Abstract
Background & Aims
Development of a vaccine against hepatitis C virus (HCV) has been hindered by our limited understanding of immune correlates of protection during real-life exposure to the virus. We studied the immune response during HCV reinfection.
Methods
We analyzed blood samples from participants in the Montreal Acute Hepatitis C Injection Drug User Cohort Study who were re-infected with HCV from 2009 and 2012. Five patients spontaneously resolved their second infection while 4 developed chronic infections. We monitored the phenotypic and functional dynamics of HCV-specific memory responses of T cells from all subjects, during natural re-exposure and reinfection.
Results
Populations of CD4+ and CD8+ T cells with HCV-specific polyfunctional memory were expanded in all 5 individuals who resolved 2 successive HCV infections. We detected CD127hi HCV-specific memory CD8+ T cells prior to reinfection regardless of a subject’s ability to clear subsequent infections. Protection against viral persistence was associated with the expansion of a CD127neg, PD1lo effector memory T cells at the peak of the response. We also observed broadening of T-cell response, indicating generation of de novo T-cell responses. The 4 individuals who failed to clear their subsequent infection had limited expansion of HCV-specific CD4+ and CD8+ memory T cells and expressed variable levels of the exhaustion marker PD1 on HCV-specific CD8+ T cells. Dominant epitope regions of HCV strains isolated from patients with persistent reinfection had sequence variations that were not recognized by the pre-existing memory T cells.
Conclusions
Protection from persistent HCV reinfection depends on the magnitude, breadth, and quality of the HCV-specific memory T-cell response. Sequence homology among viruses, and ability of T cells to recognize multiple strains of HCV, are critical determinants of protective memory.
Keywords: Cytokines, protective immunity, immune regulation, viral infection
INTRODUCTION
Despite the recent introduction of potent antivirals against HCV, there is an urgent need for an effective prophylactic vaccine. A first step is defining correlates of protective immunity during real life exposure among high risk populations like IDUs1. CD4 and CD8 HCV-specific T cell responses are induced during acute HCV and mediate spontaneous resolution. An effective response leading to viral clearance is typically of high magnitude, broad, polyfunctional (i.e. producing more than one cytokine or function) and sustained2. Individuals who spontaneously resolve acute HCV develop long-lived memory T cells3, 4. Chimpanzees who have resolved one HCV infection were protected from chronic infection upon re-exposure but protection was less efficient upon heterologous viral re-challenge (reviewed in5). Accelerated viral clearance was associated with rapid recall of memory T cell responses4 and CD4 T cell help was critical to maintain an efficient memory T cell response6. In humans, high risk IDUs who have already resolved one HCV infection were less likely to be re-infected than HCV-naïve individuals1, 7. Osburn et al. demonstrated that reinfections were characterized by reduced peak and duration of viremia as compared to primary infection and associated with broadened cellular immune responses that facilitated viral clearance8. However, the detailed phenotype and function of HCV-specific T cells during reinfection in a real-life exposure setting remain poorly defined.
Here, we examined longitudinally the breadth, phenotype and effector functions of the HCV-specific memory T cell response and variations in viral sequence during HCV reinfection in a group of IDUs who have previously resolved a primary HCV infection.
MATERIALS AND METHODS
HCV RNA, Genotype and HCV-Antibody Testing
Qualitative HCV-RNA was tested using COBAS Ampliprep/COBAS Amplicor HCV Test, version 2.0 (Roche Molecular Systems, Branchburg, NJ) (). HCV genotyping was performed as previously described9. Anti-HCV antibodies were assessed by the AxSym HCV Assay (Abbott GMBH & CO, K.G.).
Peptides and HLA Class I Tetramers
Peptides were synthesized by Sheldon Biotechnology Centre, McGill University (Montreal, QC, Canada). MHC class I tetramers were synthesized by the National Immune Monitoring Laboratory (NIML), (Montréal, QC, Canada) or the NIH Tetramer Core Facility (Emory University, Atlanta, GA) and are as follows: HLA-A1 restricted HCV NS3 peptide amino acids (aa) 1436–1444 (ATDALMTGY) [A1/NS3-1436], HLA-A2 restricted HCV NS3 peptide aa 1073–1081 (CINGVCWTV) [A2/NS3-1073], HLA-B27 restricted HCV peptide NS5B peptide aa 2841-2849 (ARMILMTHF) [B27/NS5B-2841]..
Flow cytometry based assays
All assays were performed on frozen PBMCs using a standard BD LSR II instrument with FACSDiva software version 6.1.3 (BD Biosciences). Data files were analyzed using FlowJo software version 9.5 for Mac (Tree Star, Inc., Ashland, OR). Tetramer staining coupled with phenotypic analysis, intracellular cytokine staining (ICS) and CD107a degranulation assay were performed as previously described10 in response to HCV peptide pools (1 µg/ml) or HCV minimum peptide (10 µg/ml). Polyfunctionality was assessed by exporting flow cytometry standard (FCS) data as Boolean gates using FlowJo and SPICE softwares11. Carboxyfluorescein succinimidyl ester (CFSE) 6 days proliferation assays were performed as previously described10 with or without HCV minimum peptide or peptide pool.
HCV Epitope Sequencing
HCV RNA was extracted from EDTA plasma using AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen, Union City, CA), reverse transcribed, PCR amplified, cloned and sequenced as previously described10 at the Molecular Biology and Functional Genomics service of the Institut de Recherches Cliniques de Montréal (Montreal, QC).
RESULTS
Identification of HCV reinfection cases
This study was conducted among participants of the Montreal Acute Hep C IDU Cohort Study (HEPCO)12 and approved by the institutional ethics committee (Protocol SL05.014). Primary HCV infection was identified in cohort participants who were initially negative for both HCV RNA and anti-HCV antibodies for at least 6 months of follow-up, then had a positive HCV RNA and/or antibody test as previously described10, 13. Participants who have resolved their primary HCV infection or participants who tested HCV RNA negative and HCV antibody positive at recruitment were enrolled in the reinfection study and followed every 3 months. HCV reinfection was defined by an HCV-RNA positive test following two negative tests, at least 60 days apart. The day of the first positive RNA test was defined as day zero post detection of reinfection (p.ri.). This study includes nine cases of reinfection identified between 2009 and 2012 for whom clinical data documenting their primary infection and longitudinal blood samples during the reinfection episode were available. Samples before the reinfection episode were available for 6 patients. Samples from time points during or right after clearance of the primary infection were available for 4 patients. Five patients spontaneously resolved their second infection while four patients became chronically infected, referred to hereafter as the SR/SR group and the SR/CI group, respectively. Patients’ demographics, clinical characteristics and infection history are listed in Supplementary Table S1.
Spontaneous resolution of HCV reinfection is associated with an increase in the magnitude and breadth of the HCV-specific T cells
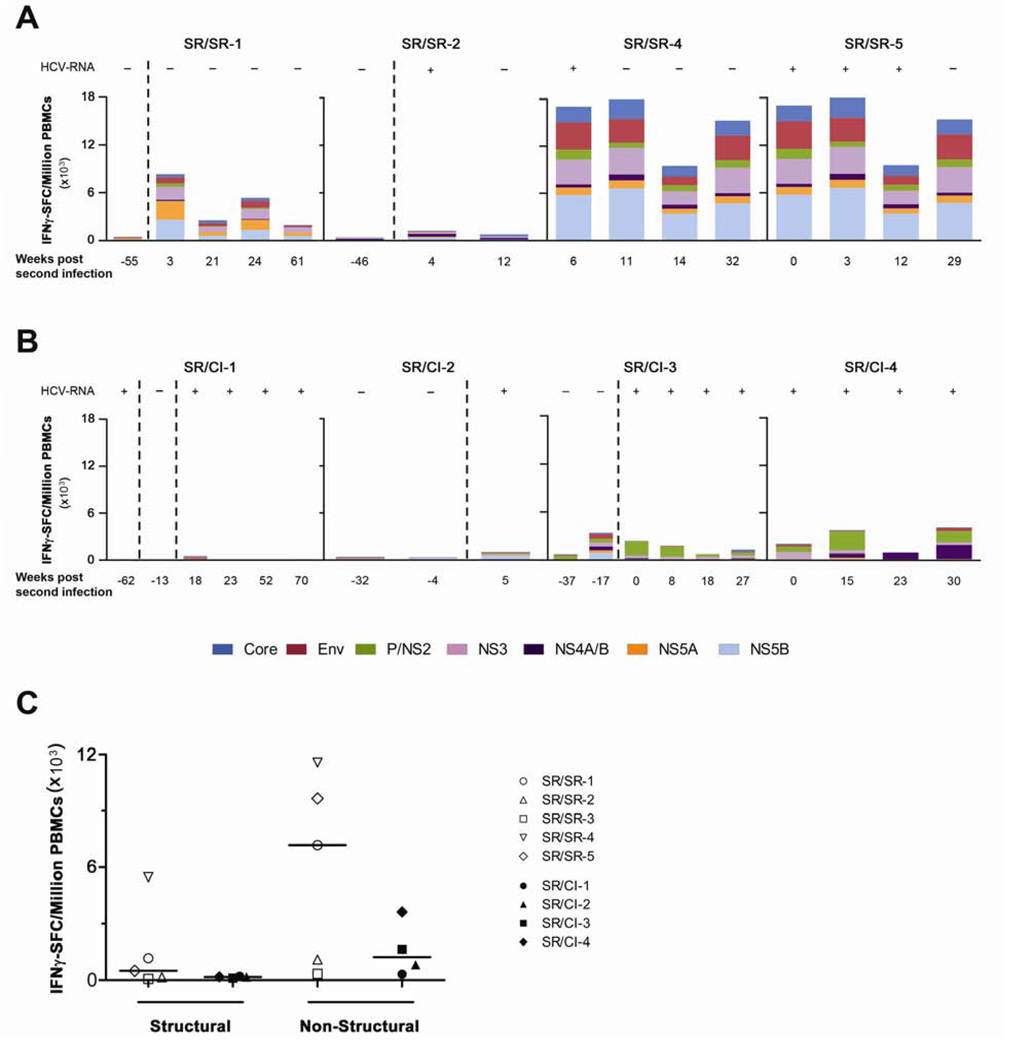
We examined the magnitude and breadth of the HCV-specific T cells using an IFNγ enzyme-linked immunospot (ELISPOT) assay against overlapping peptide pools representing the HCV genotype 1a H77 polyprotein. We chose this reference sequence for the preliminary screening since all second infections were of genotype 1 and subtype 1a is the most common subtype in our cohort. The genotype of the primary infecting HCV viral strain could not be determined in many subjects because of rapid viral clearance, low viral loads or unavailability of samples. Wherever samples were available, we monitored the ELISPOT response before and during the second infection in the SR/SR (Figure 1A) and SR/CI (Figure 1B) patients. The magnitude of the HCV-specific T cell response during reinfection was higher in the SR/SR group as compared to the SR/CI group. The average frequency at the earliest time point tested following reinfection (Mean: 8 weeks) was 7440 spot forming cells (SFC)/million PBMCs in the SR/SR group versus 1760 SFC/million PBMCs in the SR/CI group. The response to pools representing the structural and non-structural regions of HCV was higher for the SR/SR group (Mean: 1470 and 5970 SFC/million PBMCs, respectively) as compared to the SR/CI group (Mean: 160 and 1600 SFC/million PBMCs) (Figure 1C). The breadth of the immune response, measured by the number of peptide pools targeted by the immune response, was also higher in the SR/SR group at the earliest time point during reinfection. An average of 10 (out of 11) peptide pools were targeted in the SR/SR group, versus 7 pools for the SR/CI group (Figures 1A, 1B and data not shown).
Figure 1. Higher magnitude and breadth of the HCV-specific T cell response in the SR/SR as compared to SR/CI patients.
PBMCs from the indicated time points were tested in an IFNγ ELISPOT assay against overlapping peptide pools representing the HCV (H77) polyprotein. The frequencies of IFNγ spot forming cells (SFC) per million PBMCs from four SR/SR patients (A) and four SR/CI patients (B) are shown. The dashed lines delineate the different infection episodes. (C) Frequency of cells targeting the structural versus the non-structural proteins at the earliest time point (mean 8 weeks) during reinfection in the SR/SR (open symbols) and the SR/CI (solid symbols) patients.
Expansion of the HCV-specific memory T cells was associated with viral clearance upon reinfection in the SR/SR group. One exception was patient SR/SR-3, who successfully cleared his reinfection within 4 weeks despite no change in the magnitude and breadth of the response against the HCV genotype 1a peptides (Supplementary Figure S1A). Since the infecting subtype could not be determined, we tested his response to a panel of peptides corresponding to genotype 1b J4 reference sequence and observed an increase in the overall frequency and breadth of HCV-specific T cell responses upon reinfection (Supplementary Figure S1B). We also retested patients where the reinfection subtype was undetermined using genotype 1b peptides (Supplementary Figure S1 C, D and E) and demonstrated that the response was slightly higher against the 1a peptide panel as compared to 1b at the same time points suggesting that they were probably infected with a genotype 1a virus.
Spontaneous resolution of HCV reinfection is associated with enhanced proliferation of HCV-specific CD4 and CD8 T cells
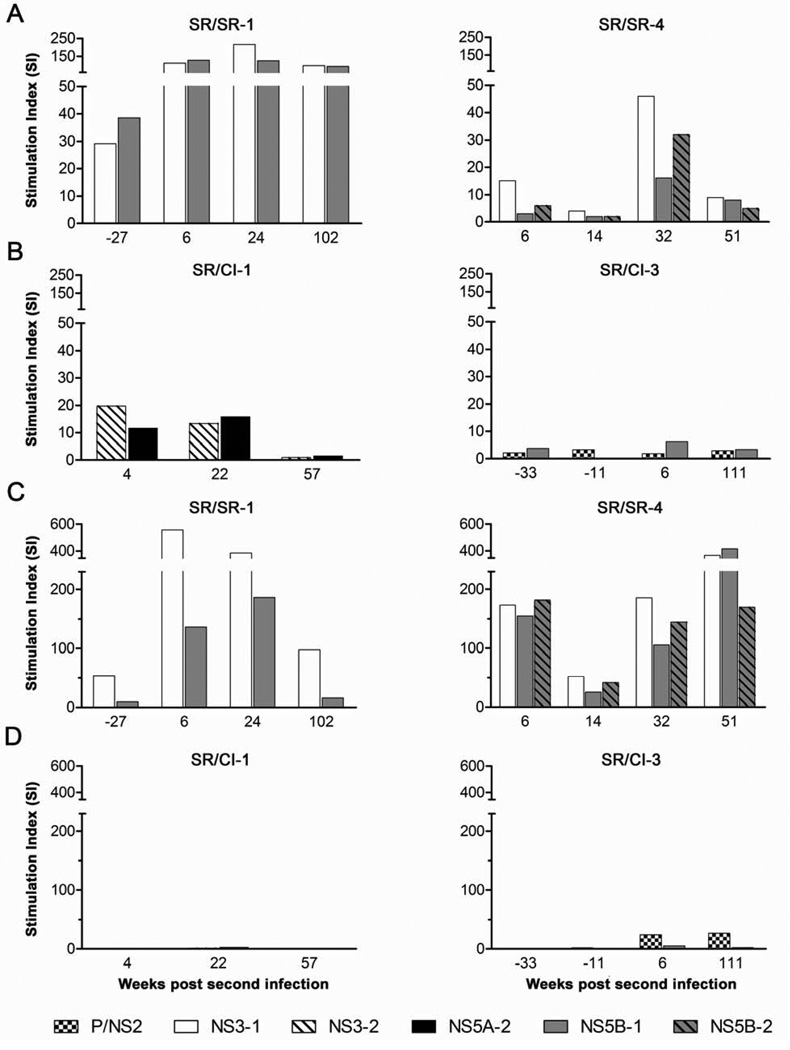
Next, we examined the proliferative capacity of HCV-specific CD4 and CD8 T cells in a CFSE dilution assay against peptide pools corresponding to the immunodominant regions of HCV (NS3 and NS5B) and/or the peptide pools showing the highest response in ELISPOT. Representative FACS data are presented in Supplementary Figure S2. Proliferation data for CD4 and CD8 T cells from two patients of each group are presented as stimulation index (SI) in Figure 2. Patient SR/SR-1 for whom samples prior to reinfection were available demonstrated high proliferation of CD4 T cells against the peptide pools tested (NS3-1 and NS5B-1) at baseline (Figure 2A). This proliferative capacity increased 3 folds at 6 weeks p.ri. Similarly, patient SR/SR-4 demonstrated some CD4 T cell proliferation at week 6 p.ri. against several pools that increased 3–5 fold by 32 weeks p.ri. (Figure 2A). In contrast, we observed very limited increase in CD4 T cell proliferation for patient SR/CI-1 and the existing proliferative response became undetectable by week 57 p.ri and patient SR/CI-3 demonstrated very limited proliferation (Figure 2B).
Figure 2. Higher proliferative capacity of HCV-specific CD4 and CD8 T cells from SR/SR patients as compared to SR/CI patients.
Proliferation of HCV-specific T cells in a 6-day CFSE proliferation assay against overlapping peptide pools spanning the HCV proteins indicated. Representative proliferation data for CD4 T cells from two SR/SR (A) and two SR/CI (B) patients. Representative proliferation data for CD8 T cells from two SR/SR (C) and two SR/CI (D) patients. Proliferating cells were identified as percent viable CFSElo, CD4+ or CD8+, CD3+ T cells and presented as stimulation index (SI).
For CD8 T cell proliferation, patient SR/SR-1 demonstrated high proliferative response prior to reinfection that increased to an SI of 560 in response to NS3 pool 1 at 6 weeks p.ri. (Figure 2C). In patient SR/SR-4 very high levels of proliferation were observed at 6 weeks against all pools tested and although they partially declined at week 14, they recovered and were sustained at week 51 p.ri. (Figure 2C). In contrast, no CD8 proliferative responses were detected in the two SR/CI patients against the peptide pools tested (Figure 2D). In summary, spontaneous resolution of second HCV infection was associated with an increase in HCV-specific CD4 and CD8 T cell proliferative responses.
Spontaneous resolution of HCV reinfection is associated with enhanced cytokine production by HCV-specific T cells
Polyfunctional CD8 T cells producing several effector cytokines and molecules are associated with spontaneous resolution of primary acute HCV10. We thus examined the polyfunctionality of HCV-specific T cells longitudinally during reinfection in two SR/SR and two SR/CI patients against peptide pools corresponding to the immunodominant regions of HCV and/or pools showing the highest response in ELISPOT. We assessed the production of the antiviral cytokines IFNγ, tumor necrosis factor alpha (TNFα) and the T cell growth factor IL-2 for CD4 and CD8 T cells. For CD8 T cells we also monitored the expression of the degranulation marker CD107a. Representative FACS plots are presented in Supplementary Figure S3.
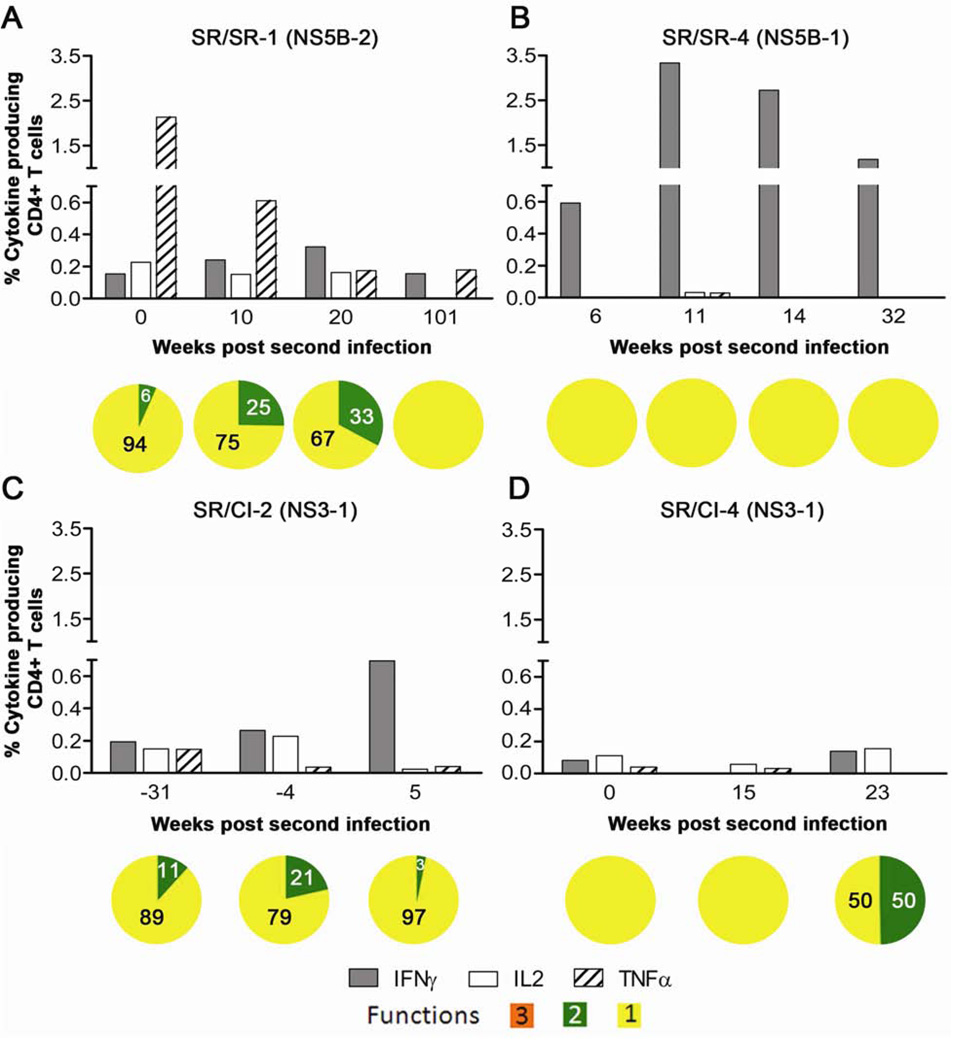
The production of the individual cytokines by CD4 T cells was higher in the two SR/SR patients (Figures 3A and 3B) when compared to the SR/CI patients (Figures 3C and 3D). IFNγ was the major cytokine produced by CD3+CD4+ T cells from the SR/SR patients reaching up to 3.33% followed by TNFα reaching up to 2.13% (Figures 3A and 3B). In contrast, production of individual cytokines was weak in the two patients from the SR/CI group reaching only 0.69% of CD3+CD4+ T cells (Figures 3C and 3D). When polyfunctionality was examined, we observed that 6–33% of the total cytokine producing cells in patient SR/SR-1 produced more than one function at different stages of reinfection as compared to 3–21% in patient SR/CI-2. Cytokines producing cells were strictly monofunctional, producing primarily IFNγ in patient SR/SR-4. In patient SR/CI-4, although 50% of cytokine producing cells produced two cytokines at 23 weeks p.ri, the low frequency of cytokine producing cells may have biased the results. These results suggest an association between the level of individual cytokine production (IFNγ or TNFα) by HCV-specific CD4 T cells rather than polyfunctionality with the protection from chronicity upon reinfection.
Figure 3. Higher production of cytokines by HCV-specific CD4 T cells from SR/SR patients as compared to SR/CI patients during reinfection.
PBMCs were stimulated with overlapping peptide pools representing HCV (NS3 and NS5B) and pools showing the highest response in the IFNγ ELISPOT then ICS was performed to measure the production of IFNγ, IL-2 and TNFα. Representative results from two SR/SR patients (A, B) and two SR/CI patients (C and D), gated on CD4+ CD3+ viable T lymphocytes. Peptide pools are indicated between brackets and represent the following regions: NS3-1 (NS3 aa: 1016–1341), NS5B-1 (NS5B; aa: 2416–2720) and NS5B-2 (NS5B; aa: 2710–3014). For polyfunctionality, orange represents triple functional cells, green represents double functional and yellow represents monofunctional cells of any combination.
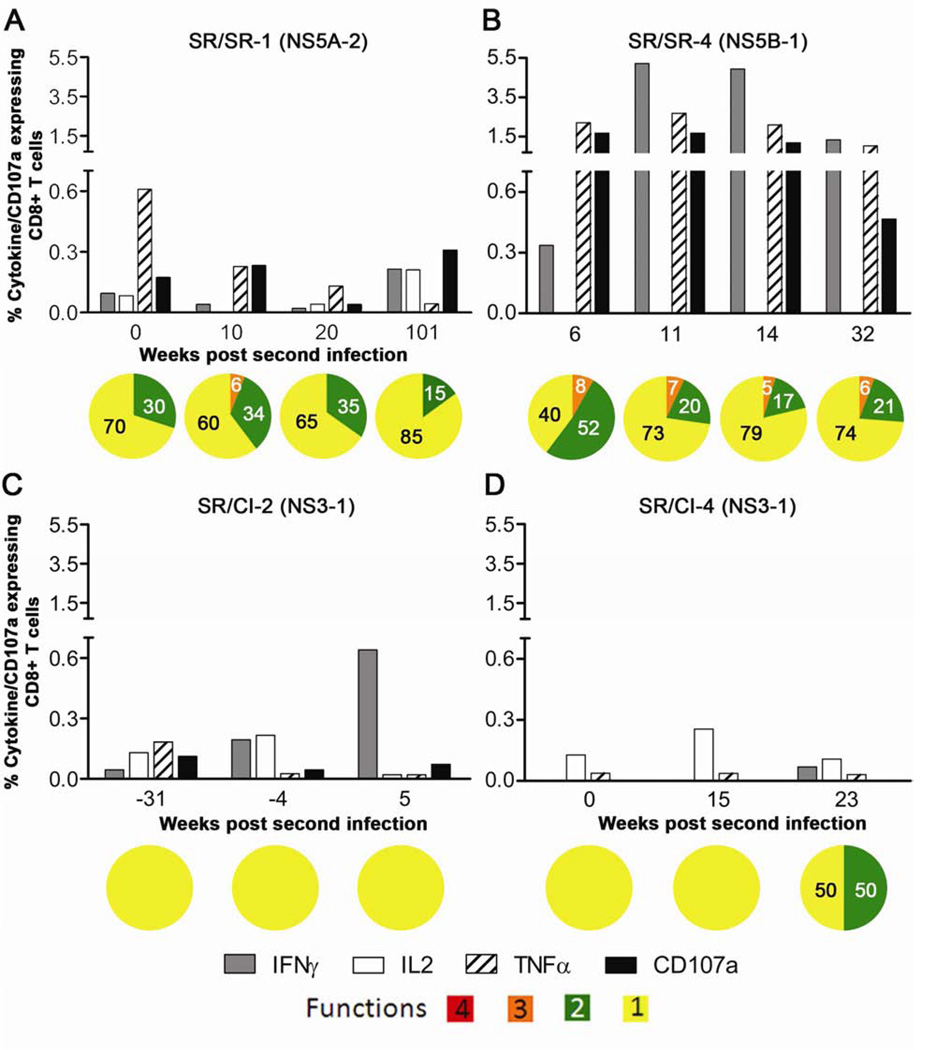
HCV-specific CD8 T cells from the SR/SR patients also expressed higher levels of the individual cytokines and CD107a as compared to SR/CI patients (Figure 4). IFNγ was the major cytokine produced by CD3+CD8+ T cells from the SR/SR patients reaching up to 5.22% followed by TNFα and the degranulation marker CD107a (Figures 4A and 4B). Individual cytokine production or CD107a expression was weak in the SR/CI group with a maximal production of 0.49% of CD3+CD8+ T cells (Figures 4C and 4D). Polyfunctionality was also higher in the SR/SR group, where we observed that ~6% of the total functional cells in the two SR/SR patients produced three functions at different stages of reinfection as compared to none in the two SR/CI patients. Furthermore, 15–52% of total cytokine/CD107a expressing cells expressed two functions in the SR/SR patients as compared to none in the SR/CI group (except the latest time point in patient SR/CI-4 where the overall frequency of functional cells was 0.2%) (Figure 4). In summary, the frequency of individual cytokine producing CD8 T cells was higher in the SR/SR group and within these functional cells the frequency of polyfunctional cells was higher. These results suggest an association between the level of individual cytokine production and polyfunctionality of HCV-specific CD8 T cells and protection from viral persistence upon reinfection.
Figure 4. Higher polyfunctionality of HCV-specific CD8 T cells from SR/SR patients in comparison to SR/CI patients during reinfection.
PBMCs were stimulated overnight with overlapping peptide pools representing HCV (NS3 and NS5B) and HCV pools showing the highest response in the IFNγ ELISPOT. ICS was performed to measure the production of IFNγ, IL-2 and TNFα and expression of CD107a. Representative results from two SR/SR patients (A, B) and two SR/CI patients (C and D), gated on CD8+ CD3+ viable T lymphocytes. Peptide pools are indicated between brackets and represent the following regions: NS3-1 (NS3 aa: 1016–1341), NS5A-2 (NS5A; aa: 2192–2426) and NS5B-1 (NS5B; aa: 2416–2720). For polyfunctionality, orange represents triple functional cells, green represents double functional and yellow represents monofunctional cells of any combination.
Spontaneous resolution of HCV reinfection is associated with higher polyfunctionality of HCV-specific CD8 T cells targeting minimal epitopes
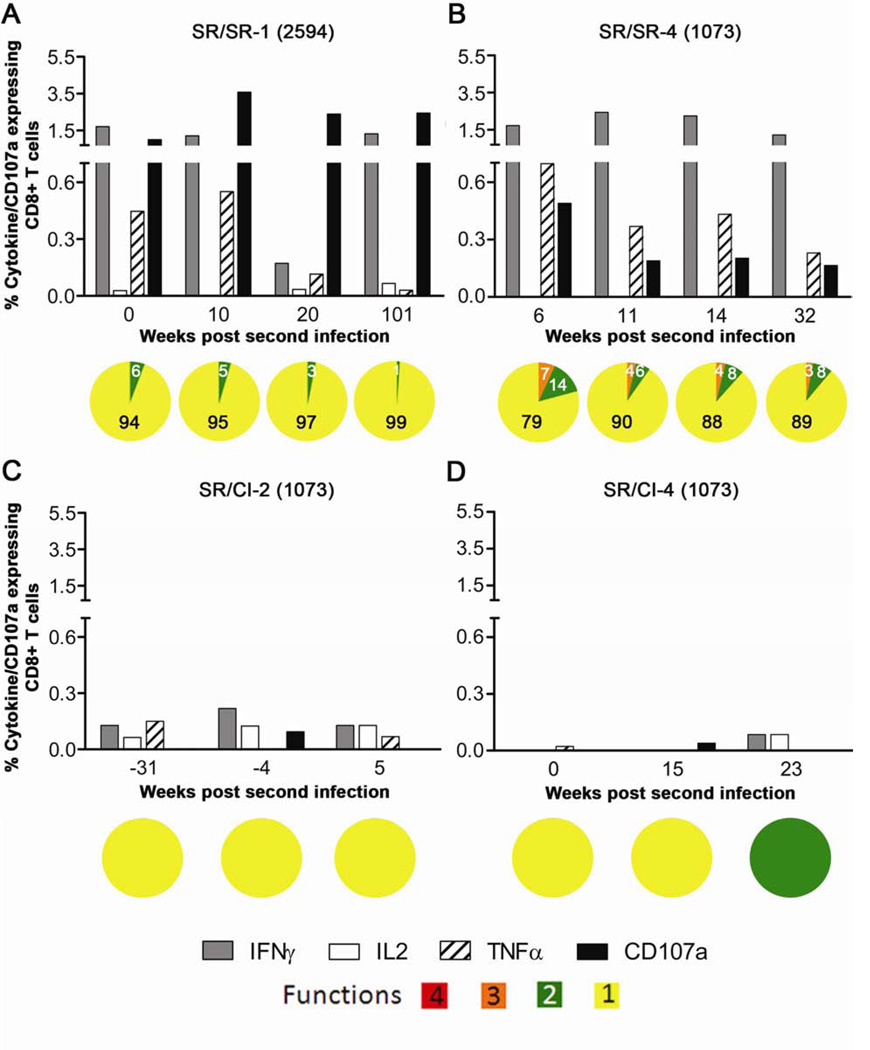
Next, we evaluated polyfunctionality of HCV-specific CD8 T cells in response to predicted minimal epitopes restricted by HLA-A2 and showing the highest response (NS5B-2594 and NS3-1073) in two patients from the SR/SR and two patients from the SR/CI group. The production of individual cytokines and/or expression of CD107a was higher in the SR/SR group when the cells were stimulated with the minimum cognate peptide reaching 3.58% (mean 0.54%) of the total CD3+CD8+ T cells (Figures 5A and 5B). In contrast, individual cytokine production and/or CD107a expression in the SR/CI group never exceeded 0.22% (mean 0.06%) (Figures 5C and 5D). The frequency of CD3+CD8+ T cells expressing two or three functions was 14% and 7%, respectively of total CD3+CD8+ functional T cells in the SR/SR patients (Figures 5A and 5B) and almost none in the SR/CI patients (Figures 5C and 5D). Similar to previous results using peptide pools, these data suggest an association between the level of individual cytokine production and polyfunctionality of HCV-specific CD8+ T cells targeting dominant epitopes with the protection from persistent viremia upon reinfection with HCV.
Figure 5. Higher polyfunctionality of HCV-specific CD8 T cells targeting minimal epitopes in SR/SR patients in comparison to SR/CI patients during reinfection.
PBMCs from the indicated time points were stimulated overnight with the minimum cognate peptide and ICS/CD107a staining was performed. Representative results from two SR/SR patients (A B) and two SR/CI patients (C and D), gated on viable CD3+CD8+ T lymphocytes. Peptides used are: 1073 = A2 restricted NS3-1073 and 2594 = A2 restricted NS5B-2594.
Spontaneous Resolution of HCV reinfection is associated with expansion of CD127lo HCV tetramer+ CD8 T cells
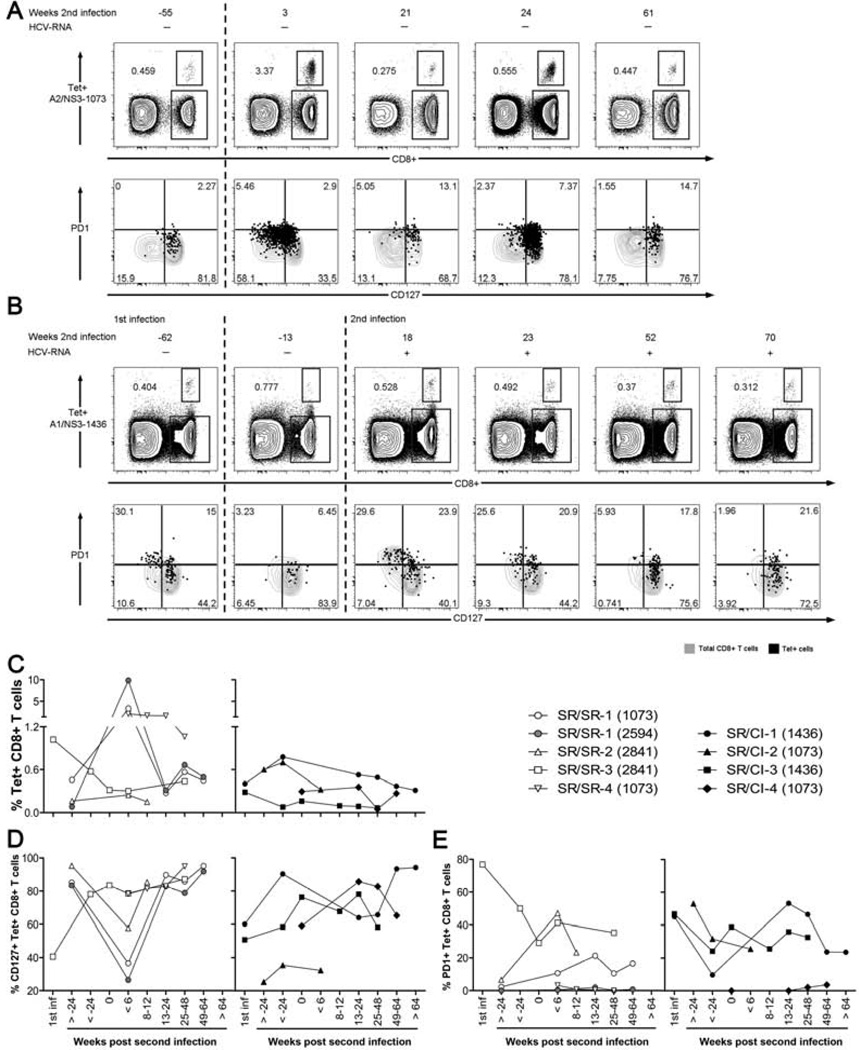
Next, we performed longitudinal phenotypic analysis of HCV-specific CD8 T cells to assess their frequency, differentiation and exhaustion status using the predicted MHC class I tetramers detailed in Supplementary Table S1. We monitored markers of T cell maturation and memory (CD27 and CD127) and T-cell activation/exhaustion (PD1, 2B4 and Tim3) on tetramer+ CD8 T cells. Representative FACS results for patient SR/SR-1, using the A2/NS3-1073 tetramer are shown in Figure 6A and Supplementary Figure S4A, and for A2/NS5B-2594 tetramer in Supplementary Figure S5. Representative data for patient SR/CI-1, using the A1/NS3-1436 tetramer, are shown in Figure 6B and Supplementary Figure S4B. Tetramer frequency data during reinfection in all subjects are represented in Figure 6C. We observed an average expansion of 42 folds at the peak of response (<6 weeks p.ri.) in patients of the SR/SR group (Figure 6C). The only exception was patient SR/SR-3, where the reinfecting HCV genotype was likely 1b rather than the 1a sequence used in the tetramer (Supplementary Figure S1A). Very limited expansion was observed in the tetramer+ population upon reinfection in all patients of the SR/CI group (Figure 6C).
Figure 6. Expansion of HCV-specific tetramer+ CD8 T cells upon reinfection and generation of a CD127lo effector T cell population in SR/SR patients.
HCV tetramer staining and phenotyping were performed directly ex vivo on PBMCs from the indicated time points. Representative dot plots for longitudinal tetramer staining (upper panels) and phenotyping (lower panels) for (A) patient SR/SR-1 with A2/NS3-1073 tetramer and (B) patient SR/CI-1 with A1/NS3-1436 tetramer. Top panels are gated on viable CD3+ T lymphocytes. Lower panels are gated on viable CD3+CD8+ T lymphocytes. Tetramer frequency is represented as percent Tetramer+ CD8+ CD3+ T cells. In lower panels, black dots represent the tetramer+ HCV-specific CD8+ T cells overlaid on grey contour plots of CD8+ CD3+ T cells. Numbers in each quadrant represent percent expression on tetramer+ CD8+ CD3+ T cells. The dashed lines delineate the different infection episodes. (C–E) Tetramer expansion and phenotyping data from all SR/SR and SR/CI patients; (C) Frequency of HCV tetramer+ CD8+ CD3+ T cells, (D) Frequency of CD127+ tetramer+ CD8+CD3+ T cells, (E) Frequency of PD1+ tetramer+ CD8+CD3+ T cells.
The most distinctive phenotypic change was expression of CD127. In one SR/SR and two SR/CI patients where samples were available during the first infection, we observed that approximately half of the HCV tetramer+ CD8 T cells were CD127hi. As patients cleared their primary HCV infection, the majority of tetramer+ CD8 T cells became CD127hi as previously described10 and consistent with a memory T cell phenotype14. A similar phenotype was observed for the tested time points prior to reinfection. At the earliest time point tested during reinfection we observed peak expansion of tetramer+ cells in SR/SR patients and the majority of these expanded cells were CD127lo, consistent with an effector phenotype (Figures 6A and 6D). In the SR/CI patients, tetramer+ cells did not expand or undergo significant changes in the CD127 expression (Figures 6B and 6D). There was no significant variation in the expression of the other markers examined (CD27, Tim3 or 2B4) at the different time points tested for the two groups of patients (Supplementary Figure S4). Finally, We observed higher expression of the activation marker PD1 on HCV tetramer+ CD8+ T cells in most patients (3/4) of the SR/CI group in comparison to SR/SR patients (Figures 6B and 6E). In summary, spontaneous resolution of a second HCV infection was associated with expansion of pre-existing memory T cells into CD127lo effectors and lower levels of PD1.
In patient SR/SR-1 who was positive to two tetramers A2/NS3-1073 and A2/NS5B-2594, we observed different levels of expansion of CD8 T cells specific to the two tetramers. The frequency of the A2/NS5B-2594 tetramer+ T cells was at a much lower level than that for NS3-1073 prior to reinfection (0.082% and 0.458%, respectively) (Figure 5A and Supplementary Figure S5), yet the A2/NS5B-2594 tetramer+ population expanded 120 folds upon reinfection (Supplementary Figure S5) compared to 7 folds only for the A2/NS3-1073 tetramer+ population (Figure 5A) (9.84% and 3.37%, respectively). This suggests a shift in the dominance of the epitopes targeted in this patient during reinfection.
Heterogeneity in targeted epitopes and reduced recognition during reinfection in the SR/CI group
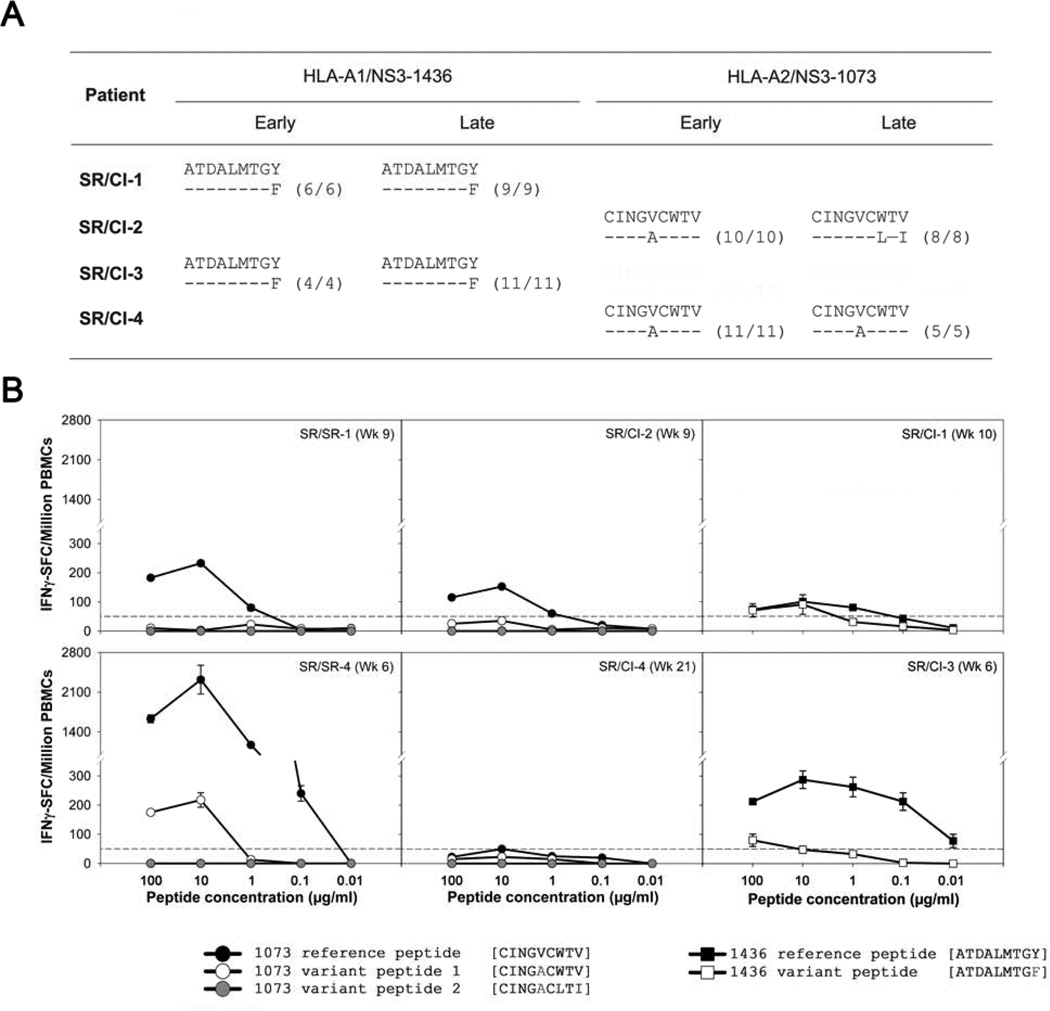
Despite the limited expansion of HCV-tetramer+ CD8 T cells in patients of the SR/CI group upon reinfection, they proliferated in response to their cognate epitope in an in vitro CFSE proliferation assay (data not shown) suggesting that they are not inherently defective in proliferation. To determine whether this lack of expansion in the SR/CI patients is due to reinfection with variant viral sequences that are not recognized by the pre-existing memory T cells, and to determine whether the change existed since the beginning or arose during reinfection, we sequenced the NS3-1436 and NS3-1073 epitopes targeted by the four patients at the earliest and the latest time points available during reinfection (Figure 7A). We detected a mismatch between the amino acid sequence of the reference peptide used in the tetramer and the autologous HLA-A1 restricted NS3-1436 epitope in patients SR/CI-1 and SR/CI-3 at the early and late time points. This 1444 Y → F substitution was previously shown to be recognized less efficiently by specific T cells15. An amino acid substitution 1077V → A was observed in the NS3-1073 for patients SR/CI-2 and SR/CI-4 at the early time point. This variant was also previously suggested to be less efficient in inducing CD8 T cell expansion and IFNγ production16, 17. This variant remained fixed in patient SR/CI-4 at the late time point but underwent three additional changes in patient SR/CI-2: 1077V → A; 1081V → I and 1078 W→ L substitution.
Figure 7. Sequence variation in targeted epitope and reduced recognition by CD8 T cells in SR/CI patients.
(A) Sequences of the epitopes targeted by tetramer+ CD8 T cells in SR/CI patients at early and late time points during reinfection. (B) Responses of total PBMCs collected during reinfection in a dose-response IFN-γ ELISPOT assay to the reference sequences versus variant autologous sequences for two SR/SR patients in response to the NS3-1073 epitope (left panels), two SR/CI patients in response to the NS3-1073 epitope (middle panels) and two SR/CI patients in response to the NS3-1436 epitope (right panels).
To determine how well the pre-existing memory T cells recognized the reference versus the autologous sequences, we tested their functional avidity by measuring dose-dependent production of IFNγ in response to stimulation by the different epitope variants during the reinfection episode in SR/SR and SR/CI patients. We observed higher functional avidity of HCV-specific T cells from SR/SR patients in response to the NS3-1073 reference epitope sequence when compared to SR/CI patients (Figure 7B). Patient SR/SR-4 had higher functional avidity than patient SR/SR-1 in response to the reference sequence and the response was >10-folds than patient SR/CI-2. Patient SR/CI-4 poorly recognized all variants. Interestingly, Patient SR/SR-4 also responded to the intermediate variant sequence of the NS3-1073 epitope indicating better cross-recognition of this variant. The reference sequence of the NS3-1436 was also better recognized than the autologous variant by patients SR/CI-1 and SR/CI-4. Altogether, these results suggest that variations in the targeted epitope could be a possible mechanism underlying the functional differences or lack of recognition in HCV-specific CD8 T cells observed between the two groups of patients.
DISCUSSION
We have performed an exhaustive phenotypic and functional characterization of the virus-specific T cell response during HCV reinfection. Although all nine patients in this study had successfully eliminated an earlier HCV infection, only five of them were able to spontaneously resolve their subsequent infection. Spontaneous resolution of reinfection was associated with an increase in both the magnitude and breadth of the total HCV-specific T cell response suggesting generation of de novo T cells responses. In addition, we observed expansion of HCV-specific memory T cells and the transient appearance of CD127low population indicative of an effector T cell phenotype. Patients who failed to clear their subsequent infection showed very limited expansion of HCV-specific T cells. Variations in the autologous sequence of the targeted epitopes were detected in all patients of this group and not recognized by the pre-existing memory T cells.
Protection from viral persistence upon HCV reinfection was associated with an increase in the magnitude and breadth of the HCV-specific T-cell response as observed by ELISPOT assays and tetramers consistent with the observations of Osburn et al.8. For the SR/SR group the increase in the magnitude and the de novo responses mostly targeted the non-structural proteins underscoring the immune dominance of this region18, 19. Furthermore, in patient SR/SR-1, we observed a shift in dominance between the two epitopes targeted. Further research with additional patients is required to elucidate whether shifting dominance may occur upon reinfection with variant viral strains or may reflect differential expansion capacity of memory T cells.
Proliferation of CD4 and CD8 T cells is predictive of the outcome of primary HCV infection20, 21. We demonstrate that protective immunity upon reinfection was associated with higher proliferative capacity for both CD4 and CD8 T cells when stimulated with peptide pools representing immunodominant regions. Nevertheless, proliferation assays using cognate epitopes demonstrated that HCV tetramer+ memory CD8 T cells in the SR/CI unprotected group proliferated to similar levels as those of the SR/SR protected group (data not shown). This suggests they are not inherently defective, and despite being present at low frequency, they could expand if stimulated with the right peptide or antigen.
Spontaneous resolution of primary acute HCV and response to vaccination were associated with generation of polyfunctional HCV-specific CD127+ CD8 T cells10, 22. Here, we demonstrate that polyfunctionality is also an important determinant of viral clearance upon reinfection. Furthermore, we extend this polyfunctionality to CD8 T cells targeting entire regions of HCV, as well as, minimal epitopes. Data from the HIV model suggest that such polyfunctional T cells are cells of the highest functional avidity23. It is also tempting to speculate that repeated reinfections with the same viral variant would selectively expand and enrich such cells. On the other hand, rechallenge with a slightly different antigenic variant may favor expansion of T cells carrying a flexible T cell receptor that can recognize these new variants. Flexibility in the T cell repertoire correlated with control of viral escape mutants during HCV and HIV infections24–26 and would require further investigation in the reinfection setting.
Our results suggest that pre-existing variations in the viral sequence of the epitopes targeted by the memory immune response could be a possible reason for persistence upon reinfection. We demonstrated higher functional avidity in response to the reference sequence of the epitope in the SR/SR group. In addition, three SR/CI patients had higher response to the reference sequence than the autologous sequence. No mutations were observed during the course of the reinfection, except for the NS3-1073 epitope in patient SR/CI-2 where the epitope underwent three changes during the reinfection episode, suggesting that it may have been under immune selection pressure. The fact that this intermediate variant was recognized by patient SR/SR-4 suggests that this variant can be recognized by some individuals. However, those changes in the targeted epitopes do not explain the general decrease in the frequency of the immune response observed in ELISPOT analysis. Thus, the association between the magnitude of the T cells and the degree of homology in the entire HCV genome between the infecting viruses in both episodes of infection remains to be assessed.
The capacity to detect HCV reinfection is dependent on the testing interval, so we cannot rule out that we have missed shorter low level infections that may have contributed to exhaustion of HCV-specific T cells or expansion of HCV-specific regulatory T cells (Tregs) that may suppresses T cell responses against subsequent primary acute infection27. Since the patients studied here have all cleared a previous primary acute HCV infection with systemic viremia and that the memory T cells responded efficiently to in vitro stimulation with their cognate peptides, it is unlikely that inhibition by Tregs was a major player. These issues need to be addressed by closer follow-up intervals and improved patient questionnaires that can identify high risk exposures, with tools to detect low level viremia or potential subclinical infections. Similarly, the contribution of other CD4 T cell subsets like Th17 cells in the maintenance of memory CD8 T cells, as well as the cross regulation between Tregs and Th17 will require additional investigations13.
Osbourn et al. demonstrated that spontaneous resolution upon HCV reinfection correlates with generation of neutralizing antibodies. We attempted to examine the neutralizing capacity of longitudinal plasma samples against a panel of cell-cultured HCV (HCVcc) representing the most common HCV genotypes in Canada (Genotypes 1a, 1b and 3a). However, no distinct difference could be detected between the SR/SR and SR/CI group which may be due to the low number of patients in each group (data not shown).
In conclusion, we demonstrate that protection against HCV persistence upon re-exposure is possible. Although we could not assess whether there was sterilizing immunity or long-term protection in our cohort, our results and results from other groups suggest that protection from viral persistence is higher upon subsequent exposure than primary infection. We also demonstrate that changes in the breadth of the immune response and immune dominance are possible, suggesting generation of de novo T cell responses and offering hope for vaccine development. Immunization and boosting with different HCV genotypes may afford a broader and more effective protection.
Supplementary Material
Acknowledgments
Grant Support : This work is supported by grants from the Canadian Institutes for Health Research (CIHR) (MOP-106468), Alberta Innovates-Health Solutions, Fonds de recherche du Québec – Santé (FRQS) AIDS and Infectious Disease Network (Réseau SIDA-MI) and the National Institute on Drug Abuse (NIDA) funded International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) study (R01DA031056). MSA received doctoral fellowships from the Université de Montréal, the National CIHR Training Program on Hepatitis C (NCRTP-Hep C) and CIHR. NHS and JB received Chercheur Boursier salary awards from the FRQS. The funders had no influence on the study design and data interpretation.
Abbreviations used in this paper
- ELISPOT
Enzyme-linked immunospot assay
- HCV
Hepatitis C virus
- ICS
Intracellular cytokine staining
- IDUs
Injection drug users
- p.ri
Post reinfection
- SFC
spot forming cell
- TNFα
Tumor necrosis factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
Author’s contributions: MSA performed most of the experiments and participated in the design of the study, data analysis and writing the manuscript. NB performed some of the experiments and provided overall technical support for the study. DM performed HCV genotyping assays and viral quantification. JB recruited and provided clinical follow-up for study participants, she also participated in the study design, data analysis and writing the manuscript. NHS supervised the whole study including design, data analysis and writing the manuscript.
REFERENCES
- 1.Grebely J, Prins M, Hellard M, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis. 2012;12:408–414. doi: 10.1016/S1473-3099(12)70010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker CM. Adaptive immunity to the hepatitis C virus. Adv Virus Res. 2010;78:43–86. doi: 10.1016/B978-0-12-385032-4.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takaki A, Wiese M, Maertens G, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat Med. 2000;6:578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 4.Shoukry NH, Grakoui A, Houghton M, et al. Memory CD8+ T Cells Are Required for Protection from Persistent Hepatitis C Virus Infection. J. Exp. Med. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Hakeem MS, Shoukry NH. Protective Immunity against Hepatitis C: Many Shades of Grey. Frontiers in Immunology. 2014;5 doi: 10.3389/fimmu.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grakoui A, Shoukry NH, Woollard DJ, et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 7.Mehta SH, Cox A, Hoover DR, et al. Protection against persistence of hepatitis C. Lancet. 2002;359:1478–1483. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 8.Osburn WO, Fisher BE, Dowd KA, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2010;138:315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy DG, Willems B, Deschenes M, et al. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences. J Clin Microbiol. 2007;45:1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badr G, Bedard N, Abdel-Hakeem MS, et al. Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells. J Virol. 2008;82:10017–10031. doi: 10.1128/JVI.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of postcytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebely J, Morris MD, Rice TM, et al. Cohort profile: the International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) Study. Int J Epidemiol. 2013;42:1649–1659. doi: 10.1093/ije/dys167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kared H, Fabre T, Bedard N, et al. Galectin-9 and IL-21 mediate cross-regulation between Th17 and Treg cells during acute hepatitis C. PLoS Pathog. 2013;9:e1003422. doi: 10.1371/journal.ppat.1003422. Epub 2013 Jun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaech SM, Tan JT, Wherry EJ, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. Epub 2003 Nov 16. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Hakeem MS, Bedard N, Badr G, et al. Comparison of immune restoration in early versus late alpha interferon therapy against hepatitis C virus. J Virol. 2010;84:10429–10435. doi: 10.1128/JVI.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbani S, Amadei B, Cariani E, et al. The impairment of CD8 responses limits the selection of escape mutations in acute hepatitis C virus infection. J Immunol. 2005;175:7519–7529. doi: 10.4049/jimmunol.175.11.7519. [DOI] [PubMed] [Google Scholar]
- 17.Kasprowicz V, Kang YH, Lucas M, et al. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J Virol. 2010;84:1656–1663. doi: 10.1128/JVI.01499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauer GM, Barnes E, Lucas M, et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Smyk-Pearson S, Tester IA, Lezotte D, et al. Differential antigenic hierarchy associated with spontaneous recovery from hepatitis C virus infection: implications for vaccine design. J Infect Dis. 2006;194:454–463. doi: 10.1086/505714. [DOI] [PubMed] [Google Scholar]
- 20.Lechner F, Wong DK, Dunbar PR, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thimme R, Oldach D, Chang KM, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SH, Shin EC, Capone S, et al. Successful vaccination induces multifunctional memory T-cell precursors associated with early control of hepatitis C virus. Gastroenterology. 2012;143:1048–1060. doi: 10.1053/j.gastro.2012.06.005. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida JR, Price DA, Papagno L, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer-Olson D, Shoukry NH, Brady KW, et al. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iglesias MC, Almeida JR, Fastenackels S, et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ladell K, Hashimoto M, Iglesias MC, et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 27.Park SH, Veerapu NS, Shin EC, et al. Subinfectious hepatitis C virus exposures suppress T cell responses against subsequent acute infection. Nat Med. 2013;19:1638–1642. doi: 10.1038/nm.3408. Epub 2013 Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.