Abstract
Electronic medical records (EMR) and treatment plans are used in research on patient outcomes and radiation effects. In many situations researchers must remove protected health information (PHI) from EMRs. The literature contains several studies describing the anonymization of generic Digital Imaging and Communication in Medicine (DICOM) files and DICOM image sets but no publications were found that discuss the anonymization of DICOM radiation therapy plans, a key component of an EMR in a cancer clinic. In addition to this we were unable to find a commercial software tool that met the minimum requirements for anonymization and preservation of data integrity for radiation therapy research. The purpose of this study was to develop a prototype software code to meet the requirements for the anonymization of radiation therapy treatment plans and to develop a way to validate that code and demonstrate that it properly anonymized treatment plans and preserved data integrity. We extended an open-source code to process all relevant PHI and to allow for the automatic anonymization of multiple EMRs. The prototype code successfully anonymized multiple treatment plans in less than 1 minute per patient. We also tested commercial optical character recognition (OCR) algorithms for the detection of burned-in text on the images, but they were unable to reliably recognize text. In addition, we developed and tested an image filtering algorithm that allowed us to isolate and redact alpha-numeric text from a test radiograph. Validation tests verified that PHI was anonymized and data integrity, such as the relationship between DICOM unique identifiers (UID) was preserved.
Keywords: Digital Imaging and Communication in Medicine (DICOM), anonymize, radiation oncology, protected health information
1. Introduction
The biological effect of ionizing radiation on humans has been researched intensively for more than a century. Some effects may occur years or even decades after exposure and may include an increase in the risk for developing cancer, cognitive deficits, fertility problems, and other chronic health issues. [1, 2] Despite monumental research efforts and considerable progress, our knowledge of effects of radiation in humans is incomplete. To some extent, one may bridge the gaps in knowledge by extrapolating from experimental results from animals, invitro cell cultures, and subcellular structures. [3] However, the validity of such extrapolations to effects in humans is difficult to establish with certainty. An attractive alternative approach is to conduct clinical trials and epidemiological studies of populations of patients who received radiation exposures from diagnostic or therapeutic medical procedures. [4]
In radiation epidemiology studies, the process of reconstructing radiation dose from abstracted paper medical records introduces substantial uncertainties in the estimates of radiation dose. [5] This may involve the translation of patient records from foreign languages, transcription of handwritten records, and dealing with incomplete or missing data on the patient’s anatomy and radiation treatment fields. In recent years, great strides have been made in standardizing the reporting of radiotherapy treatments, including terminology. [6–8] Recently, internationally standardized methods have emerged for the electronic storage and exchange of medical data for diagnostic radiology, such as the Digital Imaging and Communication in Medicine (DICOM) standards committee [9] and by Integrating the Healthcare Environment (IHE) group. [10, 11] The standards include capabilities specifically for radiation oncology. [12, 13]
In the future, investigations of radiation effects will increasingly utilize electronic medical records (EMRs) containing protected health information (PHI). For ethical and legal reasons, researchers are required to anonymize patient data before they can be made available to the public. In the United States this means complying with the Health Insurance Portability and Accountability Act of 1996 (HIPAA). [14] To date several works have discussed techniques and methods for anonymizing DICOM image sets and generic DICOM files.[15–21] While DICOM Working Group 18 published a comprehensive list of tags to be anonymized, [22] no publications are available discussing the anonymization of treatment plans for radiation therapy. In addition to this we were unable to find a commercial software product that met our requirements for the anonymization of treatment plans. These requirements included automatic anonymization of multiple EMRs and the anonymization of DICOM tags listed in DICOM supplement 142, [22] which is an extension of the DICOM standard specifically related to de-identification of patient records for the clinical trials. It arose due to the determination that DICOM confidentiality profile PS 3.15 did not sufficiently protect identities for patient records used in clinical trials.
The objective of this work is to develop and test a prototype software code to anonymized EMRs for patient, while maintaining the integrity of the files and the data within them. We modified an existing open source DICOM anonymizer [23] following the recommendations from DICOM Supplement 142 for the anonymization of radiation therapy data. The software was validated for compliance with HIPAA and verified that the integrity of the data was maintained.
2. Methods and materials
2.1 Classifications of research involving private information
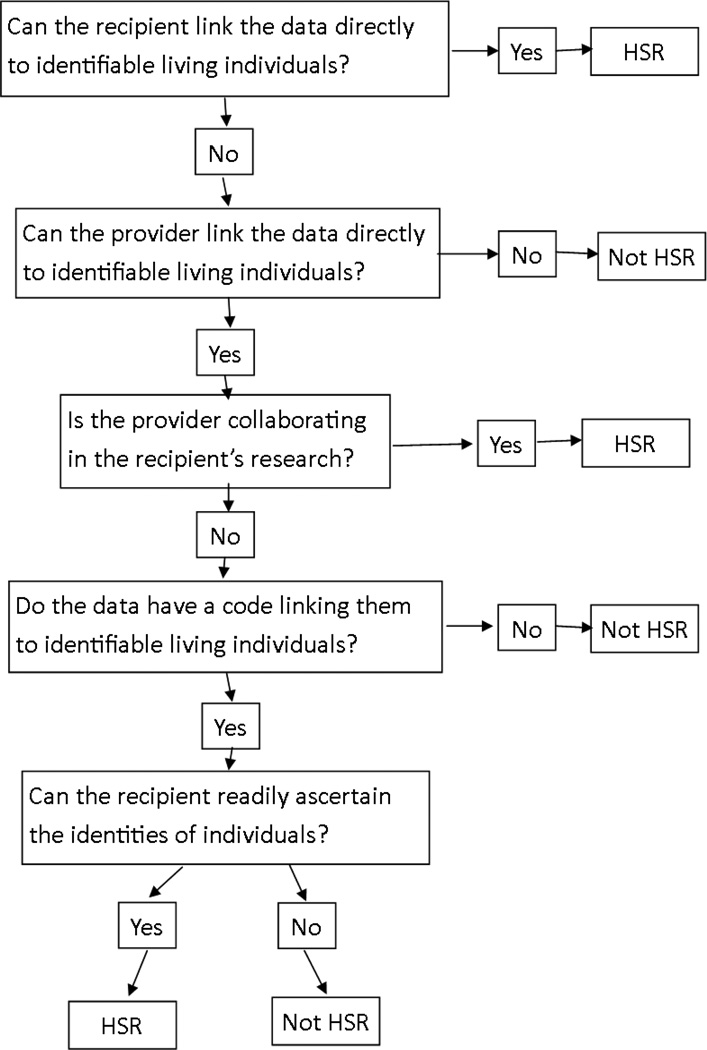
The processing of private electronic health information will vary with the intended use and recipients of the data. For example the NIH classifies research involving human as Human Subjects Research (HSR) or Not Human Subjects Research (NHSR) using a decision making flowchart. [24] Figure 1 shows a flowchart that was based on the DHSS flowchart and simplified to focus on the objectives of this study. In general, the HSR requires more processing and administrative oversight than NHSR and consequently may be more complicated, computationally expensive, and time consuming. Therefore, in many cases in which it is not required, it will be preferable to anonymize the data in a manner that allows the investigation to be classified as NHSR.
Fig. 1.
Decision making flowchart for classifying human data as human-subjects research (HSR) or non human-subjects research (NHSR). The flowchart is for classifying EMRs to be used in radiation therapy research and was adapted from a more comprehensive guidance document [24].
2.2 Protected health information (PHI) of relevance to anonimization
Table 1 lists the protected health information (PHI) that were deleted, overwritten, or created during the anonymization process. The list is a minimum set of attributes to be processed as recommended in DICOM Supplement 142. [22] Depending on institutional practices, attributes which did not contain PHI at our institution could contain PHI at another institution. For example a comment field could contain the name of a patient or clinician. Additionally records generated by other radiotherapy information systems may use attributes that were not used here and they may have additional data stored in private tags (User defined attributes that are, by definition, not included in DICOM specifications).
Table 1.
DICOM fields containing protected health information of relevance to anonymization in our data sets and the needed action to those information (d: delete, w: overwrite).
| Attribute Name | Tag | Action |
|---|---|---|
| Instance Creator UID | (0008,0014) | w |
| SOP Instance UID | (0008,0018) | w |
| Accession Number | (0008,0050) | w |
| Institution Name | (0008,0080) | d |
| Institution Address | (0008,0081) | d |
| Referring Physician’s Name | (0008,0090) | d |
| Referring Physician’s Address | (0008,0092) | d |
| Referring Physician’s Telephone Numbers | (0008,0094) | d |
| Station Name | (0008,1010) | d |
| Study Description | (0008,1030) | d |
| Series Description | (0008,103E) | d |
| Institutional Department Name | (0008,1040) | d |
| Physician(s) of Record | (0008,1048) | d |
| Performing Physicians’ Name | (0008,1050) | d |
| Name of Physician(s) Reading Study | (0008,1060) | d |
| Operators’ Name | (0008,1070) | d |
| Admitting Diagnoses Description | (0008,1080) | d |
| Referenced SOP Instance UID | (0008,1155) | w |
| Derivation Description | (0008,2111) | d |
| Patient’s Name | (0010,0010) | w |
| Patient ID | (0010,0020) | w |
| Patient’s Birth Date | (0010,0030) | w |
| Patient’s Birth Time | (0010,0032) | w |
| Patient’s Sex | (0010,0040) | w |
| Other Patient Ids | (0010,1000) | w |
| Other Patient Names | (0010,1001) | w |
| Patient’s Age | (0010,1010) | w |
| Patient’s Size | (0010,1020) | w |
| Patient’s Weight | (0010,1030) | w |
| Medical Record Locator | (0010,1090) | d |
| Ethnic Group | (0010,2160) | d |
| Occupation | (0010,2180) | d |
| Additional Patient’s History | (0010,21B0) | d |
| Patient Comments | (0010,4000) | d |
| Device Serial Number | (0018,1000) | w |
| Protocol Name | (0018,1030) | d |
| Study Instance UID | (0020,000D) | w |
| Series Instance UID | (0020,000E) | w |
| Study ID | (0020,0010) | w |
| Frame of Reference UID | (0020,0052) | w |
| Synchronization Frame of Reference UID | (0020,0200) | w |
| Image Comments | (0020,4000) | d |
| Request Attributes Sequence | (0040,0275) | d |
| UID | (0040,A124) | w |
| Content Sequence | (0040,A730) | d |
| Storage Media File-set UID | (0088,0140) | w |
| Referenced Frame of Reference UID | (3006,0024) | w |
| Related Frame of Reference UID | (3006,00C2) | w |
2.3 DICOM-RT method for storing private information on patients and their treatments
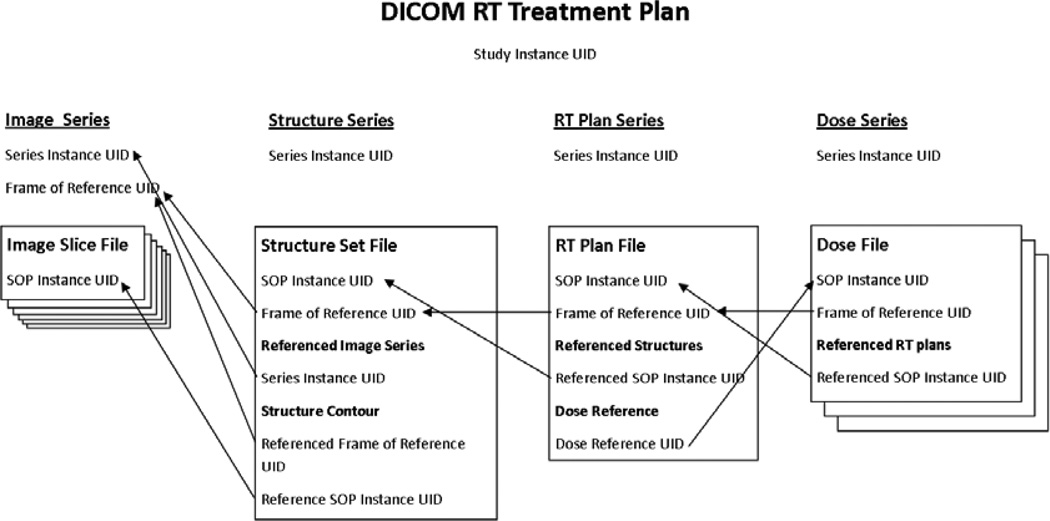
DICOM radiation therapy treatment plans typically consist of an image series, a treatment plan file, one or more dose files, and a structure set file. DICOM utilizes unique identifiers (UIDs) to identify and describe the relationship between files. Figure 2 illustrates the various relationships between UIDs in DICOM treatment records. Instance UIDs are given to each individual file as well as each series and study (treatment plan). Reference UID tags are used to describe which files are meant to be associated with a particular tag. For example a structure set file contains contours for anatomical structures used in the treatment plan. These are stored in the form of coordinates that outline the structure on each image slice. Each DICOM file has a Service Object pair (SOP) instance UID which identifies that file. In the case of structure in the structure set file the referenced SOP instance UID is used to identify the image slice the coordinates are meant to correspond with. Anonymization codes should modify the instance UIDs to ensure that the data will not clash in a picture archiving and communications system (PACS), and also to ensure anonymity. [17] Importantly, the corresponding referenced UIDs must also be changed consistently across an entire EMR to preserve the relationship between various data files.
Fig. 2.
Relationships between various UIDs used in DICOM RT treatment plans.
2.4 Software to anonymize DICOM-RT private information
We enhanced the DVTK anonymizer [23], which was already capable of anonymizing many PHI data items. We extended the code to process additional PHI and UIDs. Specifically we added processing of the attributes noted in table 1 including the following UIDs: Frame Of Reference UID, Referenced Frame Of Reference UID, and Referenced SOP Instance UID. All of the attributes listed in table 1 were overwritten or removed. For a given session (e.g., one execution of the code on a directory containing the medical record for one patient), the UIDs and references to those UIDs were anonymized but kept consistent. The code was also extended to automatically operate on a directory tree, meaning records for multiple patients were anonymized in one session. We also added object definition files, which includes DICOM-RTION related SOP Classes. The object definitions provided in the “standard installation package” for the DVTK Anonymizer lacked these definitions.
2.5 Validation of anonymization software
The anonymization software was validated using DICOM-RT and DICOM-RT-ION treatment plans exported from a commercial radiotherapy treatment planning system (Eclipse; Varian Medical Systems, Inc., Palo Alto, CA). After each treatment plan was anonymized it was tested in three ways. First, it was re-imported into the treatment planning system to ensure the integrity of data and relationships between various data were preserved. For each plan, we checked key treatment beam parameters, the dose prescription, and dose values at 10 anatomical locations to verify that they had not changed during the anonymization process. This test also ensures that the treatment planning system does not re-associate the anonymized treatment plan with the original treatment plan, which would result in the re-identification of the anonymized treatment plan. Second, a script was used to output the metatag and name for each element that was anonymized. This list was compared to the list in table 1 to ensure that elements containing PHI were anonymized. Finally, the anonymized EMRs were converted into text files and electronically searched for PHI to verify that all of the attributes in table 1 had been anonymized.
2.6 Electronic medical records used for validation
The EMRs used in this study were taken from NIH funded studies involving treatments for prostate cancer, medulloblastoma, and Hodgkin lymphoma. The prostate cancer EMRs consisted of proton therapy treatment plans for 13 patients as described by Fontenot et al. [25] The medulloblastoma records consisted of 1 conventional photon therapy plan, 1 IMRT plan, and 1 proton therapy plan each for 2 patients treated with craniospinal irradiation for medulloblastoma, as described by Newhauser et al. [26] and Howell et al. [27]. The Hodgkin lymphoma records consisted of 1 IMRT treatment plan, 1 mantle field treatment plan, and 1 anterior posterior (AP)-posterior anterior (PA) treatment plan each for 1 patient. All plans were created according to the prevailing standard of care at The University of Texas MD Anderson Cancer Center (UT MDACC).
2.7 Detection, Recognition, and Removal of Textual PHI in Image
In addition to the anonymization of encoded textual DICOM elements, one may need to detect and/or anonymize textual PHI which is burned into medical images. We implemented the mature and widely used open-source Tesseract optical character recognition (OCR) engine sponsored and hosted by Google [28] to explore this problem. This approach presented two challenges. First, the algorithm operated only on bi-tonal images, whereas our images were grayscale. This necessitated a binarization process, in which each pixel was converted to either white or black depending on a predefined threshold brightness value. The second challenge was that the OCR algorithm was optimized for typical office correspondence documents that are 8.5”×11” (letter) or 8.5”×14” (legal) sizes. Typically these documents are scanned at a resolution between 200 and 400 dpi, resulting in an image that is about 1700×2200 pixels. Text in an 8 point font scanned at this resolution would be approximately 22 pixels tall. In contrast typically CT images are typically 512×512 pixels.
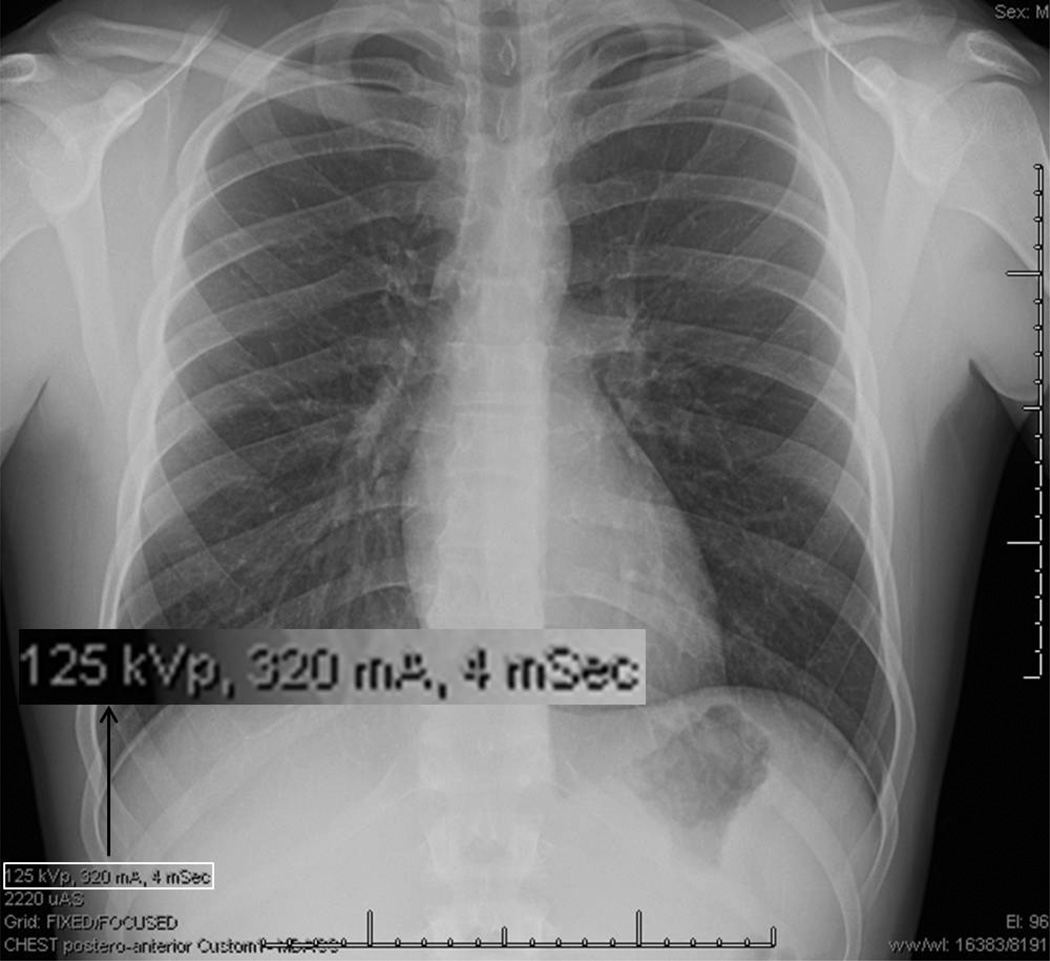
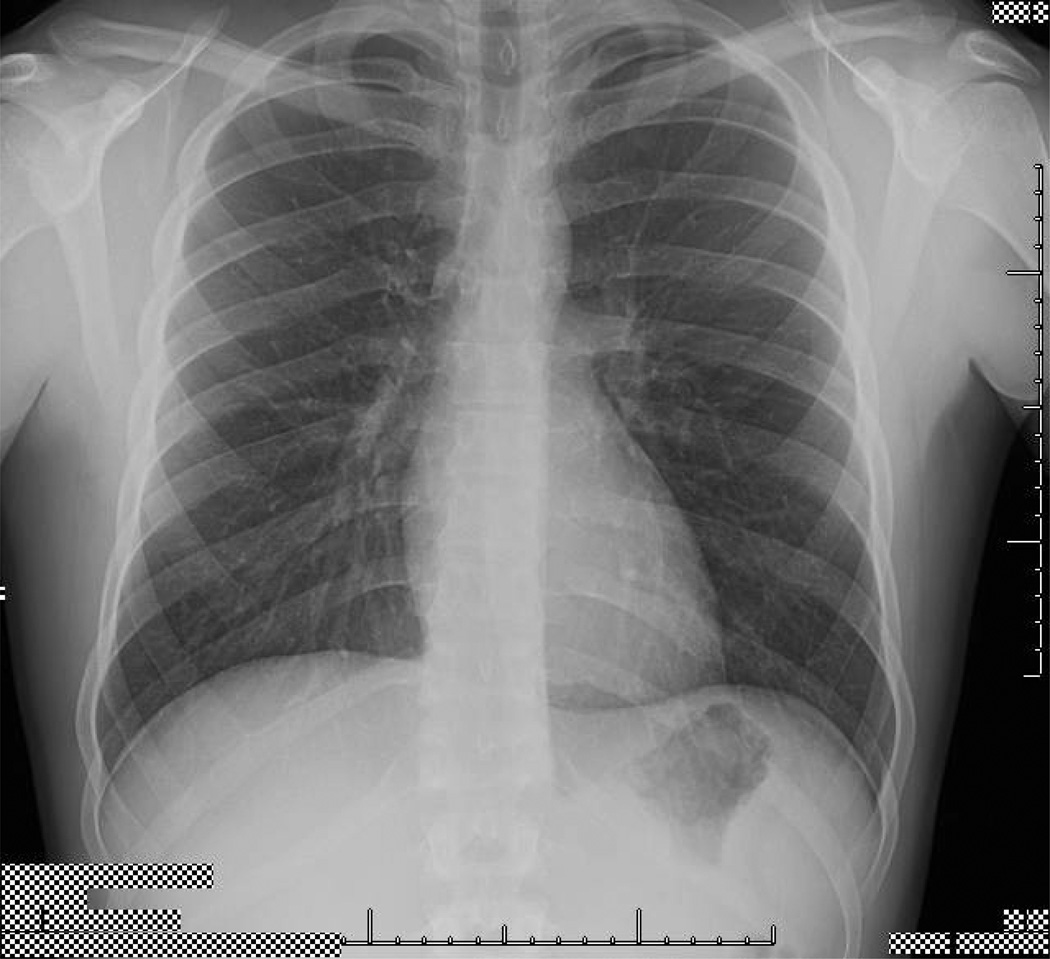
The performance of the OCR algorithm was tested on a sample radiographic chest exam image with burned in text from UTMDACC (cf. Fig 3). This image was 1465 × 800 pixels and the text height was approximately 7–9 pixels tall. This resolution is significantly less than the neural network in the OCR algorithm was designed to operate on, e.g., with accuracy dropping off rapidly below 8 pt at 300 dpi and at a text height of below about 8 pixels, the algorithm will remove most of the text during the noise removal process. Indeed, at this low resolution, the OCR algorithm was unable to reliably identify the text characters burned in our test image, indicating the need for alternative and/or additional image processing methods.
Fig. 3.
Chest radiograph used to test methods described in the text to detect burned-in alphanumeric characters. The inset is an enlarged example of alpha-numeric information that must be detected, bounded, and redacted.
The first strategy we investigated comprises two key additional steps; detecting a subregion of the image that contains suspected text then resampling the subregion at a high resolution (corresponding to a character cap height of 31 pixels), i.e., at the resolution the OCR algorithm was designed to operate. To accomplish this, we used the ImageJ software [29] from the National Institutes of Health because it provided many image processing functions and a scripting capability that were of relevance to this work. Specifically, we successfully isolated the text from the image by setting the brightness threshold to a lower value of 1 and upper value of 255, with a white background. We then resampled the image using bicubic interpolation to a total size of 5448 × 2975 pixels, corresponding to a 5-fold increase in the number of pixels per character height. This can be thought of as effectively amplifying the signal and noise, and preserving the signal-to-noise ratio (SNR). However, each of the characters in the interpolated image still contained defects (cf. Fig 4b). Specifically, undesired features were present in various strokes, including gaps in the spine of the letter s; the vertical strokes of k, p, and 4; angled strokes of 1, 5, k, V, 2, A, and 4; the arches of m; the closed rounded stroke (bowl) of p; and the trailing outstrokes of k. In addition, there were openings in closed counters of A, 0, and e; the ears of p and m were missing, and other defects. However, using an interpolated image facilitated experimentation aimed at repairing pixel defects in the individual characters. Specifically, we used with additional standard image preprocessing techniques, such as dilation (i.e., thickening by adding pixels to the edge of an object), erosion (thinning by removing pixels), smoothing, sharpening, despeckling and other mathematical image filtering algorithms. This overall method will be subsequently referred to as the interpolation-OCR method.
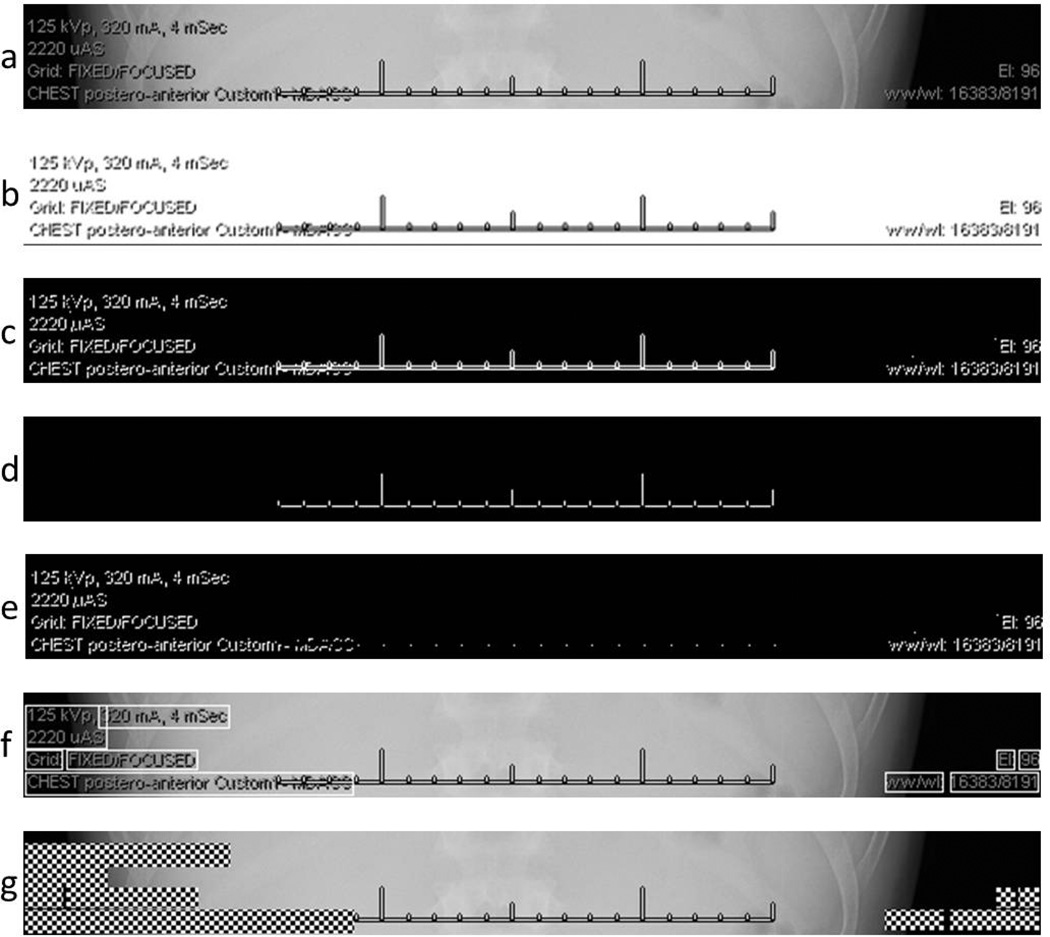
Fig. 4.
Intermediate and final results of the PHI detection and redaction processes. (a) The subregion of the original un-altered radiograph as described in Fig. 3. (b) The original image after application of the interpolation-OCR method, involving a brightness threshold filtering followed by bicubic interpolation. This image was tested with OCR algorithms without success. The threshold-redaction algorithm was as follows, with the step letter indicating the corresponding sub-figure: (c) The original image after application of a low-threshold filter. (d) The original image after application of a high-threshold filter. (e) An image obtained after performing a single pixel expansion of image 4d and subtract it from image 4c. (f) Determination of bounding boxes overlaid on the original image. (g) Redacted of alpha-numeric information inside the bounding boxes suing a checkerboard pattern applied to the original image.
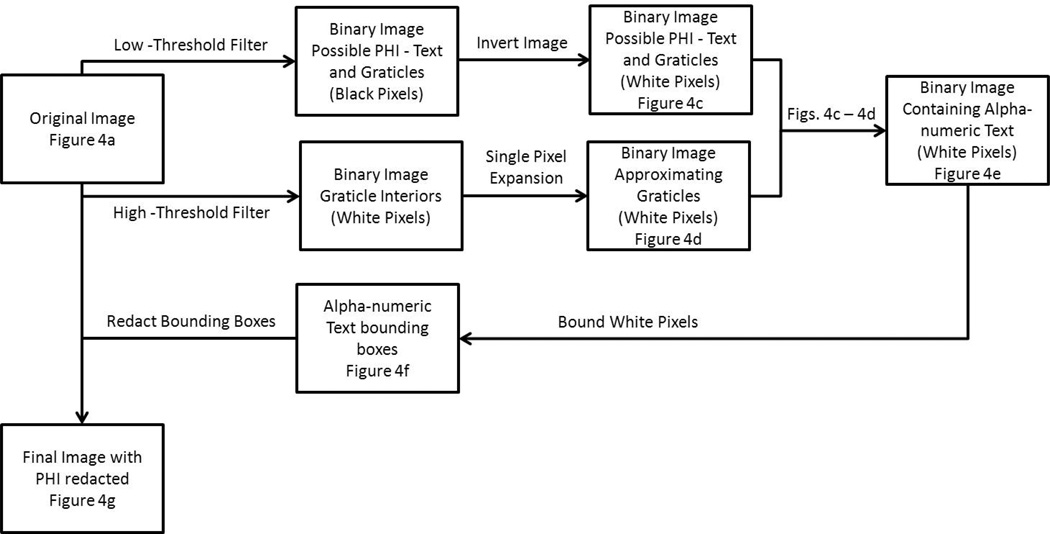
The second strategy we tested was to detect sub-regions of the image possibly containing one or more alpha-numeric characters. We redacted (erased) these sub-region region(s) to remove PHI by reassigning the pixel values of a sub-region. This approach had the advantage that PHI could be detected and removed without the necessity to perform OCR. To implement this strategy, we began by filtering the original image and determining the bounding boxes of sub-regions containing alpha-numeric characters. Only the pixels inside the bounding boxes were overwritten with a visibly obvious redaction pattern, i.e., a black and white checkerboard pattern. With this approach, the redacted image remained identical to the original image, except in the redacted regions. This method will be subsequently referred to as the threshold-redaction algorithm. Figure 5 shows the flow diagram for this threshold-redaction algorithm.
Fig. 5.
The flow diagram for threshold-redaction algorithm used in this work.
The threshold-redaction algorithm began with identifying the areas of the original image containing alpha-numeric characters. First, the low-threshold filter, which is a binarization filter that converts a grayscale image to a binary image with a threshold value of 1/255, was applied to the original image and the negative of the result was taken (Fig. 4c). This low-threshold filter isolated the non-radiographic information (i.e., alpha-numeric characters and graticules) from the original image. Then, the high-threshold filter, which has a threshold value of 244/255, was applied to the original image. The high-threshold filter isolated the completely white pixels in the original image, i.e., the graticules (cf. Fig. 4d). Since the graticules shown in Fig. 4d are the interior of the graticules shown in Fig. 4a, a one pixel expansion was applied to of all white pixels in Fig. 4d to approximate the actual size of the graticules. After applying this pixel dilation to Fig. 4d, the resulting image was subtracted from that in Fig. 4c, creating a binary image (Fig. 4e) with white pixels where the original image contained alpha-numeric information.
After identifying the pixels of interest, bounding boxes were generated to encapsulate these pixels. However, two image reduction steps were applied to Fig. 4e before the bounding boxes were determined. All lone white pixels were removed and the remaining white pixels underwent a one pixel expansion, which filled in the spaces between words and created fewer bounding boxes. Then, bounding boxes were determined via a built-in Matlab function (“regionprops”) that can be used to identify regions of connected white pixels in a binary image (Fig. 4f). Finally, after the bounding box regions were identified, the corresponding regions of the original image were redacted with a checker board pattern (Fig. 4g). The results of the threshold-redaction algorithm applied to the entire original image are shown in Fig. 6.
Fig. 6.
The final anonomized radiographic image, with PHI information redacted (checkerboard pattern) while preserving the integrity of the original radiographic information, including anatomy and burned-in graticules.
3. Results
3.1 Automated anonymization of PHI in DICOM-RT in EMRs
With the prototype code described in this work, we anonymized 17 EMRs. The tags listed in table 1 in DICOM files were deleted or overwritten in the treatment plans. The code was capable of anonymizing a maximum of 7 patients per session. The average anonymization time was less than 1 minute per patient, which showed the anonymization method is suitable for large population-based research studies.
3.2 Validation of data integrity and removal of PHI
All anonymized treatment plans were successfully imported into the treatment planning system without errors or re-association with the original treatment plans, indicating that the UIDs were changed and the relationships between them were maintained. The dose prescription, beam parameters and dose values were identical to the original non-anonymized treatment plans. When the lists of anonymized tags for each DICOM files were compared to the list in table 1 of this document, all of the recommended tags had been anonymized. No PHI was found when searching the textual version of the DICOM files, indicating that no PHI was present in tags which were not anonymized.
3.3 Detection, Recognition, and Redaction of Text in Images
The OCR algorithm was unable to reliably recognize the text characters burned in our original test image a resampled copy of it.. However, the threshold-redaction algorithm worked well (cf. Fig. 5). The prototype script demonstrated that detection and removal of burned-in text can be automated with Matlab, an important aspect when anonymizing large numbers of images.
4. Discussion
Our results show that the prototype software successfully removed the PHI listed in DICOM supplement 142 while maintaining the integrity of the data and data relationships. While the OCR method evaluated was not able to identify text in a sample radiology image, the removal of text from the CT images in the EMRs we used was not necessary because they contained no burned-in text. OCR, when accurately and reliably implemented, will enable interrogation of the recognized text for PHI content. PHI can be then be redacted while preserving other potentially valuable information, such as information about the imaging technique used and the image itself. Our attempts at OCR revealed some of the technical obstacles and may help to inform future attempts to overcome them. Our experience suggests that finding and validating solutions will be non-trivial; we speculate that solutions will be found. While the anonymization of full DICOM RT treatment plans has similar requirements as the anonymization of DICOM image sets, this was to our knowledge the first attempt in the literature to address the additional concerns and test compatibility with a radiotherapy planning system which is sensitive to changes to UIDs. All of the commercial DICOM anonymizers we investigated did not attempt to anonymize the UIDs in the DICOM files.
While a UID is not generally considered patient identifiable information, we chose to anonymize these because in some early attempts the treatment planning system automatically linked the anonymized data to the original data, unintentionally re-identifying the data.
The major limitation of this study is that we used only one radiotherapy planning system at one institution. Other systems and institutions are potentially similar but it is unknown whether or not the results would be applicable to other groups. In particular, the DICOM tags listed are only the subset of tags in DICOM supplement 142 which were being used by our radiotherapy planning system. If the prototype anonymization software was to be used at another institution or with a different radiotherapy planning system, similar tests would need to be done to verify compatibility with other radiotherapy planning systems and to insure that patient information is not stored in other text fields or in private tags. We also expect that digital images in medical records will vary greatly in terms of PHI content, methods used to represent it, and image quality. For example, we have seen some electronic medical records that contain digitized images of noisy scanned films containing hand written PHI in cursive. Clearly, this is an extreme example, but for the field of radiation epidemiology this is a highly relevant issue. Hence, we expect a large degree of variability in the images to be interrogated, which may require additional algorithm development and validation testing. The other limitation is that we used multiple existed software programs instead of developing an integrated software. However, the main value of our study is the information regarding what anonymization involves. A robust, integrated code would be desirable. Unfortunately, the development of a generally applicable code that is suitable for routine clinical use is well beyond our scope. Mainly this is due to constraints on our time and resources available for this kind of work.
Our finding support the premise that it is possible to automatically de-identify full DICOM RT treatment plans, while maintaining the integrity of the data and data relationships. Our findings may be useful to other research groups that conduct treatment planning studies with radiotherapy treatment plans (software is available upon request).
Highlights.
We extended an open-source code to process multiple EMRs automatically.
We tested commercial optical character recognition (OCR) algorithm for the detection of burned-in text on a test image.
OCR was unable to recognize the burned-in text reliably.
We also developed and tested an image filtering algorithm to redact burned-in text from the test radiograph.
Validation tests verified that PHI was anonymized and data integrity was preserved.
Acknowledgments
The authors are grateful to Drs. Walter Bosch, Kenneth Homann, and Phillip Taddei for helpful discussions. This work was supported in part by the National Cancer Institute (award 1 R01 CA131463-01A1) and Northern Illinois University through a subcontract of a Department of Defense contract (award W81XH-08-1-0205).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Diller L, Chow EJ, Gurney JG, Hudson MM, Kadin-Lottick NS, Kawashima TI, Leisenring WM, Meacham LR, Mertens AC, Mulrooney DA, Oeffinger KC, Packer RJ, Robison LL, Sklar CA. Chronic disease in the Childhood Cancer Survivor Study cohort: a review of published findings. J Clin Oncol. 2009;27:2339–2355. doi: 10.1200/JCO.2008.21.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robison LL, Armstrong GT, Boice JD, Chow EJ, Davies SM, Donaldson SS, Green DM, Hammond S, Meadows AT, Mertens AC, Mulvihill JJ, Nathan PC, Neglia JP, Packer RJ, Rajaraman P, Sklar CA, Stovall M, Strong LC, Yasui Y, Zeltzer LK. The Childhood Cancer Survivor Study: a National Cancer Institute-supported resource for outcome and intervention research. J Clin Oncol. 2009;27:2308–2318. doi: 10.1200/JCO.2009.22.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NRC. Health Risks from Exposure to Low Levels of Ionizing Radation: BEIR VII - Phase 2. Washington, D.C.: Nation Research Council of the National Academies; 2006. [PubMed] [Google Scholar]
- 4.Curtis RE. U.S. Dept. of Health and Human Services, National Institutes of Health. Washington, D.C.: National Cancer Institute; 2006. New malignancies among cancer survivors : SEER cancer registries, 1973–2000. [Google Scholar]
- 5.Stovall M, Weathers R, Kasper C, Smith SA, Travis L, Ron E, Kleinerman R. Dose reconstruction for therapeutic and diagnostic radiation exposures: use in epidemiological studies. Radiat Res. 2006;166:141–157. doi: 10.1667/RR3525.1. [DOI] [PubMed] [Google Scholar]
- 6.ICRU. ICRU Report 50. Bethesda, MD: ICRU; 1993. Prescribing, recording, and reporting photon beam therapy. [Google Scholar]
- 7.ICRU. Supplement to ICRU Report 50. Bethesda, MD: ICRU; 1999. ICRU Report 62 Prescribing, recording, and reporting photon beam therapy. [Google Scholar]
- 8.ICRU. ICRU Report No 78. Bethesda, MD: ICRU; 2007. Prescribing, Recording and Reporting Proton Beam Therapy. [Google Scholar]
- 9.NEMA. Digital Imaging and Communications in Medicine (DICOM) Part 1: Introduction and Overview. Rosslyn, Virginia, USA: National Electrical Manufacturers Association; 2008. [Google Scholar]
- 10.IHE. IHE Radiology Technical Framework. Supplement 2007–2008, Radiation Exposure Monitoring (REM) Integration Profile. 2008 [Google Scholar]
- 11.IHE. IHE Radiology Technical Framework - Revision 9.0 (RAD-TF V9.0) 2009 [Google Scholar]
- 12.IHE. Integrating the Healthcare Enterprise, IHE Radiation Oncology. Technical Framework. 2011;1 (RO TF-1), Revision 1.7. [Google Scholar]
- 13.NEMA. Digital Imaging and Communications in Medicine (DICOM) Part 1: Introduction and Overview. Rosslyn, Virginia, USA: National Electrical Manufacturers Association; 2011. [Google Scholar]
- 14.HIPAA. The Health Insurance Portability and Accountability Act of 1996 (HIPAA), P.L. No. 104–191, 110 Stat. 1996;1938(1996) [Google Scholar]
- 15.Aryanto KY, Broekema A, Oudkerk M, van Ooijen PM. Implementation of an anonymisation tool for clinical trials using a clinical trial processor integrated with an existing trial patient data information system. Eur Radiol. 2012;22:144–151. doi: 10.1007/s00330-011-2235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez DR, Carpenter T, van Hemert JI, Wardlaw J. An open source toolkit for medical imaging de-identification. Eur Radiol. 2010;20:1896–1904. doi: 10.1007/s00330-010-1745-3. [DOI] [PubMed] [Google Scholar]
- 17.Noumeir R, Lemay A, Lina JM. Pseudonymization of radiology data for research purposes. J Digit Imaging. 2007;20:284–295. doi: 10.1007/s10278-006-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onken M, Riesmeier J, Engel M, Yabanci A, Zabel B, Despres S. Reversible anonymization of DICOM images using automatically generated policies. Stud Health Technol Inform. 2009;150:861–865. [PubMed] [Google Scholar]
- 19.Lin L, Wang JZ. DDIT - A tool for DICOM brain Images de-identification; The 5th International Conference on Bioinformatics and Biomedical Engineering (iCBBE 2011); 2011. pp. 1–4. [Google Scholar]
- 20.Abouakil D, Heurix J, Neubauer T. Data models for the pseudonymization of DICOM data; The 44th Hawaii International Conference on System Sciences (HICSS); 2011. pp. 1–11. [Google Scholar]
- 21.Zhu YX, Singh PD, Siddiqui K, Gillam M. An automatic system to detect and extract text in medical images for de-identification. Medical Imaging 2010: Advanced Pacs-Based Imaging Informatics and Therapeutic Applications. 2010;7628:762803–762811. [Google Scholar]
- 22.NEMA. DICOM Supplement 142: Clinical Trial De-identification Profiles, DICOM Standards Commitee, Working Group 18. Virginia, USA: Rosslyn; 2011. [Google Scholar]
- 23.DVTK. 2012 Available from: http://dicom.dvtk.org/modules/wiwimod/index.php?page=Download+DICOM+Anonymizer&cmenu=downloads. [Google Scholar]
- 24.DHSS. Research Involving Private Information or Biological Specimens. USDHSS; 2006. http://grants.nih.gov/grants/policy/hs/PrivateInfoOrBioSpecimensDecisionChart.pdf.[cited 2012]. [Google Scholar]
- 25.Fontenot JD, Lee AK, Newhauser WD. Risk of secondary malignant neoplasms from proton therapy and intensity-modulated x-ray therapy for early-stage prostate cancer. Int J Radiat Oncol Biol Phys. 2009;74:616–622. doi: 10.1016/j.ijrobp.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newhauser WD, Fontenot JD, Mahajan A, Kornguth D, Stovall M, Zheng Y, Taddei PJ, Mirkovic D, Mohan R, Cox JD, Woo S. The risk of developing a second cancer after receiving craniospinal proton irradiation. Phys Med Biol. 2009;54:2277–2291. doi: 10.1088/0031-9155/54/8/002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howell RM, Giebeler A, Koontz-Raisig W, Mahajan A, Etzel CJ, D'Amelio AM, Jr, Homann KL, Newhauser WD. Comparison of therapeutic dosimetric data from passively scattered proton and photon craniospinal irradiations for medulloblastoma. Radiat Oncol. 2012;7:116–127. doi: 10.1186/1748-717X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Google. Tesseract-ocr. Optical Character Recognition (OCR)) 2012 [cited 2012]; Available from: http://code.google.com/p/tesseract-ocr/. [Google Scholar]
- 29.NIH. ImageJ Java Image Processing Program. 2012 [cited 2012]; Available from: http://rsbweb.nih.gov/ij.