Abstract
Urease pre-treatment of urine has been utilized since the early 1960s to remove high levels of urea from samples prior to further processing and analysis by gas chromatography-mass spectrometry (GC-MS). Aside from the obvious depletion or elimination of urea, the effect, if any, of urease pre-treatment on the urinary metabolome has not been studied in detail. Here, we report the results of three separate but related experiments that were designed to assess possible indirect effects of urease pre-treatment on the urinary metabolome as measured by GC-MS. In total, 235 GC-MS analyses were performed and over 106 identified and 200 unidentified metabolites were quantified across the three experiments. The results showed that data from urease pre-treated samples 1) had the same or lower coefficients of variance among reproducibly detected metabolites, 2) more accurately reflected quantitative differences and the expected ratios among different urine volumes, and 3) increased the number of metabolite identifications. Overall, we observed no negative consequences of urease pre-treatment. In contrast, urease pretreatment enhanced the ability to distinguish between volume-based and biological sample types compared to no treatment. Taken together, these results show that urease pretreatment of urine offers multiple beneficial effects that outweigh any artifacts that may be introduced to the data in urinary metabolomics analyses.
Keywords: urease, urine, gas chromatography-mass spectrometry, metabolomics, statistics
1 Introduction
Urine is a commonly used biofluid for metabolite analysis and metabolomics studies in humans due to its facile and noninvasive collection and metabolite richness. It is an almost perfect matrix for identifying and monitoring biomarkers for various aspects of human health. The glomerular filtrate (the glomerulus is the filtering unit of the nephron; both kidneys of a human contain a combined ~2,400,000 nephrons) has almost exactly the same composition as blood plasma, with the exception of protein content (Guyton, 1981). Blood plasma is directly linked to the extracellular fluids in the body, and the rate of blood flow through the kidneys is ~1200 mL/min for a 70 kg man (Guyton, 1981). Thus, metabolites present in blood due to cell damage/death or excretion stand an excellent chance of appearing in the urine as biomarkers. Urinary metabolite biomarkers of interest include those used to assess drug (Wilkins, 1997) and anabolic steroid abuse (Shelby et al., 2011), identify individuals with cancer (Ganti and Weiss, 2011) and other diseases (Kussmann et al., 2006), diagnose inborn errors of metabolism (Kuhara, 2007), determine exposure to carcinogens (Hecht, 2002), pesticides (Egeghy et al., 2011), and endocrine disruptors (Meeker et al., 2009), and monitor levels of oxidative stress (Roberts and Morrow, 2000).
Current metabolite analysis and metabolomics studies rely almost exclusively on nuclear magnetic resonance spectroscopy, liquid chromatography-mass spectrometry (LC-MS), and gas chromatography-mass spectrometry (GC-MS) (Metz et al., 2007). GC in particular is unsurpassed in terms of separation peak capacity – a measure of the number of chromatographic peaks that can be baseline-resolved within the analysis time – and is used extensively in the analysis of small molecules in various matrices, including urine. High levels of urinary urea (typically on the order of 9.3–23.3 g/L (Putnam, 1971)) present a challenge in GC-MS-based metabolomics analyses and can consume the chemical derivatizing reagent, resulting in incomplete derivatization of other metabolites. Urea can be eliminated by pre-treating urine with the enzyme urease prior to metabolite extraction, derivatization, and analysis. Wells et al. (1964) first reported on the use of urease pre-treatment of urine samples in 1964, and the method has been improved by others (Shoemaker and Elliott, 1991; Matsumoto et al., 1994; Chan et al., 2011) over the years.
While urease pre-treatment of urine can completely remove urea from the sample, its effects, if any, on the measurement of the urinary metabolome are unknown. Creatine is converted to creatinine during urease pre-treatment (Shoemaker and Elliott, 1991; Matsumoto and Kuhara, 1996), due to the increase in pH of the sample from liberation of two moles of ammonia per mole of urea, and various preparations of urease can introduce contaminants such as myoinositol (Clements and Starnes, 1975) and citrate (Shoemaker and Elliott, 1991). In developing a comprehensive urinary metabolomics approach for identifying kidney cancer, Kind et al. (2007) observed decreased abundances for aconitate, acotinate, ascorbate, citrate, glycerol, hypoxanthine, succinate, and tyrosine using GC-MS, and for several unidentified compounds using LC-MS, after urease pre-treatment. However, beyond these findings, a detailed analysis of the potential metabolomic artifacts of urease pre-treatment of urine has not yet been reported. Therefore, we conducted a statistical analysis of the effects of urease pre-treatment of urine samples as measured by GC-MS over three separate experiments involving 1) a constant volume of urine, 2) varying volumes of urine, and 3) a comparison of male and female urines.
2 Materials and methods
2.1 Chemicals and reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. A mixture of fatty acid methyl esters (FAMEs) dissolved in hexane was prepared for use as a retention index standard. Even numbered FAMEs from C8 to C28 were prepared at 533.3 µg/mL, with the exception of C16 which was prepared at 733.3 µg/mL. A creatinine assay kit (DetectX Urinary Creatinine Detection Kit; catalog number K002-H1) was obtained from Arbor Assays (Eisenhower, PA). Deionized and purified water was used to prepare buffer and standard solutions (Milli-Q System Advantage A10, Merck Millipore, Billerica, MA).
2.2 Urine samples
Approval for the conduct of this programmatic research was obtained from the Pacific Northwest National Laboratory Institutional Review Board. Urine samples from consenting male and female donors (n = 20 each, Supplemental Table S1) after an overnight fast were purchased from Bioreclamation, LLC (Hicksville, NY) and received frozen on dry ice and deidentified. To create a uniform sample for Experiments 1 and 2 (see below), aliquots from each individual sample were pooled, realiquoted, and stored at −80°C until used.
2.3 Urease pre-treatment of urine samples
2.3.1 Experiment 1 – Constant Urine Volume
To initially evaluate the effects of urease pre-treatment on the urinary metabolome, we compared the urine metabolite profiles from pooled urine after pre-treatment with urease, water, or no treatment at all. For this, 100 µL aliquots of the pooled urine sample were incubated (n = 5, each) with 100 µL of a 1 mg/mL solution of urease (Sigma-Aldrich catalog number U4002) prepared in water (urease-treated; ‘UT’) or an equal volume of water alone (water-treated; ‘WT’) for 30 min at 37°C with mild shaking (500 rpm). Identical aliquots (n = 5) were not subjected to any treatment (no treatment; ‘NT’) and allowed to sit at room temperature for 30 minutes. Metabolites were then extracted with concomitant protein precipitation by addition of 1 mL of cold (−20°C) methanol with vortexing for 30 s, and precipitated proteins were removed by centrifugation at 15,000×g for 10 min at 4°C. The supernatants were transferred to glass autosampler vials and then dried in vacuo prior to chemical derivatization. If the extracts could not be immediately derivatized and analyzed by GC-MS, then they were stored at −80°C.
2.3.2 Experiment 2 – Varying Urine Volumes
To evaluate whether the effects of urease pre-treatment of urine varied with the volume of urine prepared, we compared the urine metabolite profiles from pooled urine after pretreatment with urease (UT) and after no treatment (NT) using various volumes of urine. For this, several volumes (5, 10, 25, 50, and 100 µL) of the pooled urine sample were incubated (n = 3, each) with 100 µL of a 1 mg/mL solution of urease or were not subjected to any treatment, each as described above. Metabolites were then extracted as described above.
2.3.3 Experiment 3 – Male Versus Female Urines
Finally, to evaluate whether any artifacts introduced by urease pre-treatment on the urinary metabolome interfered with the ability to distinguish between comparative samples, we compared the metabolite profiles from individual male and female urine samples after pretreatment with urease or after no treatment (previous metabolomics studies of male and female urines (Pasikanti et al., 2008; Slupsky et al., 2007; Saude et al., 2007; Psihogios et al., 2008) have reported differences in metabolite levels). For this, 50 µL aliquots of individual male and female urine samples (n = 20, each) were blocked, randomized, and then incubated with 50 µL of a 1 mg/mL solution of urease (UT) or were not subjected to any treatment (NT), each as described above. Metabolites were then extracted as described above. Creatinine was quantified in each sample using the DetectX Urinary Creatinine Detection Kit according to the manufacturer’s instructions, with the exception that a simple linear regression was used for the standard curve.
2.4 Chemical derivatization
Dried metabolite extracts were chemically derivatized using a modified version of the protocol used to create FiehnLib (Kind et al., 2009). Briefly, dried metabolite extracts were dried again to remove any residual water if they had been stored at −80°C. To protect carbonyl groups and reduce the number of tautomeric isomers, 20 µL of methoxyamine in pyridine (30 mg/mL) were added to each sample, followed by vortexing for 30 s and incubation at 37°C with generous shaking (1000 rpm) for 90 min. At this point, the sample vials were inverted one time to capture any condensation of solvent at the cap surface, followed by a brief centrifugation at 1000×g for 1 min. To derivatize hydroxyl and amine groups to trimethylsilyated (TMS) forms, 80 µL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) were then added to each vial, followed by vortexing for 10 s and incubation at 37°C with shaking (1000 rpm) for 30 min. Again, the sample vials were inverted one time, followed by centrifugation at 1000×g for 5 min. The samples were allowed to cool to room temperature and were analyzed in the same day.
For Experiment 1, the derivatized samples were split into 3 portions and transferred to 3 new autosampler vials with inserts. For Experiment 2, the derivatized samples were kept intact and each transferred to a single new autosampler vial with insert. For Experiment 3, the derivatized samples were split into 2 portions and transferred to 2 new autosampler vials with inserts.
2.5 GC-MS analysis
Samples were analyzed according to the method used to create FiehnLib (Kind et al., 2009). An Agilent GC 7890A coupled with a single quadrupole MSD 5975C (Agilent Technologies, Inc; Santa Clara, CA) was used, and the samples were blocked and analyzed in random order for each Experiment. A HP-5MS column (30 m × 0.25 mm × 0.25 µm; Agilent Technologies, Inc) was used for untargeted metabolomics analyses. The sample injection mode was splitless, and 1 µL of each sample was injected. The injection port temperature was held at 250°C throughout the analysis. The GC oven was held at 60°C for 1 min after injection, and the temperature was then increased to 325°C by 10°C/min, followed by a 5 min hold at 325°C (Kim et al., 2013). The helium gas flow rates for each Experiment were determined by the Agilent Retention Time Locking function based on analysis of deuterated myristic acid and were in the range of 0.45-0.5 mL/min. Data were collected over the mass range 50 – 550 m/z. A mixture of FAMEs (C8-C28) was analyzed once per day together with the samples for retention index alignment purposes during subsequent data analysis. All raw GC-MS data will be made available via the Metabolights metabolomics data repository (http://www.ebi.ac.uk/metabolights/index).
2.6 Data processing and metabolite identification
GC-MS raw data files from each Experiment were processed using the Metabolite Detector software, version 2.0.6 beta (Hiller et al., 2009). Briefly, Agilent.D files were converted to netCDF format using Agilent Chemstation, followed by conversion to binary files using Metabolite Detector. Retention indices (RI) of detected metabolites were calculated based on the analysis of the FAMEs mixture, followed by their chromatographic alignment across all analyses after deconvolution. Metabolites were initially identified by matching experimental spectra to an augmented version of FiehnLib (Kind et al., 2009) (i.e., the Agilent Fiehn Metabolomics Retention Time Locked (RTL) Library, containing spectra and validated retention indices for over 700 metabolites), using a Metabolite Detector match probability threshold of 0.6 (combined retention index and spectral probability). All metabolite identifications were manually validated to reduce deconvolution errors during automated data-processing and to eliminate false identifications. We propose that this approach results in a metabolite identification confidence of Level 1.5, according to the guidelines recommended by the Metabolomics Standards Initiative Chemical Analysis Working Group of the Metabolomics Society (Sumner et al., 2007). While the library used to identify metabolites was generated by an external laboratory, this library contains both retention indices and mass spectra from analyses of authentic chemical standards, and our analyses were performed using methods identical to those used to create the library. The NIST 08 GC-MS library was also used to cross validate the spectral matching scores obtained using the Agilent library and to provide identifications of unmatched metabolites (Level 2 identifications). The three most abundant fragment ions in the spectra of each identified metabolite were automatically determined by Metabolite Detector, and their summed abundances were integrated across the GC elution profile; fragment ions due to trimethylsilylation (i.e. m/z 73 and 147) were excluded from the determination of metabolite abundance. A matrix of identified metabolites, unidentified metabolite features (characterized by mass spectra and retention indices and assigned as ‘unknown’; Level 4 identifications), and their abundances was created for each Experiment for statistical analysis. Features resulting from GC column bleeding were removed from the data matrices prior to further data processing and analysis.
2.7 Statistical Pre-processing
The GC-MS data matrices from all Experiments were log10 transformed and processed similarly to remove metabolites with inadequate information for statistical analyses and identify outlier GC-MS analyses. For each Experiment, metabolites were filtered from the dataset if they did not meet the minimum requirements for a quantitative statistical test, such as t-test or Analysis of Variance (ANOVA), and qualitative test (G-test) (Webb-Robertson et al., 2010). However, the data processing of the GC-MS raw files (Section 2.6) did not return any features, identified or unidentified, with inadequate data for statistics in one or more treatment groups from any Experiment. Extreme behavior in GC-MS datasets (outliers) were identified using a combination of correlation, principal component analysis (PCA), and an approach based on a robust Mahalanobis distance (rMd) to assess the reproducibility of the distribution of metabolite abundance values across replicate runs of the same sample, as well as across related biological samples (Matzke et al., 2011). Strategies for normalization of the identified metabolite and unidentified metabolite feature abundances in each data matrix were evaluated using the Statistical Procedure for the Analyses of Normalization Strategies (SPANS) protocol (Webb-Robertson et al., 2011).
2.7.1 Experiment 1-Constant Urine Volume
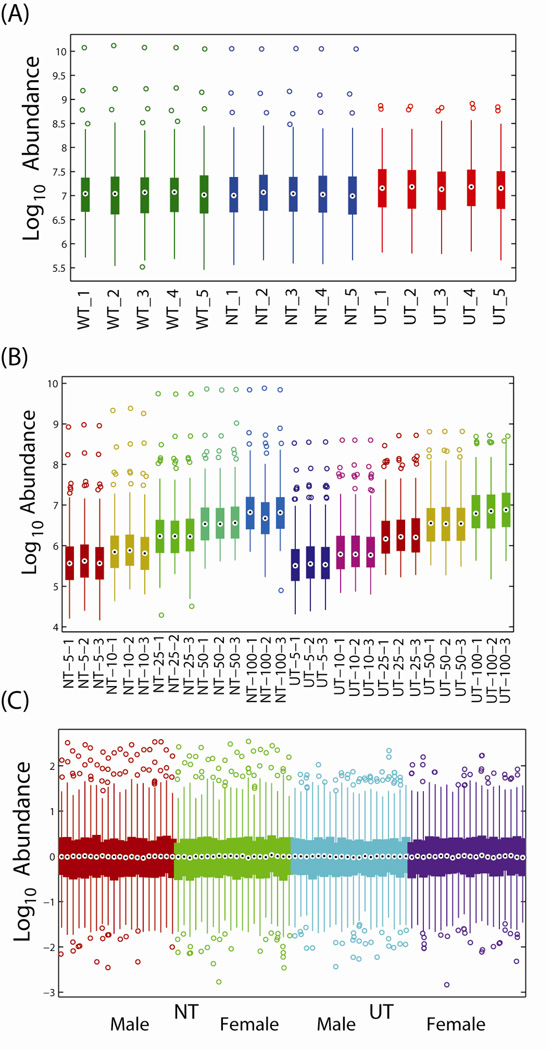
Experiment 1 resulted in the identification of 228 metabolites across 45 total GC-MS analyses (5 aliquots of urine measured in triplicate across three pre-treatment approaches). Outlier evaluation found data from one GC-MS analysis displaying extreme behavior (Supplemental Figure S1). Analysis via SPANS found that all strategies, both global and local, introduced bias into the dataset. Further exploration found that the UT group had significantly larger overall abundance profiles affecting the majority of the metabolites and that adequate metabolites with unchanging profiles were not confidently identified to normalize against (Supplemental Figure S2). However, as seen in Figure 1A, after averaging technical replicates, the variation between GC-MS datasets within each pre-treatment were minimal and therefore lack of normalization will not dramatically affect the results of subsequent statistics.
Figure 1.
Boxplots displaying the distributions of the log10 transformed abundances of all metabolites for each process or biological replicate from (A) constant volume experiment, (B) varying volume experiment and (C) male versus female experiment. The central dot represents the median, the edges of the box represent the 25-th and 75-th percentiles, and the lines extent to the 5-th and 95-th percentiles. WT: water pre-treatment; NT: no pre-treatment; UT: urease pretreatment.
2.7.2 Experiment 2 – Varying Urine Volumes
The second experiment resulted in the identification of 250 metabolites across 30 total GC-MS analyses (3 aliquots of urine measured across 5 dilutions with 2 pre-treatment approaches). Outlier evaluation found no unusual behavior. Again, due to the significant differences in the volumes of the metabolites being measured, no normalization was performed; however, the variability between replicates was low, since they were based on a common pool (Figure 1B).
2.7.3 Experiment 3 – Male Versus Female Urines
Finally, the last experiment comparing the profiles from individual male and female urines resulted in 207 total metabolite identifications across 160 GC-MS analyses (20 males and 20 females in duplicate for each of 2 pre-treatments). Processing for outliers found no unusual behavior. For direct comparison of NT to UT, no normalization was required since the samples were paired and the analysis is based on the difference between paired samples. However, to compare males versus females within each pre-treatment, subset normalization was required. We chose to not normalize the data against donor’s urinary creatinine levels as it was previously reported from a 6-year survey of >20,000 individuals that adult males have significantly greater (p < 0.0001) urinary creatinine concentrations than adult females (Barr et al., 2005), and normalizing metabolite abundances against creatinine levels would have artificially increased the metabolite abundances in female versus male urine samples. Despite the fact that there was no statistical difference (two-sample t-test; p-value ~0.3) between male and female urinary creatinine values in our study, normalization of metabolite abundances against urinary creatinine resulted in every detected metabolite being statistically significant at a p-value of 0.05 due to bias introduced by this normalization step. In particular, the means and standard deviations after creatinine normalization were 8.2 ± 3.3 and 7.3 ± 3.8 for the males and females, respectively. Performing a paired test based on the rank order of the samples was highly significant (p~4.4e-5). In lieu of creatinine normalization, we used a global median normalization, which was found to not introduce bias using the SPANS algorithm. Figure 1C shows the resulting distribution of each GC-MS dataset after normalization. In addition, since the sample sizes were large for this experiment, all p-values computed from statistical hypothesis tests were corrected across metabolites using a Bonferroni correction (Broadhurst and Kell, 2006).
3 Results and Discussion
3.1 Exploratory Analysis of Pre-treatment Approaches
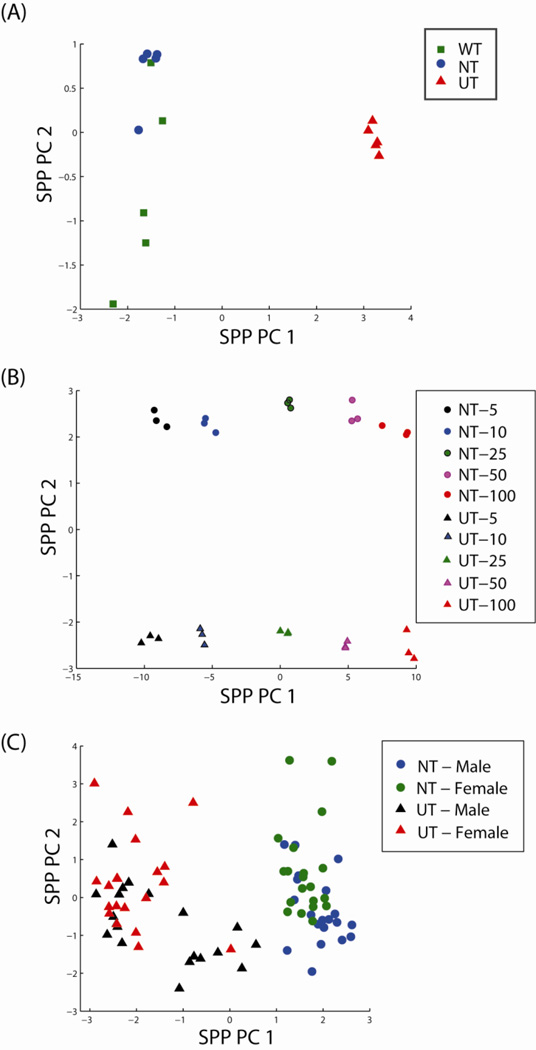
In Experiment 1, we evaluated the effects of urease pre-treatment (UT) vs water treatment (WT) or no treatment (NT) on the urinary metabolome using a constant volume (100 µL) of urine. The data matrix consisted of abundance values for 228 metabolites across 45 GC-MS analyses (Supplemental Table S2). Eighty-eight of these were identified based on matching of spectra and retention indices to entries in the Agilent Fiehn Metabolomics RTL Library, while 140 remain unidentified. Unsupervised exploratory analysis was performed using Principal Component Analysis (PCA) without imputation of missing values based on a sequential projection pursuit (sppPCA) approach (Webb-Robertson et al., 2013), meaning that when a metabolite was not identified for a sample we did not attempt to infer the mechanism by which it was missing or estimate the value of the missing observations. The sppPCA analysis shows a clear discrimination of UT from WT and NT samples in the scores plot based on the first principal component (Figure 2A). In the second dimension, some segregation of WT and NT samples is observed, and although we do not know the reason for this segregation, a separate control experiment showed that metabolite abundances from WT treated samples was consistently more variable than that from NT samples (data not shown). Due to the similarity between the WT and NT treatments both in the exploratory data analysis and in more in depth statistical analyses (see section 3.3, below), we chose only to compare UT and NT in Experiments 2 and 3.
Figure 2.
Scores plots of the first two principal components for the (A) constant volume experiment, (B) varying volume experiment and (C) male versus female experiment based on estimates from Sequential Projection Pursuit PCA (sppPCA) show large amounts of variation being driven by pre-treatment. WT: water pre-treatment; NT: no pre-treatment; UT: urease pretreatment.
To determine if the effects of urease pre-treatment on the urinary metabolome were volume dependent, Experiment 2 compared UT versus NT over five volumes (5, 10, 25, 50 and 100 µL) of urine. The data matrix consisted of abundance values for 250 metabolites across 30 GC-MS analyses (Supplemental Table S3). Ninety-six of these were identified based on matching of spectra and retention indices to entries in the Agilent Fiehn Metabolomics RTL Library, while 154 remain unidentified. The sppPCA analysis, Figure 2B, again showed a clear discrimination of NT from UT; however, in this case pre-treatment was associated with the second principal component, while the largest variation driving the first principal component was urine volume.
Lastly, to evaluate whether the effects of urease pre-treatment on the urinary metabolome confounded the ability to identify statistical differences between comparative samples, we compared UT vs NT in the analysis of fasting urine from male and female donors (n = 20, each; Supplemental Table S1). As mentioned above, previous metabolomics studies of male and female urines (Pasikanti et al., 2008; Slupsky et al., 2007; Saude et al., 2007; Psihogios et al., 2008) have reported differences in metabolite levels. The final data matrix in our study consisted of abundance values for 207 metabolites across 80 GC-MS analyses (Supplemental Table S4). Eighty-three of these were identified based on matching of spectra and retention indices to entries in Agilent Fiehn Metabolomics RTL Library, while 124 remain unidentified. We observed that the largest source of variability was based on pre-treatment with variability related to gender viewed as a lower-resolution visual separation on the second principal component (Figure 2C).
3.2 Reproducibility of Pre-treatment Approaches
The reproducibility of all experiments was evaluated using the percent coefficient of variation (CV) within replicates for the pre-treatment groups. The CV shows the level of variability in relation to the mean of each group. Since raw abundance values are always positive, we calculated CVs using the final normalized data back-transformed from the log scale to original units.
3.2.1 Experiment 1-Constant Urine Volume
Of the 226 metabolites measured in Experiment 1, 212 had sufficient observations across the technical replicates of the three pre-treatments to compute CV values. The average percent CVs for WT, NT, and UT were 16.6%, 12.5% and 12.3%, respectively. However, the median percent CVs were 7.9%, 8.5% and 7.7%, respectively (the larger average percent CV for WT was driven by several large values (outliers) for a small number of metabolites). Due to the non-normality of the CV values across the 212 metabolites, we used a Friedman’s test (Hollander and Wolfe, 1999) to compare the abundance distributions. A Friedman’s test is a non-parametric equivalent to a two factor ANOVA, treating CV as the primary factor and metabolite as a secondary factor. The Friedman’s test identified no statistically significant differences between the three pre-treatments (p-value ~0.08); however, WT was noted as having several extreme values.
3.2.2 Experiment 2 – Varying Urine Volumes
Of the 250 metabolites measured in Experiment 2, 202 metabolites had sufficient observations across the process replicates of the two pre-treatments to calculate CVs. The CVs were first computed on process replicates within each concentration. Thus, each treatment was represented by five CV values, which were then averaged, improving the overall power of the CV estimate. In this case, the comparison of UT versus NT is highly significant (p-value ~2e-20; Wilcoxon signed rank test). Comparing UT versus NT within each volume, the UT group had a significantly lower median percent CV than NT at all volumes except for 25 µL (Table 1).
Table 1.
The median percent CV for each volume and pre-treatment and a 2-sided Wilcoxon rank sum test comparing medians between the two treatments. UT: urease pre-treated; NT: no pre-treatment.
| Volume (µL) | NT CV (%) | UT CV (%) | p-value |
|---|---|---|---|
| 5 | 10.2 | 8.1 | 1e-3 |
| 10 | 9.7 | 5.1 | 6e-10 |
| 25 | 5.7 | 6.5 | 0.02 |
| 50 | 6.7 | 3.1 | 9e-9 |
| 100 | 16.7 | 6.2 | 3e-18 |
| All | 9.6 | 6.1 | 1e-20 |
3.2.3 Experiment 3 –Male Versus Female Urines
For Experiment 3, the CV was computed for each metabolite across technical replicates within each of the two pre-treatments, followed by averaging across biological replicates to obtain an average CV per metabolite. Of the 207 quantified metabolites, there were 168 with sufficient observations to calculate CV values for both pre-treatments. The median percent CV for NT was 14.1% versus 11.2% for UT, a difference that was significantly lower for UT based on a Wilcoxon signed rank test (p-value ~4e-20).
3.3 Differential Metabolite Abundances Across Pre-treatments
Finally, we analyzed the data from our study to identify any metabolites with statistical differences in abundances, which might reflect biases for or against specific metabolites or metabolite classes due to UT. In addition, for Experiment 2 we evaluated the data as they related to the experimental design; for example, the comparison of observed to expected metabolite abundance ratios and true positive separations between samples due to differences in urine volumes.
3.3.1 Experiment 1-Constant Urine Volume
The statistical trend for each metabolite quantified in Experiment 1 was calculated using Dunnett’s test and a G-test to compare the WT and NT groups to the UT group for quantitative and qualitative differences, respectively. The number of significantly different metabolites is reported for both tests based on a p-value threshold associated with a Bonferroni correction (Broadhurst and Kell, 2006) that accounts for the 228 metabolites evaluated (a corrected p-value threshold of ~0.00022). The G-test results are not affected by the Bonferroni correction, meaning there were no metabolites with marginal G-test evidence. Consequently, either the metabolite was observed nearly completely in one treatment or was absent from the other. Table 2 gives the number of metabolites that were significant and the associated trends based on decreases or increases in metabolite abundances in UT versus WT and/or NT. There were no trends where metabolite abundances in UT increased versus one treatment and decreased versus the other; these trends were thus excluded from Table 2. We observed that ~49% of the quantified metabolites have a significantly higher abundance or presence in the UT group versus either alternate treatment combined. In contrast, only 7% of metabolites were detected in lower abundance or presence in WT and/or NT versus UT.
Table 2.
The number and type of significant changes in metabolite abundance when comparing UT to WT and NT by either a quantitative Dunnett test (with a Bonferroni correction) or qualitative G-test. WT: water pre-treatment; NT: no pre-treatment; UT: urease pre-treatment.
| UT vs WT | UT vs NT | Interpretation | Number Significant Metabolites |
|---|---|---|---|
| UT > WT | UT > NT | Increase in UT versus WT and NT | 111 |
| UT > WT | Only increase of UT over WT | 13 | |
| UT > NT | Only increase of UT over NT | 11 | |
| UT < WT | UT < NT | Decrease in UT versus WT and NT | 17 |
Overall the numbers of metabolites with increases or decreases in abundances are highly similar for comparison of UT vs either WT or NT with either the Dunnett or G-test (Supplemental Figure S3) for which counts are combined in Table 2. The decreased abundances of metabolites in WT and NT versus UT samples is likely due to a combination of consumption of the derivatizing reagent by high levels of urea, as reported by Shoemaker and Elliot (1991), and to chromatographic effects from overwhelming urea concentrations. Indeed, we observed poor chromatographic peak shapes for metabolites that co-eluted with urea in WT and NT samples, such as for threonine (Supplemental Figure S4); these metabolites exhibited wide, flat peaks that could not be effectively defined by the software used to automatically process the GCMS data. Supporting the hypothesis of consumption of derivatizing reagent by high urea levels, the metabolites that were identified with higher abundances in UT versus WT and NT groups eluted for the most part after the region of urea elution (data not shown), and also were comprised of sugars and related molecules with large numbers of exchangeable protons (Supplemental Table S5). There were 4 identified (glyceric acid, hippuric acid, pyroglutamic acid, and phosphoric acid) and 11 unidentified metabolites that showed statistically significant increases in abundance in WT and NT versus UT, but their retention times were relatively uniformly distributed across the chromatograms.
Among the metabolites with statistically significant differences in abundances (Supplemental Tables S5 and S6), urea was found significant by G-test with presence in WT and NT but not in UT, as expected. Lactose showed significantly increased abundance in UT samples; however, lactose was added to the urease formulation as a stabilizer, similar to previous reports of metabolites present in other sources of urease (Clements and Starnes, 1975; Shoemaker and Elliott, 1991), and so its increase is likely an artifact of the urease pre-treatment. The manufacturer of the urease used in this study acknowledges that phosphate is also added to the formulation, and we detected trace amounts of dithiothreitol in methanol extracts of the urease. We did not detect dithiothreitol or phosphate in our analyses of urease pre-treated urine; interestingly, phosphoric acid was among 16 metabolites determined to be significantly increased in WT and NT versus UT samples based on Dunnett’s test and a G-test.
In a previous study, Kind et al. (2007) observed decreased abundances for aconitate, acotinate, ascorbate, citrate, glycerol, hypoxanthine, succinate, and tyrosine using GC-MS and for several unknown compounds using LC-MS after urease pre-treatment. In our study, we found that citrate, hypoxanthine, and aconitate (identified as trans-aconitate) were significantly increased in UT versus WT and NT samples (Supplemental Table S5). Similarly, tyrosine was significantly increased in UT versus WT but not NT samples (Supplemental Table S5). We also quantified glycerol, but its abundance was not significantly different between UT and WT/NT groups. As mentioned above, we identified glyceric acid, hippuric acid, pyroglutamic acid, and phosphoric acid, as well as 11 unidentified metabolites, as increased in abundance in WT/NT versus UT groups, but none of these metabolites overlap with the list reported by Kind et al. as decreased in abundance after urease pre-treatment. The differences in metabolite abundances between UT and NT in this study compared to that published by Kind et al. may be due to the number of process (n = 5) and technical (n = 3) replicates performed in this study, which can increase or decrease statistical significance for some metabolites. We also speculate that the differences between the two studies could be due to the sensitivity of the instruments utilized (quadrupole MS detector in this study versus time-of-flight (TOF) MS in that by Kind et al.); a quadrupole MS is less sensitive than a TOF MS and therefore requires more sample to be extracted in order to achieve similar coverage of the urinary metabolome. Finally, differences in the urine samples themselves in terms of the metabolome or the urine matrix may contribute to the quantitative and qualitative differences reported in the two studies.
3.3.2 Experiment 2 – Varying Urine Volumes
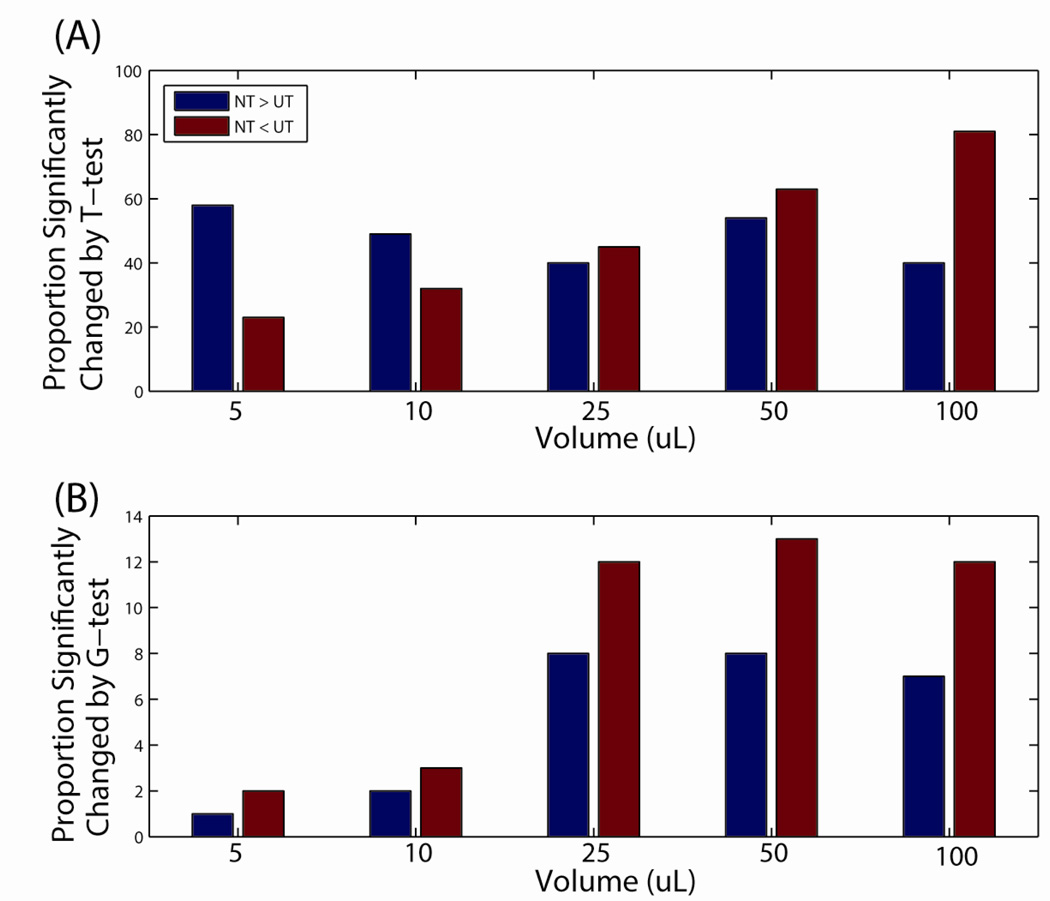
To compare differences in metabolite abundances between UT and NT, we performed both a paired t-test and a G-test on data from each urine volume (note that because n = 3, significance by G-test is only achieved when a metabolite is observed in all replicates for one treatment and in no replicates from the other treatment). Figure 3A shows the numbers of metabolites that were significantly different in abundance between the two treatments at each volume. At lower volumes (5 and 10 µL) of urine prepared, metabolites from NT treated samples were detected in higher abundance more frequently than in UT treated samples, which is consistent with the observations reported by Kind et al. (2007), who typically prepare ~10 µL of urine for GC-MS analyses (T. Kind, personal communication). However, in contrast to the previous study (Kind et al., 2007), we did not identify the same metabolites (aconitate, acotinate, ascorbate, citrate, glycerol, hypoxanthine, succinate, and tyrosine) present in lower abundance in NT vs UT. Instead, we found that 2-methyl-3-hydroxybutyrate, dehydroalanine (likely produced as an artifact of derivatization of cysteine (Kim et al., 2011)), glycine (partial derivative), hippurate, hypoxanthine, porphine, and 5 unidentified metabolites were higher in abundance in NT versus UT samples at all volumes. Again, the differences between these two studies in terms of the effects of UT could be due to experimental design, instrumentation used, or the cohort studied.
Figure 3.
The proportion of metatbolites significantly changed when comparing urine pretreatments using either a (A) t-test or (B) g-test. NT: no pre-treatment; UT: urease pre-treatment.
There were 32 metabolites found to have significantly higher abundances in NT versus UT samples at a majority (≥3) of urine volumes evaluated. Eighteen of these remain unidentified, while gluconate (25, 50, and 100 µL), glycine (complete derivative; 25, 50, and 100 µL), malate (25, 50, and 100 µL), 3-indoleacetate (10, 25, and 50 µL), fumarate (5, 10, and 50 µL), and 3-aminoisobutyrate (5, 10, 25, and 50 µL) were significantly different between the two treatments at the volumes indicated. It is difficult to identify common physical properties that might result in lower abundances for these metabolites in UT versus NT samples. As expected, urea was also consistently identified in higher abundance in NT versus UT samples. At 25 µL of urine prepared, there was roughly an equal number of metabolites detected in higher abundance in either treatment compared to the other. At 50 and 100 µL of urine, there were increasingly more metabolites detected in higher abundance in UT vs NT samples. Qualitatively, more metabolites were detected in UT versus NT samples at all volumes (Figure 3B) based on a G-test. As expected, the number of metabolites detected increased as the volume increased within each treatment (data not shown).
We also compared the data from UT samples to that from NT samples in terms of the global identification rate. There were 198 metabolites that were identified at all 5 volumes for both treatments, and 52 metabolites with missing identifications for one or more volumes and pre-treatment combinations. Table 3 shows the distribution of the identifications of these 52 metabolites across each volume/pre-treatment combination and the overlap between pretreatments within a volume. For example, at 5 µL of urine prepared there are 7 metabolites that were detected in all replicates for either NT or UT, but only 3 of these were detected in all replicates for both NT and UT. Using a z-test of proportion (Ott and Longnecker, 2008) at each volume, significant increases in complete identifications were observed for UT versus NT at volumes of 50 µL and above. As expected, we observed a clear log-linear relationship between the number of unique metabolites identified reproducibly (i.e. across all replicates) and the volume of urine prepared (Supplemental Figure S5). When the same data is plotted on a non-log scale, the increase in identifications becomes relatively flat for both treatments from 50 to 100 µL of urine. There was a higher metabolite identification rate with the UT versus NT group; however, the 95% confidence intervals for the two treatments did overlap (data not shown). Based on an F-test, there were 20 metabolites in the NT group that did not show linear behavior (p-value > 0.05; null hypothesis that the data is non-linear). In contrast, there were only 5 metabolites in the UT group that showed a non-linear relationship between abundance and urine volume.
Table 3.
The number of metabolites with complete information within each volume and pre-treatment from the 52 total metabolites with some level of missing observations in Experiment 2. UT: urease pre-treated; NT: no pre-treatment.
| Volume (µL) | NT | UT | Overlap | p-value |
|---|---|---|---|---|
| 5 | 7 | 7 | 3 | 0.5 |
| 10 | 12 | 17 | 9 | 0.14 |
| 25 | 21 | 27 | 13 | 0.12 |
| 50 | 31 | 40 | 22 | 0.03 |
| 100 | 33 | 41 | 25 | 0.04 |
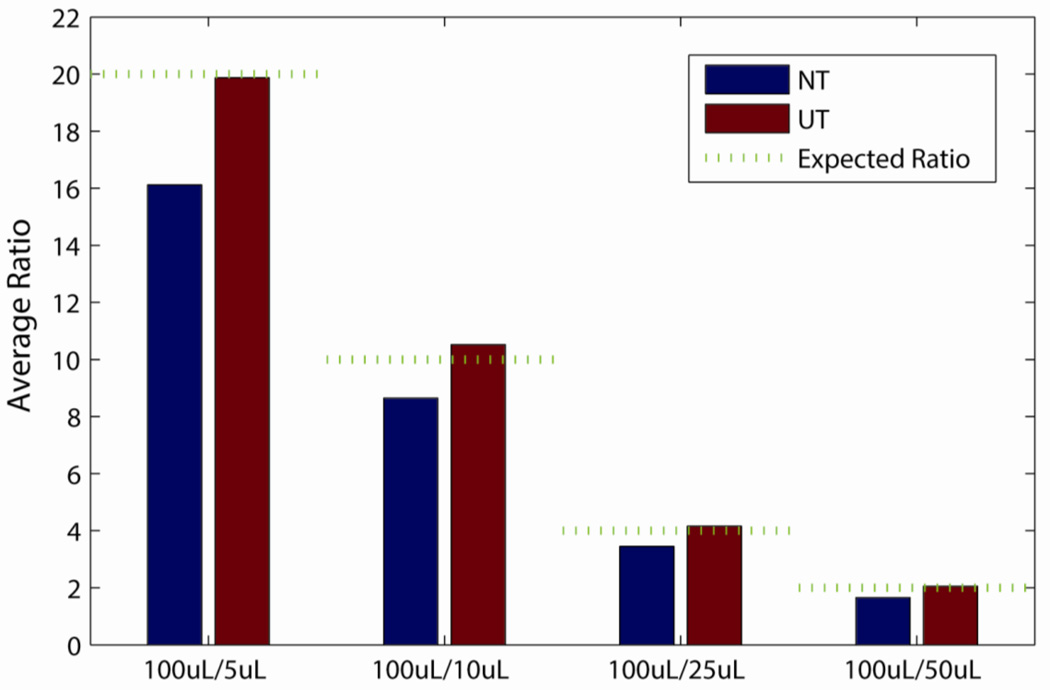
Experiment 2 data were further analyzed in the context of the expected differences in metabolite abundances due to the different urine volumes studied. Dividing the largest urine volume of 100 µL by the other volumes yields ratios of 20:1, 10:1, 4:1 and 2:1 for 5, 10, 25 and 50 µL, respectively, and Figure 4 clearly shows that the data from UT samples had experimental ratios that were closer to these expected ratios relative to that from NT samples. The observed average ratios (back-transformed from log10 scale) of the UT group were 19.9, 10.5, 4.2 and 2.1 for urine volumes of 5, 10, 25 and 50 µL, respectively. These ratios were 16.1, 8.6, 3.4 and 1.7 for the corresponding volumes in the NT group. To evaluate these differences statistically, we subtracted the expected log ratios from each of the observed ratios and performed a t-test to test the hypothesis that the ratios were drawn from a random sample with a mean of zero and an unknown variance. The p-values for UT data ranged from 0.20 to 0.91, meaning that the average ratios from this treatment were not significantly different than that expected by the design of the experiment. However, in all cases the observed ratios for NT were less than expected based on the experimental design, with p-values ranging from 2e-3 to 9e-16.
Figure 4.
The average observed ratio comparing 100 µL to 5, 10, 25 and 50 µL urine volumes for pre-treatments. The green line represents the expected ratio based on the experimental design. NT: no pre-treatment; UT: urease pre-treatment.
Lastly, we evaluated the two treatments based on the assumption that we should be able to detect increases in metabolite abundances as the volume of urine prepared increases. We performed an ANOVA within each treatment across volumes and used a Tukey’s adjusted p-value to identify the number of significantly increased metabolites as volume increased for all 10 possible volume comparisons (5 versus 10 µL, 5 versus 25 µL, 5 versus 50 µL, 5 versus 100 µL, 10 versus 25 µL, 10 versus 50 µL, 10 versus 100 µL, 25 versus 50 µL, 25 versus 100 µL and 20 versus 100 µL). This quantitative test was performed on all metabolites that were detected across all 30 samples (198 metabolites). We chose not to use a Bonferroni correction of p-values in this case due to the relatively small sample size. In all comparisons, more significantly increased metabolites were identified in the UT rather than the NT group (Supplemental Figure S6). In particular, we observed that UT does much better at identifying differences at close volumes, for example, UT identifies 179 of the 198 as significant when comparing 50 to 100 µL, but NT only identifies 149. Presumably this is related to the lower CV values for the UT group, which improves the ability to statistically discriminate between volumes.
The data presented above indicates that there are distinct relationships between the numbers of metabolite identifications and the associated metabolite abundances with the volume of urine prepared for both UT and NT as measured using GC-MS. These relationships suggest that there could be serious implications when comparing the results of GC-MS-based urinary metabolomics studies where different volumes of urine have been prepared, and caution should be exercised among the community when such comparisons are attempted. These relationships may or may not extend to metabolomics studies of other sample types using GC-MS, or to other analytical platforms
3.3.3 Experiment 3 – Male Versus Female Urines
Similar to our observations for Experiments 1 and 2, more metabolites showed a significant increase in abundance with UT (67; 32%) versus NT (24; 11%) at a Bonferroni corrected p-value (2.4e-4) in Experiment 3. For all metabolites where at least 50% of the observations were missing from either UT or NT groups, a G-test was performed to qualitatively assess the data. There were 16 metabolites that met this criterion – 8 of the 16 had significantly more observations with UT versus NT, whereas 2 had significantly more observations with NT vs UT.
We next performed a two-sample t-test on the log10 transformed abundance values of each of the 207 metabolites to determine which, if any, were significantly different between male and female urines. When applying a Bonferroni correction to the p-value threshold of 0.05 (a corrected p-value threshold of ~0.00024), we found only 1 metabolite to be statistically different between male and female urine samples in either UT (2–3-dihydroxybutyrate) or NT (Unknown 62) samples. Thus, we conclude that either there is no significant difference in the urinary polar metabolite levels of the 20 male and 20 female subjects in our study as measured by GC-MS, or that the variability in urine concentrations among these subjects was high enough to prevent the identification of significant differences. In comparative metabolomics studies of urine, it may be necessary to account for differences in metabolite concentrations by determining the specific gravity among samples and selecting the volumes of urine prepared in such a way as to ensure that the same amount of metabolites are extracted from each sample. We also conclude that urease pre-treatment of urine samples does not confound the ability to identify (or to not identify, in this case) statistical differences between comparative samples. However, in order to more effectively compare our data with previous comparisons of male and female urines, we relaxed our p-value thresholds to 0.05. Using this threshold, we determined that 43 and 41 metabolites were statistically different between male and female urine samples in either UT or NT, respectively, with an overlap of 21 metabolites. Of the metabolites that were statistically different in at least one of the treatments, 27 were identified by library matching and are shown in Table 4. The overlap in statistical significance of the identified metabolites is relatively poor – only 7 were found to be significant between the two treatments. Previous metabolomics studies of male and female urines (Pasikanti et al., 2008; Slupsky et al., 2007; Saude et al., 2007; Psihogios et al., 2008) also identified statistical differences in some of the metabolites reported here, but these studies also identified differences in metabolites that were not determined to be significant or were not detected in our study, illustrating the likely variation in both the methods and sample cohorts used.
Table 4.
Metabolites significantly different (p < 0.05, t-test) between male and female urines. UT: urease pre-treated; NT: no pre-treatment; NS: not significant; N/A: not applicable due to insufficient data. PubChem Compound numbers and IUPAC International Chemical Identifiers (InChIs) are provided in Supplemental Table S4.
| Metabolite | UT p-value | Increase in Male/Female | NT p-value | Increase in Male/Female |
|---|---|---|---|---|
| 1-methyladenosine | 0.01 | Female | 0.09 | NS |
| 1-methylhistidine# | 0.04 | Male | 0.01 | Male |
| 2,4-dihydroxybutyrate 1 | 0.01 | Female | 0.06 | NS |
| 2,3-dihydroxybutyrate 1 | 0.04 | Female | 0.09 | NS |
| 2,3-dihydroxybutyrate 2 | 4e-05 | Male | 0.02 | Male |
| 2-deoxyribonate | 0.1 | NS | 0.008 | Male |
| 3-(3-hydroxyphenyl)-3-hydroxypropionate | 0.04 | Female | 0.04 | Female |
| alpha-ketoglutarate | 0.01 | Female | 0.5 | NS |
| aminomalonate | 0.5 | NS | 0.02 | Female |
| benzoate | 0.3 | NS | 0.0006 | Female |
| citramalate | 0.0002 | Female | 0.007 | Female |
| citrate#,¥,€ | 0.01 | Female | 0.4 | NS |
| D-(+)-melezitose | 0.1 | NS | 0.02 | Female |
| dehydroalanine | 0.02 | Male | 0.9 | NS |
| D-xylose 1 | 0.002 | Female | 0.2 | NS |
| ethanolamine | 0.002 | Female | 0.05 | NS |
| fumarate¥,§ | 0.1 | NS | 0.04 | Female |
| glycerate | 0.03 | Female | N/A | NS |
| hippurate#,€ | 0.1 | NS | 0.002 | Female |
| L-(+)-lactate#,€ | 0.03 | Female | 0.009 | Female |
| L-glutamate | 0.4 | NS | 0.007 | Female |
| L-glutamine | 0.05 | NS | 0.01 | Female |
| L-threonate 2 | 0.2 | NS | 0.01 | Female |
| pantothenate | 0.003 | Female | 0.02 | Female |
| phosphate | 0.5 | NS | 0.01 | Male |
| ribose | 0.0006 | Female | 0.2 | NS |
| sucrose | 0.03 | Female | 0.04 | Female |
Also identified as significantly different in (Saude et al., 2007)
Also identified as significantly different in (Slupsky et al., 2007)
Also identified as significantly different in (Psihogios et al., 2008)
Also identified as significantly different in (Pasikanti et al., 2008)
Note: Numbers after metabolites indicate the number of TMS groups for partially derivatized molecules; otherwise, the molecule is fully derivatized.
Finally, we ran a Naïve Bayesian classifier with 5-fold cross-validation on both the complete metabolites associated with the UT and NT datasets and calculated the classification accuracy (CA) and the area under a Receiver Operating Characteristics curve (AUC) to gain a global metric of accuracy. To place confidence bounds on the metrics of accuracy the same 5-fold cross-validation subsets were evaluated for NT and UT with random 5-fold cross-validation repeated 1000 times. We observed that UT provided higher average CA and AUC compared to NT (Supplemental Table S7).
4 Concluding Remarks
Our study goal was to evaluate the effects of urease pre-treatment on the urinary metabolome as measured by GC-MS. In the three Experiments performed, we observed beneficial effects of urease pre-treatment on the resulting data, including lower CVs among process replicates, increased abundance or presence for most metabolites detected, more accurate determination of differences in metabolite abundances among different volumes, and more metabolite identifications compared to untreated samples.
The results of the volume study suggest that urease pre-treatment is particularly beneficial when working with urine volumes of 25 µL or higher; however, this observation may be biased by the relative sensitivity of the single quadrupole MS instrument used in this study, which is not as sensitive as TOF MS instrumentation and therefore requires more sample to be prepared for optimal coverage of the metabolome. A similar evaluation of urease-pretreatment and no treatment may need to be performed using a more sensitive GC-MS platform to determine if pre-treatment is as beneficial with the lower amounts of sample required for these instruments.
We feel that the benefits of pre-treatment outweigh any potential artifacts introduced to the data and recommend this protocol for all GC-MS-based studies of the urinary metabolome.
Supplementary Material
Acknowledgments
This work was funded by NIH NIDDK grant DP3 DK094343. Significant portions of the work were performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s (DOE) Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
Footnotes
Informed Consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
References
- Barr DB, Wilder LC, Caudill SP, Gonzalez AJ, Needham LL, Pirkle JL. Urinary creatinine concentrations in the U.S. population: implications for urinary biologic monitoring measurements. Environ Health Perspect. 2005;113(2):192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics. 2006;2(4):171–196. [Google Scholar]
- Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc. 2011;6(10):1483–1499. doi: 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- Clements RS, Jr, Starnes WR. An improved method for the determination of urinary myoinositol by gas-liquid chromatography. Biochem Med. 1975;12(2):200–204. doi: 10.1016/0006-2944(75)90112-x. [DOI] [PubMed] [Google Scholar]
- Egeghy PP, Cohen Hubal EA, Tulve NS, Melnyk LJ, Morgan MK, Fortmann RC, et al. Review of pesticide urinary biomarker measurements from selected US EPA children's observational exposure studies. Int J Environ Res Public Health. 2011;8(5):1727–1754. doi: 10.3390/ijerph8051727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol Oncol. 2011;29(5):551–557. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyton AC. Textbook of Medical Physiology. 6th ed. Philadelphia, PA: W. B. Saunders Company; 1981. [Google Scholar]
- Hecht SS. Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis. 2002;23(6):907–922. doi: 10.1093/carcin/23.6.907. [DOI] [PubMed] [Google Scholar]
- Hiller K, Hangebrauk J, Jager C, Spura J, Schreiber K, Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81(9):3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric Statistical Methods. Hoboken, NJ: John Wiley & Sons, Inc; 1999. [Google Scholar]
- Kim YM, Metz TO, Hu Z, Wiedner SD, Kim JS, Smith RD, et al. Formation of dehydroalanine from mimosine and cysteine: artifacts in gas chromatography/mass spectrometry based metabolomics. Rapid Commun Mass Spectrom. 2011;25(17):2561–2564. doi: 10.1002/rcm.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Schmidt BJ, Kidwai AS, Jones MB, Deatherage Kaiser BL, Brewer HM, et al. Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes. Mol Biosyst. 2013 doi: 10.1039/c3mb25598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem. 2007;363(2):185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara T. Noninvasive human metabolome analysis for differential diagnosis of inborn errors of metabolism. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855(1):42–50. doi: 10.1016/j.jchromb.2007.03.031. [DOI] [PubMed] [Google Scholar]
- Kussmann M, Raymond F, Affolter M. OMICS-driven biomarker discovery in nutrition and health. J Biotechnol. 2006;124(4):758–787. doi: 10.1016/j.jbiotec.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Matsumoto I, Kuhara T. A new chemical diagnostic method for inborn errors of metabolism by mass spectrometry - Rapid, practical, and simultaneous urinary metabolites analysis. Mass Spectrometry Reviews. 1996;15(1):43–57. doi: 10.1002/(SICI)1098-2787(1996)15:1<43::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Zhang C, Shinka T, Inoue Y, Furumoto T, Kuhara T, et al. The chemical diagnosis of the metabolic disorders 1. Chemical diagnosis of propionic acidemia. J Kanazawa Med Univ. 1994;19:213–219. [Google Scholar]
- Matzke MM, Waters KM, Metz TO, Jacobs JM, Sims AC, Baric RS, et al. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics. 2011;27(20):2866–2872. doi: 10.1093/bioinformatics/btr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker JD, Sathyanarayana S, Swan SH. Phthalates and other additives in plastics: human exposure and associated health outcomes. Philos Trans R Soc Lond B Biol Sci. 2009;364(1526):2097–2113. doi: 10.1098/rstb.2008.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz TO, Zhang Q, Page JS, Shen Y, Callister SJ, Jacobs JM, et al. The future of liquid chromatography-mass spectrometry (LC-MS) in metabolic profiling and metabolomic studies for biomarker discovery. Biomark Med. 2007;1(1):159–185. doi: 10.2217/17520363.1.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott RL, Longnecker M. An Introduction to Statistical Methods and Data Analysis. Belmont, CA: Brooks/Cole; 2008. [Google Scholar]
- Pasikanti KK, Ho PC, Chan EC. Development and validation of a gas chromatography/mass spectrometry metabonomic platform for the global profiling of urinary metabolites. Rapid Commun Mass Spectrom. 2008;22(19):2984–2992. doi: 10.1002/rcm.3699. [DOI] [PubMed] [Google Scholar]
- Psihogios NG, Gazi IF, Elisaf MS, Seferiadis KI, Bairaktari ET. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR Biomed. 2008;21(3):195–207. doi: 10.1002/nbm.1176. [DOI] [PubMed] [Google Scholar]
- Putnam DF. Composition and Concentrative Properties of Human Urine. Huntington Beach, California: National Aeronautics and Space Administration; 1971. p. 112. [Google Scholar]
- Roberts LJ, Morrow JD. Measurement of F(2)-isoprostanes as an index of oxidative stress in vivo. Free Radic Biol Med. 2000;28(4):505–513. doi: 10.1016/s0891-5849(99)00264-6. [DOI] [PubMed] [Google Scholar]
- Saude EJ, Adamko D, Rowe BH, Marrie T, Sykes BD. Variation of metabolites in normal human urine. Metabolomics. 2007;3(4):439–451. [Google Scholar]
- Shelby MK, Crouch DJ, Black DL, Robert TA, Heltsley R. Screening indicators of dehydroepiandosterone, and rostenedione, and dihydrotestosterone use: a literature review. J Anal Toxicol. 2011;35(9):638–655. doi: 10.1093/anatox/35.9.638. [DOI] [PubMed] [Google Scholar]
- Shoemaker JD, Elliott WH. Automated screening of urine samples for carbohydrates, organic and amino acids after treatment with urease. J Chromatogr. 1991;562(1–2):125–138. doi: 10.1016/0378-4347(91)80571-s. [DOI] [PubMed] [Google Scholar]
- Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79(18):6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson BJ, Matzke MM, Jacobs JM, Pounds JG, Waters KM. A statistical selection strategy for normalization procedures in LC-MS proteomics experiments through dataset-dependent ranking of normalization scaling factors. Proteomics. 2011;11(24):4736–4741. doi: 10.1002/pmic.201100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson BJ, Matzke MM, Metz TO, McDermott JE, Walker H, Rodland KD, et al. Sequential projection pursuit principal component analysis - dealing with missing data associated with new -omics technologies. Biotechniques. 2013;54(3):165–168. doi: 10.2144/000113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson BJ, McCue LA, Waters KM, Matzke MM, Jacobs JM, Metz TO, et al. Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J Proteome Res. 2010;9(11):5748–5756. doi: 10.1021/pr1005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WW, Chin T, Weber B. Quantitative Analysis of Serum and Urine Sugars by Gas Chromatography. Clin Chim Acta. 1964;10:352–359. doi: 10.1016/0009-8981(64)90066-x. [DOI] [PubMed] [Google Scholar]
- Wilkins JN. Quantitative urine levels of cocaine and other substances of abuse. NIDA Res Monogr. 1997;175:235–252. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.