A variety of PRPs based on different protein domain structures with distinct PAMP specificities have been discovered in animals, including Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), RIG-I–like receptors (RLRs), scavenger receptors, C-type lectin receptors (CLRs), peptidoglycan recognition proteins (PGRPs), Gram-negative bacteria-binding proteins (GNBPs), siglecs, beta2-integrins, collectins, and ficolins (3–7). Some PRP families serve as dedicated sensors (e.g., TLRs and RLRs), and the others are exclusive effectors (e.g., collectins and ficolins), or both (e.g., CLRs and PGRPs). Sensor PRPs can be intracellular sensors (e.g., NLRs and RLRs), extracellular sensors (e.g., PGRPs and GNBPs), or both (e.g., TLRs) (3–7). Some PAMPs are recognized outside the cells by cell-surface receptors, whereas others are recognized intracellularly after the PAMPs have entered the cell. For example, LPS is recognized by cell-surface receptor TLR4, and muramyl dipeptide (MDP) is recognized by NOD2 in the intracellular space (1, 2).
Pathogen recognition by sensor PRPs may initiate coordinated immune responses at the molecular, cellular, tissue, and organ levels (8). The NF-κB family of transcription factors plays a central role in regulating the responses triggered by many PRPs, including TLRs, NLRs, and RLRs (9, 10). TNF receptor-associated factor 6 (TRAF6) is a crucial intermediate signal transducer for activating NF-κB in response to the signaling of TLRs, NLRs, RLRs, and TNF receptors (11, 12). During signaling, TRAF6 is activated by upstream adaptors [e.g., Myeloid differentiation primary response gene (88) (MYD88) and Toll-like receptor adaptor molecules] and kinases (e.g., IL-1 receptor-associated kinases and receptor-interacting serine-threonine kinases) (10, 11) and acts as an E3 ubiquitin ligase to catalyze Lys-63–linked polyubiquitination on itself as well as on IκB kinase γ (13). TGF-beta–activated kinase 1 (TAK1) and its binding proteins TAB2 and TAB3 are then recruited to TRAF6; TAK1 then activates downstream IκB kinases, ultimately leading to NF-κB activation (14, 15). TRAF6-depedent NF-κB activation is not only important to the vertebrate immune system but is also indispensable for the Toll and Imd antimicrobial signaling pathways in Drosophila (16, 17).
Amphioxus represents the basal living chordate lineage that diverged from two other lineages (urochordates and vertebrates) ∼550 Mya (18). This chordate invertebrate underwent no whole-genome duplications, shares extensive genomic conservation with vertebrates, and is one of the best proxies for understanding the chordate ancestral immune gene system (19–22). Amphioxus has not be found to contain molecular hallmarks of adaptive immunity, including antigen receptors and MHC molecules, but it has a much stronger innate immune gene system, which contains nearly 10 times more receptor and mediator genes than in humans and is tightly regulated during immune responses (22, 23). To date, several amphioxus PRPs have been verified as antimicrobial effectors, including a CLR, a ficolin, and a C1q-like protein (24–26). Functional analyses show that amphioxus also relies on the conserved TLR–MyD88–NF-κB and TRAF6–NF–κB/IκB pathways to regulate immune responses (27–29).
Here, we report two proteins from the amphioxus Branchiostoma japonicum (bj) that contain the ApeC domain, a novel pattern recognition domain for MDP, a bacterial-specific PAMP. The proteins share high similarity with the C-terminal regions of the apextrin proteins that were first identified in sea urchins (30, 31). We designate this domain apextrin C-terminal domain (ApeC) and named them apextrin-like protein 1 and 2 (bjALP1 and bjALP2). Functional analyses showed that bjALP1 acted as an extracellular effector for bacterial agglutination, whereas bjALP2 could mediate intracellular bacterial recognition and NF-κB activation. We therefore demonstrate that these ApeC-containing proteins represent a previously unidentified family of immune response proteins that have important roles in antimicrobial defense.
Results
ALPs and the Novel ApeC Domain.
In this study, we cloned two full-length cDNA sequences from amphioxus B. japonicum (bj), encoding a 504-aa protein (bjALP1) and a 462-aa protein (bjALP2). The two proteins share 54% sequence identity and both consist of a signal peptide, an N-terminal spacer region, and a C-terminal region of ∼200 aa (Figs. S1 and S2). A database search revealed that putative ApeC-containing proteins were presence in many invertebrate species, but no clear or unambiguous homologs were found in Caenorhabditis elegans, fruit flies, urochordates, or vertebrates (Fig. S3A). Thus, it seems that bjALP1 and 2 are membersof a group of ApeC-containing proteins that are uniquely prevalent in the invertebrate phyla. Phylogenic analysis shows that there are no cross-(sub)phylum orthologs found between ALPs (Fig. S3B).
Gene Expression Patterns of bjALP1 and bjALP2.
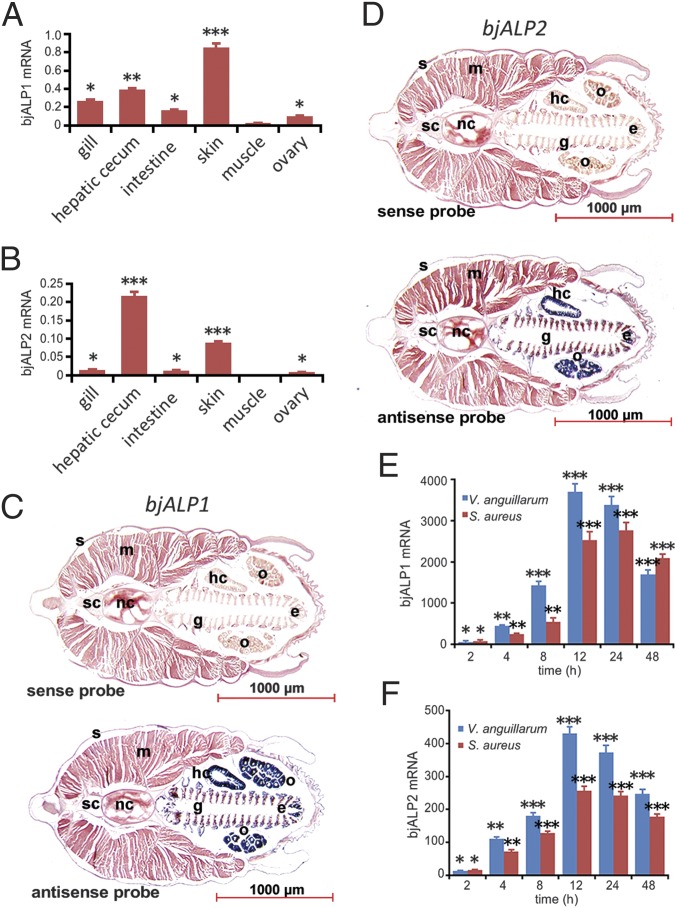
A quantitative real-time PCR (qRT-PCR) analysis showed that in adult animals bjALP1 mRNA was more highly expressed than bjALP2 in all six tissues examined, including gill, hepatic cecum, intestine, skin, muscle, and ovary (Fig. 1 A and B). Both genes showed the highest expression in the hepatic cecum and skin, whereas bjALP1 also had substantial expression in the gill and intestine. Section in situ hybridization confirmed the mRNA distribution of both genes in the digestive tract, skin, and ovary and the higher expression level of bjALP1 than of bjALP2 (Fig. 1 C and D). We further found that in the gut of adult animals the mRNA expression of both ALP genes was highly unregulated during acute antibacterial responses. BjALP1 showed an astounding 3,700-fold induction at 12 h after Gram-negative Vibrio anguillarum infection and 2,759-fold induction at 24 h after Gram-positive Staphylococcus aureus infection, whereas the peak up-regulation of bjALP2 was approximately 10 times weaker than that of bjALP1 (Fig. 1 E and F). These data suggest that bjALP1 is more strongly expressed and induced compared with bjALP2 and that both genes might be involved in gut mucosal immunity.
Fig. 1.
Expression patterns of amphioxus ALPs. (A and B) Expression of bjALP1 and bjALP2 mRNA in different tissues was determined by qRT-PCR analysis. The data are expressed as a ratio to the mRNA expression level of β-actin and are plotted as the means ± SD. (C) In situ hybridization of bjALP1 in adult female sections. (D) In situ hybridization of bjALP2 in adult female sections; the sense probe was used as the negative control. e, endostyle; g, gill; hc, hepatic cecum; i, intestine; m, muscle; nc, notochord; o, ovary; s, skin; sc, spinal cord. (E and F) The relative expression level of ALP mRNA in hepatic ceca and intestine after challenge with V. anguillarum and S. aureus was determined by qRT-PCR analysis. The data are shown as a ratio to the ALP mRNA expression level after the injection of PBS and are plotted as the means ± SD; *P < 0.05, **P < 0.01, ***P < 0.001.
Bacterial Agglutination Mediated by bjALP1 and bjALP2.
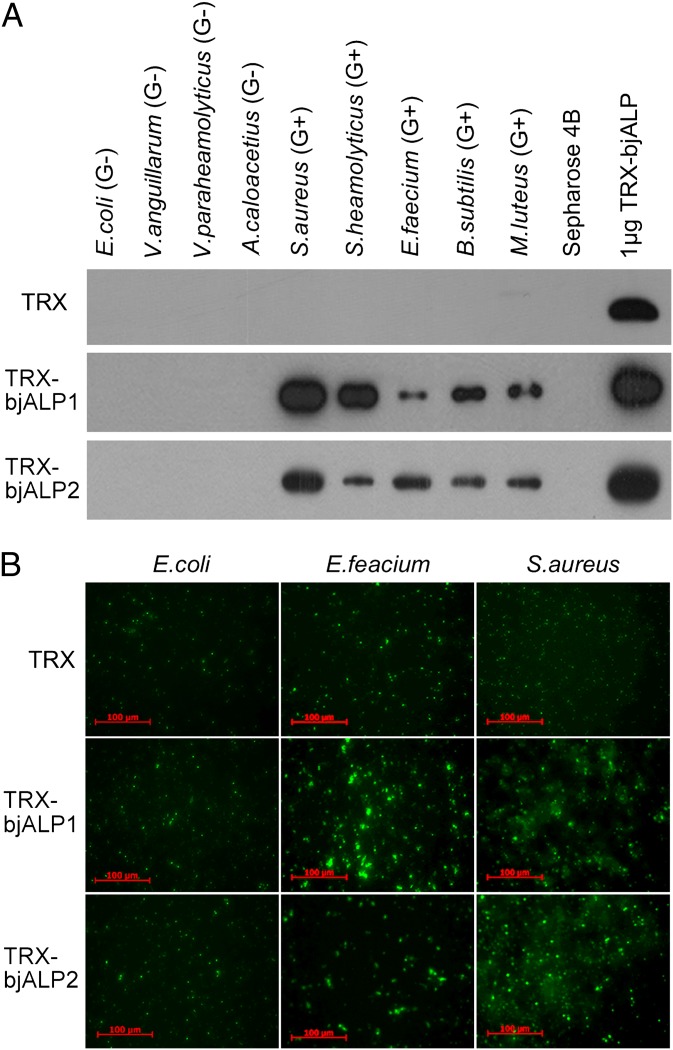
This extremely high expression and induction during infection led us to suspect that bjALP1 encodes a secreted mucosal effector. We purified recombinant His-tagged bjALP1 and bjALP2 proteins and incubated them with 10 different bacterial strains. After incubation, the bacterial pellets were washed and analyzed by Western blotting with anti-His monoclonal antibodies, which showed that both recombinant proteins could bind to Gram-positive bacteria with different affinities but not to Gram-negative bacteria (Fig. 2A). We then incubated the bjALP1 and bjALP2 recombinant proteins with FITC-labeled bacteria to determine the bacterial aggregation effect. The results showed that the recombinant proteins caused significant aggregation of the Gram-positive bacteria Enterococcus faecium and S. aureus, but not Gram-negative E. coli (Fig. 2B). Quantification of the diameter of green bacterial puncta suggested that the aggregation effect was stronger for S. aureus than for E. faecium (Fig. S4A). Intriguingly, however, we could not detect a significant bactericidal activity from recombinant bjALP1 (Fig. S4 B and C).
Fig. 2.
Amphioxus ALPs bind and aggregate microbes. (A) The binding of microorganisms by recombinant ALP proteins. Living microbial strains were incubated with ALP proteins in PBS buffer, and the washed pellets were subjected to SDS/PAGE and detection by Western blotting with an anti-His antibody. (B) Aggregation of microbes by amphioxus ALPs. The targeted proteins were incubated with FITC-labeled E. faecium, S. aureus, or E. coli (1 × 107 cfu/mL) in PBS for 1 h at room temperature and were examined using fluorescence microscopy.
bjALP1 and bjALP2 Bind PGN Through the ApeC Domain.
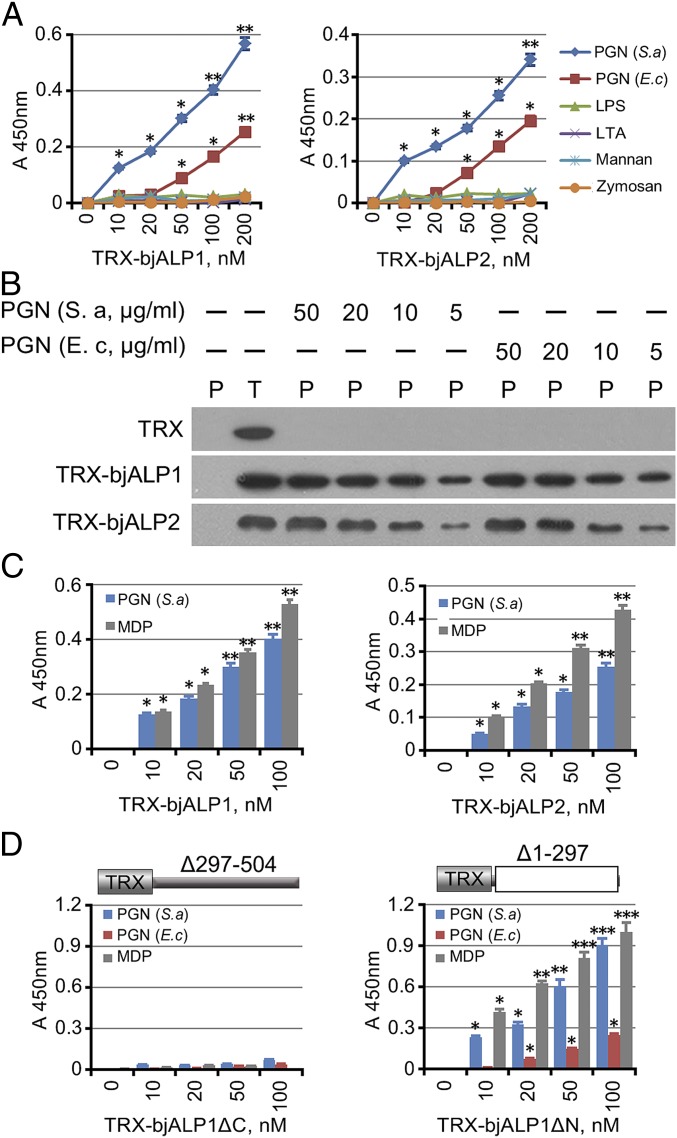
We used enzyme-linked immunosorbent assay (ELISA) to evaluate the interaction between the bjALP1 and bjALP2 recombinant proteins and different bacterial cell wall components. Both bjALP1 and bjALP2 could interact with soluble PGN from S. aureus and E. coli but not with soluble LPS, LTA, zymosan, or mannan (Fig. 3A). Pull-down assays confirmed that both recombinant proteins directly bound with insoluble PGN from S. aureus and E. coli in a dose-dependent manner (Fig. 3B). Indeed, both bjALP1 and 2 could bind specifically to MDP, the minimal PGN motif (Fig. 3C). The interaction was investigated further by using mutant bjALP1 or 2 that either lacks the ApeC domain (bjALP1ΔC, bjALP2ΔC) or the N-terminal spacer domain (bjALP1ΔN, bjALP2ΔN). The results showed that only the bjALP1ΔN and bjALP2ΔN recombinant proteins could bind with PGN and MDP (Fig. 3 D and Fig. S5), suggesting that ApeC is required for PGN/MDP recognition. Taken together, we concluded that both bjALP1 and bjALP2 are capable of agglutinating Gram-positive bacteria via interaction between the ApeC domain and the bacterial cell-wall component PGN/MDP. However, because the MDP moieties are exposed at the cell surface of Gram-positive but not of Gram-negative bacterium (32), bjALP1 or bjALP2 could only bind efficiently to Gram-positive and not to Gram-negative bacterium (Fig. 2A).
Fig. 3.
Amphioxus ALPs directly interacted with PGN. (A) ELISA analysis of the interaction between recombinant ALP proteins and microbial components. Plates were coated with 20-μg components then were washed and incubated with bjALPs at 4 °C overnight followed by detection with an anti-His antibody. One representative experiment of six is shown. Background absorbance without protein was subtracted. (B) Pull-down analysis of the binding of ALP proteins to PGN from S. aureus and E. coli. P, pellet protein; T, total protein. (C) ELISA analysis of the interaction between recombinant ALP proteins and MDP. One representative experiment of six is shown. Background absorbance without protein was subtracted. (D) ELISA analysis of the interaction between the truncated bjALP1 mutants and PGN. One representative experiment of six is shown. Background absorbance without protein was subtracted. *P < 0.05, **P < 0.01, ***P < 0.001.
Distribution of the Endogenous bjALP1 and bjALP2 Proteins.
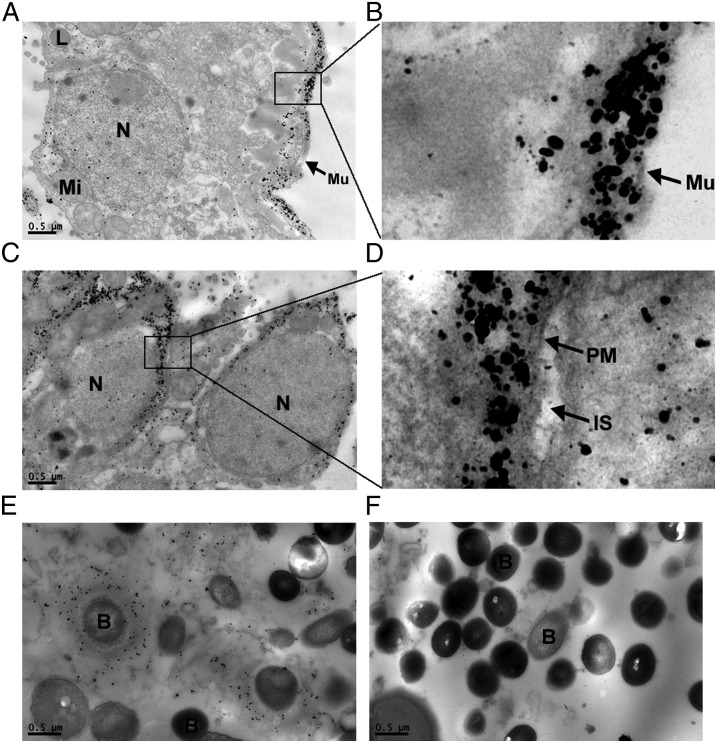
We produced anti-bjALP1 and anti-bjALP2 monoclonal antibodies (mAbs) using the peptides 412GEVPDGNYDR421 and 375DGIYNRNTEI384, respectively (Fig. S6). We performed immunogold analysis of hepatic ceca of amphioxus with these antibodies and found that gold particles localize bjALP1 mainly in mucus, with some scattered signals in cytoplasm (Fig. 4 A and B), whereas for bjALP2 most of the gold particles were distributed near plasma membrane in the cytoplasm side (Fig. 4 C and D and Fig. S7 A and B). In the excrement of these animals, which mainly consisted of mucus and S. aureus, we observed a large amount of bjALP1 surrounding the bacteria (Fig. 4E). In contrast, no bjALP2 was detected in the excrement (Fig. 4F). Western blotting confirmed the presence of bjALP1 and the absence of bjALP2 in the excrement of these bacterially fed animals (Fig. S7C).
Fig. 4.
Immunogold analysis of bjALPs. (A and B) Immunogold electron microscope (EM) localization of bjALP1 in the hepatic cecum of amphioxus. (C and D) Immunogold EM localization of bjALP2 in the hepatic cecum of amphioxus. (E) Immunogold EM localization of bjALP1 in the excrement of S. aureus-fed amphioxus. (F) Immunogold EM localization of bjALP2 in the excrement of S. aureus-fed amphioxus. B, bacteria; L, lysosome; Mi, mitochondria; Mu, mucus; N, nucleus; PM, plasma membrane.
BjALP1 Protected Amphioxus from the Harmful Effect of Bacterial Infections.
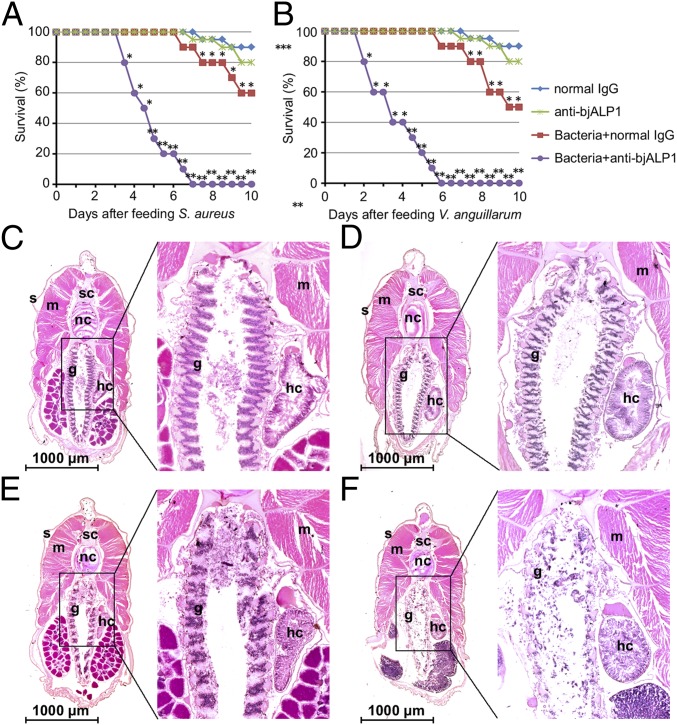
Considering the abundance of bjALP1 in the feces of infected animals, the massive induction during bacterial infection, and the bacterial agglutination effect, we suspected that bjALP1 could be an indispensable extracellular effector of gut mucosal immunity. The anti-bjALP1 mAb is specific for the ApeC domain (412–421 aa) of bjALP1 and may interfere with the interaction between bjALP1 and PGN owing to steric hindrance. ELISA verified that the addition of the anti-bjALP1 mAb did reduce the binding of recombinant bjALP1 to MDP in a dose-dependent fashion (Fig. S8). Therefore, anti-ALP1 mAb can be used to block the function of endogenous bjALP1 proteins in the gut of amphioxus. As shown in Fig. 5 A and B, oral feeding of the anti-ALP1 mAb significantly decreased the survival rates of adult amphioxus after bacterial infection. Tissue section examination showed that without the protection of extracellular bjALP1 the gill of infected amphioxus was completely destroyed by S. aureus infection within 72 h (Fig. 5 C–F).
Fig. 5.
BjALP1 plays an important role in the anti-bacterial immune response of amphioxus. (A) Survival analysis of amphioxus following feeding with S. aureus with or without the anti-bjALP1 antibody. (B) Survival analysis of amphioxus following feeding with V. anguillarum with or without the anti-bjALP1 antibody. Purified IgGs from preimmunized mouse serum were used as the control. (C–F) Section H&E staining of amphioxus after feeding with normal IgG (C), the anti-bjALP1 antibody (D), S. aureus and normal IgG (E), and S. aureus and anti-bjALP1 antibody (F). All animals were collected at 72 h after oral infection. Ten animals were used for each treatment. The representative results of three experiments are shown. g, gill; hc, hepatic cecum; m, muscle; nc, notochord; o, ovary; s, skin; sc, spinal cord; sp, spermary. *P < 0.05, **P < 0.01.
bjALP2 Could Function As an Intracellular Pattern Recognition Receptor.
Although bjALP2 proteins were present within gut epithelial cells, they could not be detected in the excrement of infected amphioxus. Also, a full-length bjALP2-enhanced GFP (EGFP) fusion protein, when ectopically expressed in HEK293T cells, was detected within the cells but not in the culture medium (Fig. S7D). The signal peptide of bjALP2 is clearly functional because fusing the bjALP2 signal peptide to the EGFP protein resulted in the secretion of EGFP (Fig. S7D). Nonetheless, replacing bjALP2’s original signal peptide with the signal peptides of Igκ and preprotrypsin also failed to induce bjALP2 secretion (Fig. S7E). Therefore, it seems that certain feature(s) of bjALP2 render its secretary signal peptide ineffective, leading to its cytoplasmic retention despite having a signal peptide.
bjALP2 Could Modulate the Activation of the TRAF6–NF-κB Pathway.
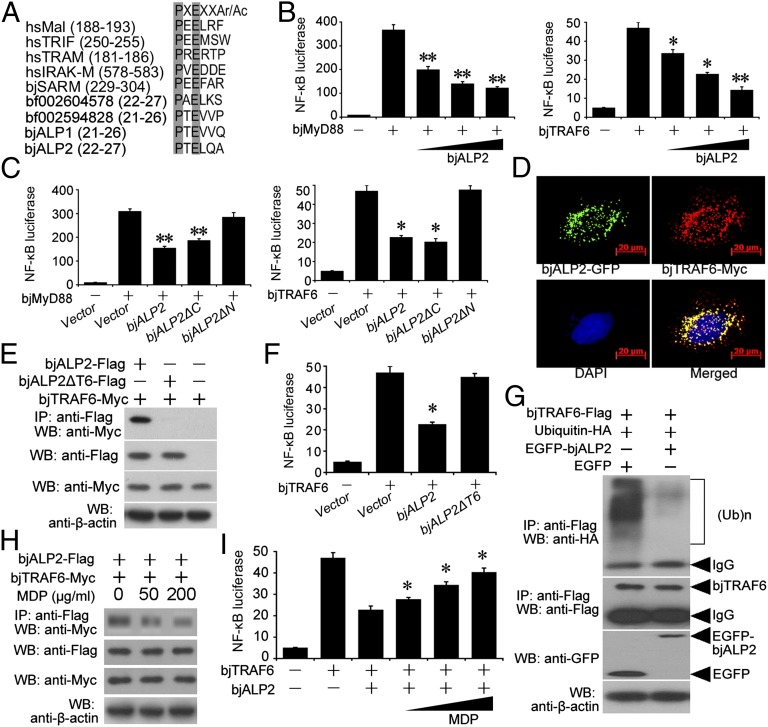
A sequence analysis revealed a potential TRAF6-binding motif in the N-terminal spacer region of bjALP2 (Fig. 6A). We suspected that when retained in the cytoplasm bjALP2 might interact with amphioxus TRAF6 (bjTRAF6). Using luciferase reporter assays in HEK293T cells, we found that full-length bjALP2 strongly inhibited NF-κB activation induced by amphioxus MyD88 (bjMyd88) and bjTRAF6 in a dose-dependent manner (Fig. 6B). Using two mutant constructs, bjALP2ΔC (lacking the ApeC domain) and bjALP2ΔN (lacking the N-terminal spacer region), we further showed that the NF-κB inhibitory activity of bjALP2 is mainly determined by its N-terminal spacer region (Fig. 6C).
Fig. 6.
BjALP2 interacts with bjTRAF6 via the TRAF6-binding motif and inhibits the ubiquitination of TRAF6 to suppress NF-κB activation. (A) Putative TRAF6-binding motifs identified in amphioxus ALPs. (B) Luciferase reporter assay showing the BjALP2 inhibition of NF-κB activation induced by bjMyD88 and bjTRAF6. (C) The influence of truncated bjALP2 mutants on NF-κB activation as analyzed by luciferase reporter assay. (D) BjALP2 colocalizes with amphioxus TRAF6 in HeLa cells. HeLa cells were cotransfected with EGFP-fused bjALP2 and myc-tagged pCMV-bjTRAF6 and were stained with an anti-myc antibody and an Alexa Fluor 532 secondary antibody. (E) Co-IP assay showing that bjALP2, but not the TRAF6-binding motif-deleted mutant, interacts with amphioxus TRAF6 when overexpressed in 293T cells. (F) Luciferase reporter assay showing that the TRAF6-binding motif-deleted mutant of bjALP2 did not inhibit NF-κB activation. (G) Co-IP assay showing that bjALP2 suppressed the ubiquitination of amphioxus TRAF6 when coexpressed in 293T cells. The data are representative of at least three independent experiments. (H) Co-IP assay showing that MDP diminished the binding of bjALP2 with bjTRAF6. (I) Luciferase reporter assay showing that MDP relieved the inhibition by bjALP2 of bjTRAF6-induced NF-κB activation. *P < 0.05, **P < 0.01.
We then analyzed the subcellular location of bjALP2 and bjTRAF6. When coexpressed with the myc-tagged bjTRAF6 protein in HeLa cells, the bjALP2-EGFP fusion protein colocalized with myc-tagged bjTRAF6 in the cytoplasm (Fig. 6D). We then constructed bjALP2ΔT6, a mutant without the TRAF6-binding motif. When overexpressed in HeLa cells, the subcellular localizations of bjALP2ΔT6 and bjALP2 were indistinguishable (Fig. S9 A and B), suggesting that the TRAF6-binding motif has no obvious impact on the subcellular location of bjALP2. However, bjALP2ΔT6-EGFP failed to colocalize with myc-tagged bjTRAF6, suggesting that bjALP2 could not interact with bjTARF6 without the TRAF6-binding motif (Fig. S9C). Indeed, coimmunoprecipitation (Co-IP) assays confirmed that bjALP2 interacted with bjTRAF6 when overexpressed in 293T cells (Fig. 6E). Furthermore, mutation of the TRAF6-binding motif not only abolished the interaction between bjALP2 and bjTRAF6 but also relieved the inhibition of bjALP2 on NF-κB activation (Fig. 6 E and F).
The self-ubiquitination of TRAF6 is key event in NF-κB activation (13, 15). Thus, to determine whether bjALP2 can interfere with the self-ubiquitination of bjTRAF6, we created two plasmid constructs containing Flag-tagged bjTRAF6 and HA-tagged bjUbiquitin, respectively. When these two constructs were cotransfected into HEK293T cells with the EGFP-tagged bjALP2 construct we recorded a significant reduction of the ubiquitination level of bjTRAF6 (Fig. 6G).
MDP Could Relieve the Inhibition of bjALP2 on bjTRAF6.
In mammals, whereas MDP is a PAMP that is recognized by NOD2, an intracellular pattern recognition receptor, MDP cannot cross the plasma membrane of mammalian cells efficiently through diffusion (33). Interestingly, S. aureus, a microbial pathogen for amphioxus, could get into the intracellular space of the amphioxus gut epithelium shortly after being administered orally (34). Together, these observations have suggested that MDP enters the cells as part of an intact microbe and hence serves as an excellent indicator for microbe intracellular invasion in amphioxus. In this context, our finding that bjALP2 also physically interacted with bjTRAF6 prompted us to speculate whether the interaction of bjALP2 with bjTRAF6, and accordingly the effect of TRAF6-mediated NF-κB activation, could be affected by MDP. Because we showed that bjALP2 is able to recognize PGN/MDP through its C-terminal ApeC domain, we sought to ascertain whether MDP can prevent bjALP2 from suppressing bjTRAF6. We therefore performed Co-IP assays with the presence of MDP (during plasmid transfection) in 293T cells, which showed that the binding between bjALP2 and bjTRAF6 was weakened by the addition of MDP in a dose-dependent manner (Fig. 6H). Using luciferase reporter assays, we observed that the presence of MDP did reverse the inhibitory effect of bjALP2 on bjTRAF6-dependent NF-κB activation in a dose-sensitive fashion (Fig. 6I). In summary, our functional data indicate that the recognition of PGN/MDP by cytoplasmic bjALP2 freed bjTRAF6 from the binding with bjALP2 and released bjTRAF6, which subsequently activated the downstream NF-κB pathway.
Discussion and Conclusions
Apextrin and its homologs have been repeatedly documented since 1998, but their structures and functions have remained elusive (30, 31, 35–42). Here, we demonstrate that ALPs contain ApeC, a novel protein domain structure that is conserved in several invertebrate lineages and capable of recognizing the bacterial cell wall component PGN and its moiety MDP. Without the crystal structure of ApeC we cannot rule out the possibility that there are distant ApeC homologs in vertebrates and several other invertebrates (C. elegans, Drosophila, and urochordates). Regardless, ApeC is an ancient domain type that can be dated back to the common ancestor of eumetazoa.
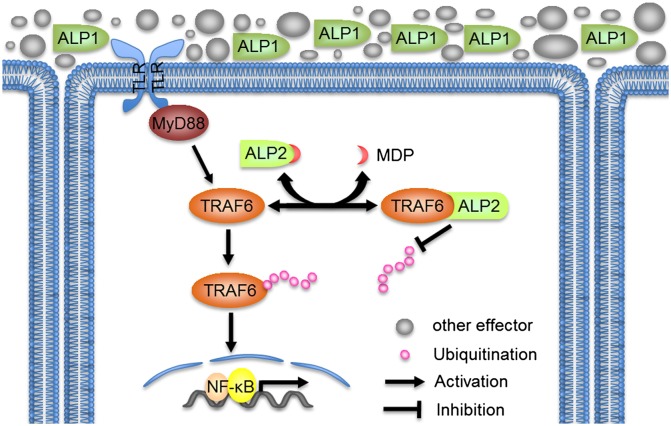
Comprehensive functional analyses revealed the role of two amphioxus ALPs and their ApeC domains in gut mucosal immune responses (Fig. 7). Briefly, a huge amount of ALP1 is secreted into the gut lumen to protect the epithelium, together with other extracellular effectors. ALP1 aggregates and immobilizes Gram-positive bacteria and hence most likely facilitates the killing and excreting of these bacteria. ALP2, however, is retained within epithelial cells by an unknown mechanism that likely arose through adaptive evolution. ALP2 binds with TRAF6 and prevents TRAF6 from self-ubiquitination and hence from NF-κB activation. Endoplasmic PGN/MDP competes with TRAF6 for ALP2, which releases a sufficient amount of free TRAF6 to activate the NF-κB pathway.
Fig. 7.
Proposed model of the synergic function of BjALP1 and BjALP2. During the immune response a large amount of BjALP1 proteins are secreted to form a mucosal cushion with other mucosal effectors to protect the epithelium surface, whereas intracellular BjALP2 regulates the immune response and homeostasis by monitoring the PGN/MDP concentration.
Based on the current information, we can conceive a possible interplay between the two amphioxus ALPs. During bacterial infection in the gut a large amount of PGN and MDP may enter the cytoplasm of gut epithelial cells via the pinocytosis of PGN/MDP or phagocytosis of the bacteria. PGN/MDP binds with ALP2 and releases a sufficient amount of free TRAF6 to activate the NF-κB pathway. In return, NF-κB most likely induces the expression of ALP1. If so, ALP1 will help to quickly clear out the bacteria in the gut lumen and hence reduce the amount of PGN/MDP that ALP2 can access. ALP2 will then rebind TRAF6 to suppress NF-κB. As low NF-κB activity reduces the expression of ALP1, new homeostasis will be achieved at the end of the antibacterial response.
In conclusion, the discovery of the novel ApeC domain (Pfam accession no. PF16977) and the functional information of two amphioxus ALPs define ALPs as a previously unidentified type of PRP. Furthermore, we demonstrate that amphioxus ALPs can serve as extracellular effector PRPs and intracellular sensor PRPs, which provides new insight into the importance and diversified function of ALPs in different animal phyla or subphyla.
Materials and Methods
Twenty amphioxi were maintained in 100 mL filtered seawater, and S. aureus and V. anguillarum were added to the seawater to a final concentration of ∼1 × 107 cfu/mL with or without 0.5 μg/mL anti-bjALP1 antibody. Dead amphioxi were counted at the indicated time points after bacterial infection. Purified IgGs from preimmunized mouse serum were used as the control. All primers used in this study are shown in Table S1.
Supplementary Material
Acknowledgments
This work was supported by project 2013CB917800 of the Major National Scientific Research Project, project 2013CB835300 of the National Basic Research Program (973), projects 2008AA092601 and 2012AA092201 of the State High-Tech Development Project (863), key projects 91231206 and 30730089 and projects 31171193 and 30901305 of National Natural Science Foundation, and projects from the Commission of Science and Technology of Guangdong Province and Guangzhou City and from Sun Yat-sen University Science Foundation. The data analysis was also supported by the Guangdong Province Key Laboratory of Computational Science and the Guangdong Province Computational Science Innovative Research Team.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KM017614 and KM017615) and the Pfam database (accession no. PF16977, which will be available in the v28.0 release).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405414111/-/DCSupplemental.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449(7164):819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Kim YS, et al. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem. 2000;275(42):32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 4.Areschoug T, Gordon S. Pattern recognition receptors and their role in innate immunity: Focus on microbial protein ligands. Contrib Microbiol. 2008;15:45–60. doi: 10.1159/000135685. [DOI] [PubMed] [Google Scholar]
- 5.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7(8):232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway—its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay S, Gordon S. The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology. 2004;209(1-2):39–49. doi: 10.1016/j.imbio.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. NF-κB in immunobiology. Cell Res. 2011;21(2):223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2(10):725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 10.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 11.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8(1):7. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayden MS, Ghosh S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamothe B, et al. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J Biol Chem. 2007;282(6):4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 15.Kanayama A, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15(4):535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Cha GH, et al. Discrete functions of TRAF1 and TRAF2 in Drosophila melanogaster mediated by c-Jun N-terminal kinase and NF-kappaB-dependent signaling pathways. Mol Cell Biol. 2003;23(22):7982–7991. doi: 10.1128/MCB.23.22.7982-7991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25(1):697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 18.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439(7079):965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 19.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18(7):1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453(7198):1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 21.Louis A, Roest Crollius H, Robinson-Rechavi M. How much does the amphioxus genome represent the ancestor of chordates? Brief Funct Genomics. 2012;11(2):89–95. doi: 10.1093/bfgp/els003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18(7):1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S, et al. The evolution and regulation of the mucosal immune complexity in the basal chordate amphioxus. J Immunol. 2011;186(4):2042–2055. doi: 10.4049/jimmunol.1001824. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, et al. Functional characterization of a ficolin-mediated complement pathway in amphioxus. J Biol Chem. 2011;286(42):36739–36748. doi: 10.1074/jbc.M111.245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, et al. A novel C1q family member of amphioxus was revealed to have a partial function of vertebrate C1q molecule. J Immunol. 2008;181(10):7024–7032. doi: 10.4049/jimmunol.181.10.7024. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, et al. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol. 2007;179(12):8425–8434. doi: 10.4049/jimmunol.179.12.8425. [DOI] [PubMed] [Google Scholar]
- 27.Yuan S, et al. The archaic roles of the amphioxus NF-κB/IκB complex in innate immune responses. J Immunol. 2013;191(3):1220–1230. doi: 10.4049/jimmunol.1203527. [DOI] [PubMed] [Google Scholar]
- 28.Yuan S, et al. Genomic and functional uniqueness of the TNF receptor-associated factor gene family in amphioxus, the basal chordate. J Immunol. 2009;183(7):4560–4568. doi: 10.4049/jimmunol.0901537. [DOI] [PubMed] [Google Scholar]
- 29.Yuan S, et al. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-kappaB pathway via MyD88. Mol Immunol. 2009;46(11-12):2348–2356. doi: 10.1016/j.molimm.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 30.Haag ES, Raff RA. Isolation and characterization of three mRNAs enriched in embryos of the direct-developing sea urchin Heliocidaris erythrogramma: Evolution of larval ectoderm. Dev Genes Evol. 1998;208(4):188–204. doi: 10.1007/s004270050173. [DOI] [PubMed] [Google Scholar]
- 31.Haag ES, Sly BJ, Andrews ME, Raff RA. Apextrin, a novel extracellular protein associated with larval ectoderm evolution in Heliocidaris erythrogramma. Dev Biol. 1999;211(1):77–87. doi: 10.1006/dbio.1999.9283. [DOI] [PubMed] [Google Scholar]
- 32.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 34.Yang P, et al. Origin of the phagocytic respiratory burst and its role in gut epithelial phagocytosis in a basal chordate. Free Radic Biol Med. 2014;70:54–67. doi: 10.1016/j.freeradbiomed.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Miller DJ, et al. The innate immune repertoire in cnidaria—ancestral complexity and stochastic gene loss. Genome Biol. 2007;8(4):R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayward DC, et al. Differential gene expression at coral settlement and metamorphosis—a subtractive hybridization study. PLoS ONE. 2011;6(10):e26411. doi: 10.1371/journal.pone.0026411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grasso LC, et al. Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics. 2008;9:540. doi: 10.1186/1471-2164-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer E, Aglyamova GV, Matz MV. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol. 2011;20(17):3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x. [DOI] [PubMed] [Google Scholar]
- 39.David E, Tanguy A, Pichavant K, Moraga D. Response of the Pacific oyster Crassostrea gigas to hypoxia exposure under experimental conditions. FEBS J. 2005;272(21):5635–5652. doi: 10.1111/j.1742-4658.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 40.Estévez-Calvar N, Romero A, Figueras A, Novoa B. Involvement of pore-forming molecules in immune defense and development of the Mediterranean mussel (Mytilus galloprovincialis) Dev Comp Immunol. 2011;35(10):1017–1031. doi: 10.1016/j.dci.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Dheilly NM, Haynes PA, Bove U, Nair SV, Raftos DA. Comparative proteomic analysis of a sea urchin (Heliocidaris erythrogramma) antibacterial response revealed the involvement of apextrin and calreticulin. J Invertebr Pathol. 2010;106(2):223–9. doi: 10.1016/j.jip.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Huang G, et al. Profile of acute immune response in Chinese amphioxus upon Staphylococcus aureus and Vibrio parahaemolyticus infection. Dev Comp Immunol. 2007;31(10):1013–1023. doi: 10.1016/j.dci.2007.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.