Abstract
Objectives
Congenital diarrhea disorders are a group of genetically diverse and typically autosomal recessive disorders that have yet to be well characterized phenotypically or molecularly. Diagnostic assessments are generally limited to nutritional challenges and histologic evaluation, and many subjects eventually require a prolonged course of intravenous nutrition. Here we describe next-generation sequencing techniques to investigate a child with perplexing congenital malabsorptive diarrhea and other presumably unrelated clinical problems; this method provides an alternative approach to molecular diagnosis.
Methods
We screened the diploid genome of an affected individual, using exome sequencing, for uncommon variants that have observed protein-coding consequences. We assessed the functional activity of the mutant protein, as well as its lack of expression using immunohistochemistry.
Results
Among several rare variants detected was a homozygous nonsense mutation in the catalytic domain of the proprotein convertase subtilisin/kexin type 1 gene. The mutation abolishes prohormone convertase 1/3 endoprotease activity as well as expression in the intestine. These primary genetic findings prompted a careful endocrine reevaluation of the child at 4.5 years of age, and multiple significant problems were subsequently identified consistent with the known phenotypic consequences of proprotein convertase subtilisin/kexin type 1 (PCSK1) gene mutations. Based on the molecular diagnosis, alternate medical and dietary management was implemented for diabetes insipidus, polyphagia, and micropenis.
Conclusions
Whole-exome sequencing provides a powerful diagnostic tool to clinicians managing rare genetic disorders with multiple perplexing clinical manifestations.
Keywords: enteroendocrine cell, Neurogenin-3, PC1/3, proprotein convertases
Congenital diarrheal disorders are uncommon yet frequently devastating chronic conditions that are secondary to a diverse group of autosomal recessive mutations. They can be classified into those that selectively impair the transport or hydrolysis of single nutrients or electrolytes and those that attenuate the assimilation of all forms of nutrients (1). They may also be grouped as either malabsorptive or secretory in nature, and by an array of histologic features, including changes within the enterocytes or their migration along the crypt–villus axis.
Regardless, children presenting shortly after birth with severe diarrhea—given their rarity and heterogeneity—are frequently misdiagnosed. Failure to diagnose such patients quickly and accurately can undermine their tenuous hold on life. When correctly diagnosed, subjects with impairment of selective nutrient assimilation generally do well on a lifelong nutrient-specific restricted diet; however, those presenting with malabsorption of multiple forms of nutrients (proteins, carbohydrates, and fats) generally have an adverse clinical course that includes lifelong or prolonged total intravenous (parenteral) nutrition, and/or intestinal and occasionally concomitant liver transplantation (1). Although these disorders are frequently fatal without proper dietary and nutritional modifications, present state-of-the-art therapeutic modalities are primitive and are associated with extremely significant morbidity and mortality, as well as daunting medical care costs (2).
As a group, generalized malabsorptive diarrheal disorders are frequently idiopathic, and their physiologic basis is poorly understood (3). These limitations serve as the impetus to a general search for the molecular basis of these disorders, thus propelling the use of recently feasible pluripotent and somatic stem cell technologies to discover alternative therapeutic approaches (4).
In some cases, the histology and nutrient absorption characteristics may point to the possibility of dysfunction in known genes (Neurogenin-3 [NEUROG3], SGLT1, EPCAM, MYO5B, SPINT2, TTC37, SKIV2L, ADAM17), which can then be directly sequenced to identify likely causative mutations (5–11). Often, however, clinical evidence may be insufficient to implicate known genes or sequencing of candidate genes fails to reveal damaging mutations resulting from genetic heterogeneity. These aspects, as is typical of all rare disorders, greatly impede efficient and timely diagnosis. Recent advances in sequencing technology now make it possible to sequence the coding portion and essential splice sites of approximately 95% of all protein-coding bases of all known genes (the “exome”) at a cost comparable to clinical sequencing tests of a single gene (12,13). Thus, an unbiased scan of the exome can discover known and novel mutations in known genes and also mutations in hitherto unsuspected genes in a manner that efficiently directs clinical care. Here we show an example of the use of whole-exome sequencing to identify the causative mutation in a child with congential diarrhea.
Prohormone convertase 1/3 (PC1/3) is a calcium-dependent serine endoprotease that converts proinsulin and other prohormones into active forms (14). PC1/3 is highly expressed in the small intestine. PC1/3 deficiency, resulting from the mutations in the pro-protein convertase subtilisin/kexin type 1 (PCSK1) gene, can prevent enteroendocrine cells from producing functional hormones and cause generalized malabsorption and a variety of systemic endocrinopathies that develop in an age-dependent fashion (15). The mechanism by which PC1/3 deficiency causes malabsorption is not well understood, but it may be that a novel peptide, or multiple redundant peptides, processed by PC1/3 enhance nutrient assimilation.
METHODS
DNA Sequencing
We prepared an exon-enriched sequencing library following Agilent Technologies (Santa Clara, CA) SureSelect Target Enrichment System for the Applied Biosystems (Foster City, CA) SOLiD System protocol (version 1.5.1). Briefly, genomic DNA extracted from patient saliva with an Oragene DNA Collection kit (DNA-Genotek, Kanata, Canada) was ultrasonically sheared (Covaris, Woburn, MA) into ~125 bp fragments. After fragment end repair, ligation of adapters, and gel-size selection for ~175 bp product, the library was nick translated and amplified by 12 polymerase chain reaction (PCR) cycles. The library was hybridized in solution to RNA probes from the 3-Mb SureSelect All Human Exon kit (Agilent G3361), covering 1.22% of the human genome containing the exons of the Consensus CDS genes (16). The exon-enriched library was selected by magnetic bead separation, further amplified by 12 cycles of PCR, and clonally amplified on beads by emulsion PCR (Applied Biosystems SOLiD 4 System Templated Bead Preparation Guide [March 2010]). Fragment sequencing by ligation was performed on a SOLiD 4 System (Applied Biosystems SOLiD 4 System Instrument Operation Guide [March 2010]), which yielded ~108 million 50-base reads.
Bioinformatics Analysis
We aligned the sequenced reads to build GRCh37 of the human genome (17) with Novoalign (http://www.novocraft.com) to obtain ~31 million uniquely aligned 50-base reads (~1.5G bases) after removing PCR duplicates. ~1.2G bases were within the targeted exome with a mean coverage of ~33× at each targeted position, achieving at least 10× coverage of 67% of the coding sequence of annotated protein-coding genes per Ensembl (Wellcome Trust Sanger Institute, Hinxton, UK) (18). Base quality scores were recalibrated to improve accuracy by analyzing the covariation among reported quality score, position within read, dinucleotide, and probability of mismatching the reference genome using the Genome Analysis Toolkit (GATK) (19,20). The GATK Unified Genotyper was used to genotype single-nucleotide variants (SNVs) and indels. The GATK variant quality score recalibration module was used to assign probabilities to each variant call. In addition to variants that passed quality checks, for improved sensitivity, we retained for downstream analysis a tranche of variants that had a likelihood ≤1% of being false-positives. Variant consequences were determined by the Ensembl Variant Effect Predictor (20), and the extent of protein damage was estimated with SIFT (Sorting Intolerant From Tolerant) (21–25), PolyPhen (Polymorphism Phenotyping) (26–28), and Condel (Consensus Deleteriousness) (29).
Development of Wild-Type and Mutant PC1/3 Expression Vectors
Human PC1/3 complement DNA (cDNA) was generated from RNA isolated from human pancreatic carcinoid (BON) cells. The wild-type PC1/3 was amplified by using an oligonucleotide that contained a KpnI site, and 20 nucleotides of PC1/3 from the translational start site. A 3′ oligonucleotide contained an XhoI site, and a FLAG amino acid (DYKDDDDK) sequence introduced at the stop site of the wild-type and Y343X mutant cDNA. The wild-type cDNA was subcloned into the KpnI and XhoI restriction-digested pcDNA3 vector and clones were screened. A single clone, designated WT PC1/3, was used to generate the Y343X mutant clone, and the entire clone was sequenced to verify that only the targeted variant was altered.
Transient Transfection of Expression Vectors
HEK293 cells at a density of 2 × 105 cells per well in 24-well plates were transfected with plasmids encodingWTPC1/3 or Y343X in triplicate wells. Cells were transfected with 200 ng of plasmid DNA per well using Lipofectamine Reagent (Invitrogen, Carlsbad, CA). Five hours posttransfection, 1 mL of growth medium was added to each well and incubation continued for an additional 24 hours. Postincubation, cells were washed with phosphate-buffered saline and 0.3 mLof Opti-MEM (Invitrogen) containing 100-µg/mL bovine aprotinin was added to each well. Cells were incubated for an additional 24 hours before conditioned medium and cells were harvested. Conditioned medium was analyzed first by enzyme assay; both cells and medium were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting. Expression in both types of samples was assessed using primary antiserum against the amino terminus of mature PC1/3 followed by horseradish peroxidase–coupled secondary antiserum (30). Immuno-reactive protein visualization was accomplished using the Super-Signal West Femto Maximum Sensitivity Substrate kit (Thermo Scientific, Rockford, IL).
Enzyme Assay
Enzymatic activity of secreted recombinant PC1/3 proteins present in conditioned medium obtained from transiently transfected HEK293 cells was measured in triplicate 50-µL reactions in a 96-well polypropylene plate containing 25 µL of conditioned medium, 200 µmol/L substrate (pyr-RTKR-amc [7-amino-4-methylcoumarin]), 100 mmol/L of sodium acetate, pH 5.5, 2 mmol/L of CaCl2, 0.1% of Brij 35, and a protease inhibitor cocktail (final concentrations 1 µmol/L of pepstatin, 0.28 mmol/L of tosyl phenylalanyl chloromethylketone, 10 µmol/L of E-64, and 0.14 mmol/L of Nα-tosyl-Lys-chloromethylketone). Reaction mixtures were incubated at 37°C and fluorescence measurements (380 nm excitation, 460 emission) were taken under kinetic conditions every 20 seconds for 1 hour in a Fluoroskan fluorometer (Thermo Scientific). Maximum rates were calculated.
Histology and Immunohistochemistry
Small bowel and colonic mucosal biopsies were stained with hematoxylin and eosin and immunohistochemistry (IHC) with antiserum to chromogranin A (CGA) as previously described (31). Briefly for IHC, sections were cut at 2 to 3 µm, deparaffinized, and endogenous peroxidase activity was quenched using 0.5%hydrogen peroxide. Heat-induced epitope retrieval was performed. Slides were placed on a Dako Autostainer and then incubated sequentially in primary anti-CGA (DakoCytomation, Campbellfield, Australia) for 30 minutes in rabbit anti-mouse immunoglobulin, followed by Envision+ (DakoCytomation). Diaminobenzidine and hydrogen peroxide were used as the substrates for the peroxidase enzyme. Similarly, PCSK1 staining was performed manually using a human anti-PCSK1 mouse monoclonal antibody (Novus/Biologicals, Littleton, CO); this antiserum is directed against the C-terminus and would be expected to be lost in a C-terminal truncation mutant. Antigen retrieval was performed in citrate buffer pH 6.0 at 95°C for 30 minutes in a steamer. Tissue was stained with the primary antibody (1:250) at room temperature for 45 minutes. Secondary antibody was used as obtained directly from the company and samples incubated at room temperature for 45 minutes (DakoCytomation). Diaminobenzidine incubation time was 10 minutes at room temperature.
RESULTS
Clinical History Before Exome Sequencing
The patient was initially assessed at 3 weeks of age for recurrent diarrhea and associated metabolic acidosis. He was a 41-weeks’ gestational age male infant born to a 17-year-old gravida 1 female and a biological father said to be the mother’s father’s first cousin. There was no history of substance abuse, prenatal infections, or other complications during the pregnancy. The baby was born at the appropriate size for gestational age with normal Apgar scores. At 6 days of age, he was transferred to the intensive care nursery because of poor peripheral perfusion and indirect hyperbilirubinemia, with an initial bicarbonate level of 8 and an anion gap of 13. Other liver enzymes (aspartate aminotransferase, alanine transaminase, and alkaline phosphatase) were normal, and urine was trace positive for reducing substances. The infant was treated for possible sepsis with antibiotics. Blood and urine cultures were subsequently negative.
Because of the initial presentation, metabolic laboratory values were sent twice. These included urine organic acids, serum amino acids, acylcarnitine profile, lactate and pyruvate as well as serum ammonia levels. These tests were normal. He was treated in the interim with carnitine supplementation. Two newborn state metabolic screens were normal for congenital adrenal hyperplasia, hypothyroidism, and disorders of amino acid, organic acid, and fatty acid oxidation. When feedings were stopped, the diarrhea ceased, but feeds were subsequently resumed using a standard milk protein–based formula.
Because of the extremely early onset of the diarrhea, congenital disorders were considered and stool was evaluated for reducing substances, pH, qualitative fat, and elastase-1 level, as well as for white and red blood cells. These were normal or negative. DNA genotyping of CFTR for cystic fibrosis was negative for the 97 mutations most commonly observed. Serum immunoreactive trypsinogen, serum α1-antitrypsin levels, phenotyping, and an ultrasound of the liver and gallbladder were normal.
Nephrology consultation for possible renal tubular acidosis resulted in brief treatments with oral bicarbonate replacement, but this was stopped when it was believed that no renal tubular issue was present. The patient received amoxicillin prophylaxis for 1 urinary tract infection during his neonatal intensive care unit course and grade 2 bilateral vesicoureteral reflux on a voiding cystoure-throgram study.
Both upper and lower gastrointestinal (GI) endoscopies revealed no gross or microscopic abnormalities in duodenal, gastric, and colonic biopsies, and electron microscopy of small bowel biopsies showed no ultrastructural abnormalities. Disaccharidase levels were normal (more detailed information can be found at http://links.lww.com/MPG/A254 and http://links.lww.com/MPG/A255).
The patient had several bouts of acute-onset acidosis requiring several boluses of sodium bicarbonate and fluids. He was discharged home at 3 months of age. Upon discharge, he was placed on Elecare (amino acid–based formula, Abbott Nutrition) and gaining weight adequately despite multiple interruptions of his feeding schedule for intolerance and diarrhea and multiple stool tests were positive for Clostridium difficile toxin. Even on a caseinhydrolyzed formula, he had gross blood in his stool, which dissipated on Elecare. His medications upon discharge included amoxicillin for urinary tract infection prophylaxis, multivitamin with iron, and metronidazole to complete a course for the stool C difficile.
Five weeks after discharge, he presented to a different hospital with a reported 3-day history of diarrhea and was found to be in hypovolemic shock with profound metabolic acidosis and an initial bicarbonate level of only 4.1.His serum sodium level was 163, and his chloride was 138. Before hydration, it was noted that he had decreased 420 g in weight from his neonatal intensive care unit discharge weight of 4620 g. He was transferred to another children’s medical center for admission, had his first central venous (Broviac) catheter placed, and started receiving total parenteral nutrition. At 6 months of age, he had significant failure to thrive with length <5%, and a weight of 5.1 kg (Z − 3.75).During this prolonged hospitalization, he was transferred to UCLA Medical Center for 1 month for additional evaluation and was returned to the transferring facility, where he remained hospitalized for an additional 3 weeks. Among various tests that were performed, the serum pancreatic polypeptide level was extremely elevated (>1600 pg/mL, normal <519) and serotonin was low (34 ng/mL, normal range 50–220); however, serum substance P (540 pg/mL, normal <1780), chromogranin A (26.2 ng/mL, normal <36.4), and vasoactive intestinal peptide (28.6 pg/mL, normal <50) levels were normal for age. A proinsulin level was, unfortunately, not obtained at that time.
He was readmitted to local community hospitals 19 times during the subsequent 31 months. Nine of these were emergency department visits, 7 were inpatient stays, and 3 were simply outpatient contacts for testing. He was subsequently placed into foster care because it was believed that many of his admissions were because of inadequate care of his central venous line by his biological parents or because of lack of appropriate outpatient follow-up.
He subsequently had multiple problems with central venous catheter occlusions and was diagnosed as having heparin-induced thrombocytopenia and later as having a plasminogen inhibitor deficiency, which were believed to result in multiple deep venous thrombi, for which he was treated with enoxaparin and later warfarin. Given these significant thrombotic events, his central venous catheter was removed and a percutaneous gastrostomy tube was placed to aid in the transition from parenteral to enteral nutrition support. Repeat upper endoscopy at that time revealed mild chronic gastritis and lactase deficiency on tissue analysis for disaccharidase levels (lactase activity 1.4, normal 24.5 ± 8.0). An upper GI and small bowel follow-through x-ray were normal, including normal transit time.
There was also an admission for pneumonia and respiratory distress. During that admission, he was noted to exhibit excessive thirst and hyperglycemia, with glucose levels running in the high 100s. He was hypokalemic and acidotic, requiring intravenous bicarbonate infusions as well as baking soda enterally. He also had evidence of left ventricular dysfunction requiring Lasix, enalapril, Aldactone, and potassium supplementation. As part of his evaluation for heart failure, fluorescence in situ hybridization studies for Williams syndrome were negative.
Sequencing/Bioinformatics Results
The initial dataset contained 21,804 nonreference variants (20,129 SNVs) and 1675 small insertions/deletions [indels]) (Table 1). In addition, another 17,172 SNVs were in the 1% false-positive tranche, meaning there was a 1% likelihood that the actual genotype at a given locus was wild-type. This unusually large number of false-positive tranche alleles appeared to be an artifact of the SOLiD platform’s quality score assignment algorithm as well as the fact that we had only a small number (n = 7) of sequencing experiments from the same platform available for analysis by the GATK Unified Genotyper’s Gaussian mixture model. We used custom data analysis software, based in part on the Ensembl Variant Effect Predictor (20) and next-generation sequencing-single nucleotide polymorphism (32) and implemented on a Microsoft SQL Server database system, to identify potentially causative alleles. We limited the search to variants within the coding region and flanking intronic essential splice site of protein-coding genes in the Ensembl dataset. Under the hypothesis that the disorder was rare, and therefore the causative allele(s) would not be common, we filtered out variants that were in the dbSNP (33), 1000 Genomes (34), National Heart, Lung, and Blood Institute (NHLBI) (35), or National Institute of Environmental Health Sciences (NIEHS) (36) datasets with an allele frequency ≥0.01 in any population. We also excluded variants observed in 74 locally sequenced exomes from unrelated individuals. Six of these exomes were sequenced on the SOLiD platform and were particularly useful to remove systemic bias and false-positives. We thus reduced the number of variants to 1043 SNVs and 11 indels that were sufficiently rare in multiple populations to be consistent with a rare disorder. Analysis of the consequences on protein-coding transcripts for significant adverse effects (nonsense, missense, or essential splice site mutations) reduced the candidate variant list to 467 SNVs and 3 indels.
TABLE 1.
Exome sequencing statistics
| Called | Filtered | Raw | |||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Known | Novel | All | Known | Novel | All | Known | Novel | |
| Called loci | 10,2961 | 85,413 | 17,548 | 82,876 | 19,608 | 63,268 | 18,5837 | 10,5021 | 80,816 |
| Ref loci | 81,157 | 66,133 | 15,024 | 61,104 | 16,843 | 44,261 | 14,2261 | 82,976 | 59,285 |
| Variant loci | 21,804 | 19,280 | 2524 | 21,772 | 2765 | 19,007 | 43,576 | 22,045 | 21,531 |
| SNVs | 20,129 | 18,291 | 1838 | 21,095 | 2639 | 18,456 | 41,224 | 20,930 | 20,294 |
| Insertions | 775 | 465 | 310 | 148 | 30 | 118 | 923 | 495 | 428 |
| Deletions | 900 | 524 | 376 | 529 | 96 | 433 | 1429 | 620 | 809 |
| Hets | 13,172 | 10,863 | 2309 | 21,330 | 2389 | 18,941 | 34,502 | 13,252 | 21,250 |
| Hom ref | 57,506 | 46,193 | 11,313 | 42,576 | 8661 | 33,915 | 10,0082 | 54,854 | 45,228 |
| Hom var | 8632 | 8417 | 215 | 442 | 376 | 66 | 9074 | 8793 | 281 |
| Het/hom ratio | 1.53 | 1.29 | 10.74 | 48.26 | 6.35 | 286.98 | 3.8 | 1.51 | 75.62 |
| Ti/Tv ratio | 2.64 | 3.11 | 0.69 | 0.54 | 1.27 | 0.48 | 1.15 | 2.73 | 0.49 |
Metrics for variants identified by exome sequencing. Called = variants that passed all quality control filters; Filtered = variants that did not pass all QC, including variants with a 1% likelihood that the actual genotype is wild-type; Hets = heterozygous genotypes; Hom var = homozygous nonreference genotypes; Hom ref = homozygous reference genotypes (wt); Known/novel = variants in/not in dbSNP137; Ref loci = =loci that match GRCh37 reference genome; SNVs = =single nucleotide polymorphisms; Ti/Tv ratio = ratio of transition (purine to purine or pyrimidine to pyrimidine) to transversion (purine to/ from pyrimidine) variants; Variant loci = loci that differ from the reference.
Under a recessive model, we searched the autosomes and sex chromosomes in this set of variants for homozygous and compound heterozygous mutations (the latter defined as 2 variants in the same transcript). We required that the variants be likely to have a deleterious effect on protein structure as predicted by at least 1 of SIFT (21–25), PolyPhen (26–28), or Condel (29)). Initially, we limited the search to variants that fully passed all of the data quality filters, but this resulted in no homozygous variants being selected, a surprising finding given the stated consanguineous parental relationship. By including the 1% false-positive tranche homozygous variants, we identified a single homozygous variant in PCSK1 and 6 compound heterozygous variants in 3 other genes (Table 2). The PCSK1 variant was within a 7.5-Mb homozygous interval, identified by 49 polymorphous dbSNP markers, consistent with inbreeding.
TABLE 2.
Genes with homozygous or compound heterozygous variants likely to affect protein
| Model | Gene | Variant_human_g1k_v37 | Variant_cDNA | Variant_protein | OMIM_disorder |
|---|---|---|---|---|---|
| Hom | PCSK1 | chr5:95746544G>C | ENST00000311106.3:c.1029C>G | ENSP00000308024.2:p.Tyr343X | 600955: Obesity with impaired prohormone processing |
| 2-Het | NOL9 | chr1:6601892A>G | ENST00000377705.5:c.1073T>C | ENSP00000366934.5:p.Leu358Pro | |
| chr1:6601893G>C | ENST00000377705.5:c.1072C>G | ENSP00000366934.5:p.Leu358Val | |||
| 2-Het | BAMBI | chr10:28971098A>C | ENST00000375533.3:c.551A>C | ENSP00000364683.3:p.Gln184Pro | |
| chr10:28971100G>T | ENST00000375533.3:c.553G>T | ENSP00000364683.3:p.Asp185Tyr | |||
| 2-Het | SPTBN4 | chr19:40996047T>A | ENST00000352632.2:c.387T>A | ENSP00000263373.2:p.Phe129Leu | |
| chr19:41025432G>A | ENST00000352632.2:c.3028G>A | ENSP00000263373.2:p.Val1010Met |
Seven rare (allele frequency ≤0.01) homozygous (hom) or compound heterozygous (2-het) mutations were found in the patient and absent from 74 control exomes.
The Tyr343X mutation in PCSK1 was highlighted by the fact that the protein would be truncated by a premature stop codon within its catalytic domain and by PCSK1’s bioinformatics annotation as the only 1 of the 4 genes having a known association with human disorders.
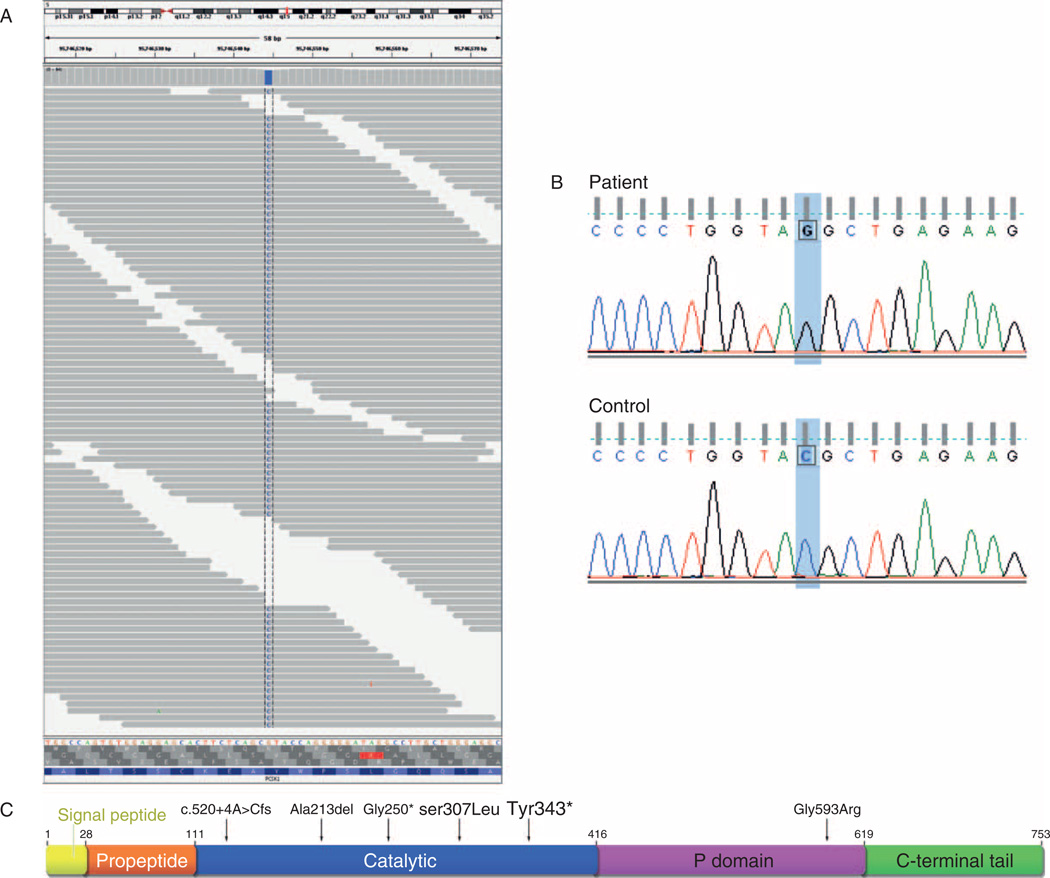
Defects in PCSK1 are the cause of PC1/3 deficiency (MIM:600955), which had previously been identified in 3 subjects and is characterized by obesity, hypogonadism, hypoadrenalism, and reactive hypoglycemia, as well as significant small-intestinal absorptive dysfunction (37–40) (Fig. 1C). Given the subject’s history of diarrhea, we confirmed the presence of the mutation in 71 sequenced fragments (Fig. 1A) and by Sanger sequencing (Fig. 1B), and assessed further whether the mutation of the PCSK1 gene could alter the protein function and account for the subject’s medical problems.
Figure 1.
Exome sequencing results. A, Aligned pileup of sequenced fragments at chr5:95746544 with 30 forward strand reads and 41 reverse strand reads of the C variant and 2 low-quality reads of the reference G allele on fragment 3′ ends. Visualized by the Integrative Genomics Viewer (39). B, Sanger sequencing validation of variant. C, Structure of PC1/3 showing locations of previously identified mutations and the novel Y343X. Adapted from (38) and (40).
Functional Analysis and In Vitro Assessment
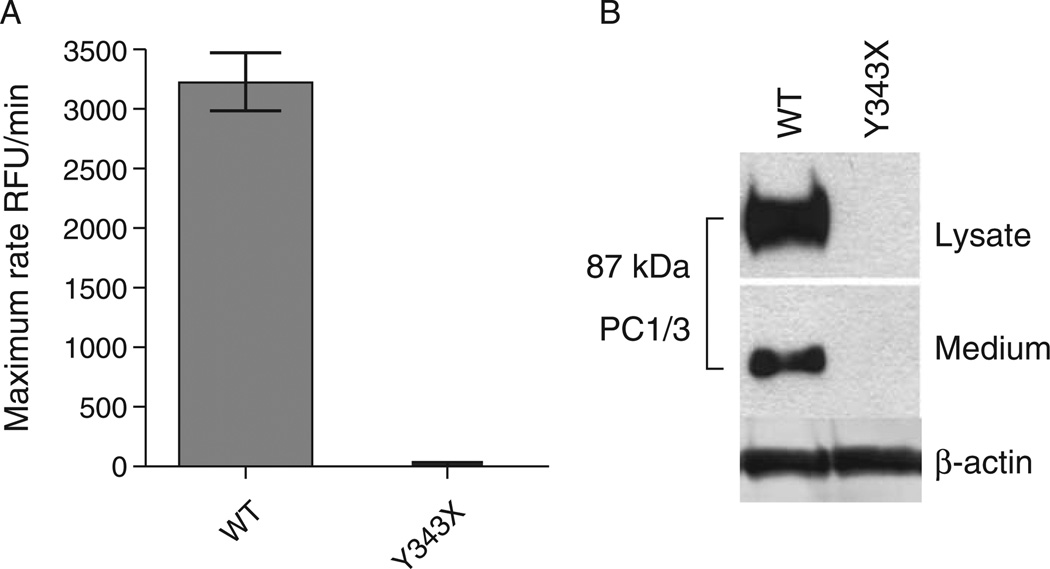
The Y343X mutation eliminates the final 410 amino acids of the protein, which includes the entire C-terminal and P domains and a portion of the catalytic domain (41). Loss of even 1 residue of the P domain is known to block enzyme expression (42). In vitro assessment allows for confirmation of the deleterious effect of the Y343X mutation on PC1/3 catalytic activity (30). The nonsense mutation rendered the Y343X gene product undetectable in either cells or media, most likely because of rapid intracellular degradation (Fig. 2A). As expected, the absence of detectable Y343X PC1/3 protein in the conditioned medium resulted in a total lack of enzyme activity (Fig. 2B).
Figure 2.
Functional characterization and visualization of wild-type (WT) prohormone convertase 1/3 (PC1/3) complementary DNA and PC1/3 containing the Y343X nonsense mutation. HEK293 cells were transiently transfected with empty pcDNA3 (not shown), WT PC1/3, and Y343X PC1/3. A, Enzymatic activity of secreted recombinant PC1/3 proteins in conditioned medium was compared by measuring maximum cleavage rates using the fluorogenic substrate pyr-RTKR-amc during a 1-hour kinetic assay. Three replicates per condition were assayed in triplicate and are shown as the mean ± standard deviation, P<0.0017 (2-tailed). B, Western blot of cell lysates and media from transfected HEK293 cells, using amino terminal–directed PC1/3 primary antiserum for detection of recombinant PC1/3 proteins. The data shown represent 1 of 3 independent experiments. β-actin shows equivalent loading of cell extracts.
Histologic Assessment
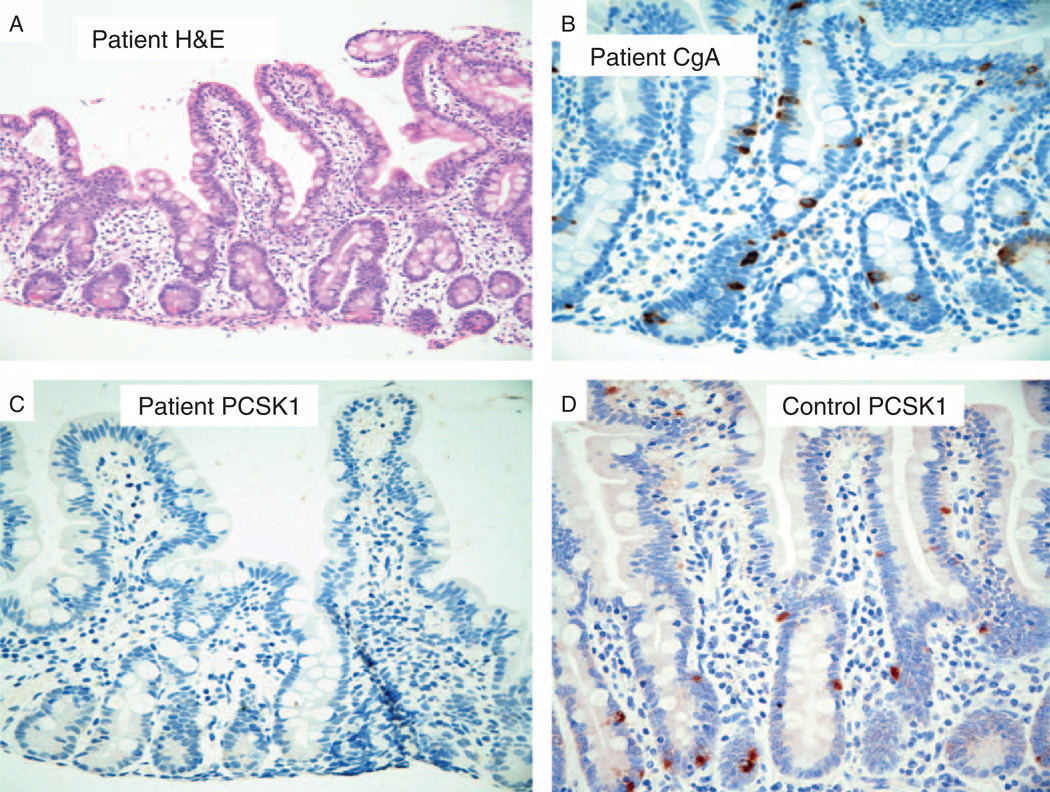
The small and large bowel mucosa from the subject was histologically normal in all respects except for the loss of PC1/3-positive enteroendocrine cells (Fig. 3). By hematoxylin and eosin staining, the architecture, immune cell complement, and epithelium were indistinguishable from normal mucosa (Fig. 3A). By IHC, the appearance and number of chromogranin A – positive enteroendocrine cells were normal (Fig. 3B); however, IHC for PC1/3-expressing enteroendocrine cells was negative relative to wild-type controls in the small (Fig. 3C, D) and large bowel (data not shown). PC1/3 IHC should decorate a subset of enteroendocrine cells in colonic and small bowel mucosa.
Figure 3.
Absence of prohormone convertase 1/3 (PC1/3)-positive enteroendocrine cells in small bowel mucosa. A, Hematoxylin and eosin (H&E)–stained proprotein convertase subtilisin/kexin type 1 mutant showing normal morphology; original magnification ×200. B, Chromogranin A immunohistochemistry of mutant tissue showing a normal pattern of enteroendocrine cells; original magnification ×400. C, PC1/3 immunohistochemistry showing complete absence of PC1/3-positive enteroendocrine cells in the mutant tissue; original magnification ×400. D, PC1/3 immunohistochemistry showing normal PC1/3 enteroendocrine cells in a healthy control; original magnification ×400.
Clinical History Following Genetic Testing
A more comprehensive and focused evaluation was prompted by identification of the genetic mutation. The adoptive parents were contacted to obtain an update of the subject’s clinical status to determine whether known clinical manifestation of PC1/3 deficiency described in the other probands was observed in this child (15). The subject had been managed at several local community facilities and was not seen at the major referral institution for >4 years. The family reported evidence suggestive of polydipsia, polyuria, enuresis, and polyphagia, but he had not been evaluated for diabetes insipidus. The subject was urgently assessed locally and found to have an undetectable serum vasopressin level. Intranasal desmopressin (DDAVP) improved significantly the severity of the polydipsia and enuretic episodes.
An endocrine analysis was performed at the referral center when the subject was 4 years, 5 months old. Before starting DDAVP, he had significant polyuria and polydipsia, drinking approximately 4 L of water per day. When parents restricted water, he would drink from the toilet, fish tank, or outdoor faucet. He experienced temper tantrums when water intake was limited. He also had evidence of severe pica and attempted to eat intravenous tubing, wood from his crib, and paper products. He was toilet trained, but still went through 1 bag of diapers per day. His appetite was described as excessive, eating more than his teenage siblings combined, and his parents keeping the refrigerator and pantry locked. His examination was otherwise normal, with the exception of a small penis: approximately 2.8 cm in stretched length (mean 5.7 cm, Z of −2.5 is 3.5 cm). Blood tests, including insulin, were generally normal except for dramatically elevated proinsulin, slightly elevated thyroid-stimulating hormone, and low serum insulin-like growth factor 1 and insulin-like growth factor–binding protein-3 (more detailed information can be found at http://links.lww.com/MPG/A254 and http://links.lww.com/MPG/A255).
DISCUSSION
Our laboratory began a systematic assessment of several distinct kindreds with various putatively genetic forms of congenital diarrhea, using exome sequencing, with the hope of identifying the molecular basis of the disorders. This article describes our analysis of a single patient, and nicely illustrates how this research technique was used to understand a challenging patient with an inherited disorder and multiple medical problems, and led to a more definitive phenotypic workup that greatly altered clinical management.
Our finding in this single case confirms the value and efficiency of exome sequencing as a primary diagnostic mode to identify mutations of genes associated with rare clinical conditions. This case pointedly illustrates how multiple hospitalizations and a barrage of various indirect, redundant, and expensive tests are frequently required—and sometimes fail—to establish medical diagnoses. Whole-exome sequencing is a transformative technology that should alter the clinician’s approach to the evaluation of such patients (43). It is conceivable that portions of the standard metabolic panels and other urine, blood, and radiographic tests will be used less frequently once exome sequencing becomes fully implemented into clinical practice. More directed phenotypic evaluation will be possible rather than the shotgun approaches typically used in children with rare genetic conditions. This phenotypic evaluation is called a “diagnostic odyssey” and is often years in duration, to the disadvantage of the patient and at great cost to family and society.
In a relatively short period of time, high-throughput sequencing has solved the mystery of numerous novel inherited disorders and has been used to identify common variants associated with various primary tumors (44,45). As costs continue to decline, this technology promises to identify the genetic basis of numerous clinical diagnoses, especially those, such as congenital diarrhea, that can be causally related to a large number of possible genes, thereby bypassing the traditional approach of targeted candidate gene sequencing (46).
Given the possibility of inbreeding, we considered the approach of using a high-density microarray to locate regions of homozygosity in the patient’s genome, then developing a set of custom-capture probes to select that region for deep sequencing, or to sequence all of the exons in the region with the traditional Sanger method. We ultimately decided on whole-exome sequencing as being more cost-effective. Furthermore, without knowledge that other family members were affected, we did not have a high previous likelihood that the mutation would be in a large region of homozygosity that could be detected by a microarray. Even though we may hypothesize that this child’s disease was caused by a homozygous mutation, there was essentially no additional cost or time needed to analyze the data for both homozygous and compound heterozygous mutations. Finally, we wished to develop an unbiased analytical pipeline that could be used for many other genetic patterns.
Others have reported on the use of next-generation sequencing to identify mutations in a gene (SLC26A3) known to be associated with chronic diarrhea; however, unlike our approach, sequencing was limited to regions that were homozygous by consanguineous descent (46a). The general efficiency of wholeexome sequencing favors the generation of whole-exome data on virtually all such patients as the most efficient and thorough means of genetic evaluation.
Severe mutations of the PCSK1 gene are rare. The Y343X mutation is novel and was not identified in the 7252 chromosomes that comprise the publicly available 1000 Genomes, NHLBI, and NIEHS datasets, suggesting an allele frequency of <0.00014 and an inferred incidence of homozygous individuals in the population of <1 in 52 million, which is consistent with the rarity of PC1/3 deficiency disorders. This mutation broadens the phenotypic consequences of PC1/3 deficiency disorders. The first PCSK1-mutant proband was a middle-aged woman who was evaluated for postprandial hypoglycemia and was found to have obesity, hypogonadotropic hypogonadism, hypoadrenalism, and elevated proinsulin levels (47,48). A second case of PC1/3 deficiency was established in an infant with generalized malabsorptive diarrhea who expired at 18 months of age (37). A third proband was described as a 6-year-old boy with a diarrheal condition that resembled the previous case, and intestinal biopsies from both children were described as a persistent enteropathy with patchy villous atrophy (38). In contrast, the subject reported here had no histopathological evidence of enteropathy and had perfectly normal crypt-villus axis without a pathologic inflammatory component (Fig. 3). We have recently presented findings on additional cases with PCSK1 mutations that we identified by Sanger sequencing (15). Retrospective questioning of the primary proband confirmed similar diarrheal symptoms that were certainly worse during early childhood (37). In our patient, as in the other 3 patients, the proinsulin level was significantly elevated. These data would suggest that serum proinsulin levels and sequencing of the PCSK1 gene could be used to establish the diagnosis; however, given the breadth of phenotypic presentations, molecular diagnosis is likely to remain challenging without implementation of broader approaches such as whole-exome sequencing.
To develop a sense of the mutational load of rare variants on PCSK1 in the whole population, we examined all 913 PCSK1 variants reported in dbSNP and the 1000 Genomes, NHLBI, and NIEHS datasets, and found 47 variants causing nonsynonymous codons or more serious consequences and having minor allele frequencies ≤0.01 (supplementary Table S1, http://links.lww.-com/MPG/A255). Interestingly, common variants in the coding region of PCSK1 are also associated with common forms of obesity (N221D, Q665E, S690T) and type 2 diabetes mellitus (Q665E, S690T) (49,50). A recent study suggests that PC1/3 deficiency is dependent on the dosage of PCSK1, and rare heterozygous mutations can cause obesity (51). From the population data, rare protein-altering mutations will be homozygous or compound heterozygous, resulting in substantial loss of PC1/3 activity in ~86 individuals per million, and these individuals would be predicted to be at risk for a life-threatening PC1/3 deficiency. Additionally, an estimated 1 in 20 individuals may harbor a modest PC1/3 deficiency, which may contribute to PC1/3 deficiency–related obesity.
Exome sequencing will typically generate upwards of 20,000 candidate variants from the reference genome in a given individual. A challenge for diagnosis by exome sequencing is to filter out variants that cannot possibly cause the disease in question. Alleles, such as synonymous or intronic variants, can be eliminated with slight risk that they are false-negatives, as can alleles that are too frequent in the population to be consistent with the incidence of the disorder. Beyond that, prediction of the functional consequences of novel mutations remains a daunting task.
Enteric anendocrinosis is another inherited intestinal endocrinopathy that has clinical features that resemble the early stages of PC1/3 deficiency, including a generalized form of malabsorption (MIM:610370) (5). Homozygous mutations of NEUROG3 were described in 3 probands, the intestines of which were devoid of enteroendocrine cells, yet had an otherwise normal-appearing intestine. NEUROG3 is a basic helix-loop-helix transcriptional factor that is necessary and sufficient to drive endocrine cell development in the pancreas and intestine (5). Although 2 of the 3 cases in the initial report did not develop insulin-dependent diabetes mellitus until preadolescent age, 2 recent cases describe the onset of diabetes during the neonatal period (52). Unlike patients with PC1/3 deficiency, children with enteric anendocrinosis do not appear to develop hypothalamic, pituitary, adrenal, thyroid, or gonadal insufficiencies.
Establishing the precise diagnosis of a congenital diarrheal condition requires an intestinal biopsy and a thoughtful approach to dietary challenges. A differential diagnosis of this condition, when presenting with seemingly histologically normal intestinal mucosa, is mostly limited to specific defects of nutrient assimilation (digestive enzymes or transport proteins) or enteroendocrinopathies. Enteroendocrinopathies are histologically subtle, are generally only discovered when specifically sought, and require immunohistochemical confirmation (31). Exome sequencing will certainly be used in the coming years to identify the inherited basis of novel diarrheal disorders and will likely be the standard of practice for genotype testing of established disorders. In our case, exome sequencing provided a diagnosis that resulted in immediate changes in patient care and an improved ability to predict clinical progression, based on previous cases of PC1/3 deficiency.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (#DK083762), and the California Institute of Regenerative Medicine RT2-01985, to M.G.M. and DA05084 to I.L.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
The authors report no conflicts of interest.
REFERENCES
- 1.Martin MG, Wright EM. Congenital intestinal transport defects. In: Walker WA, Goulet O, Kliegman RM, editors. Pediatric Gastrointestinal Disease. 4th ed. Hamilton, Canada: BC Decker; 2004. pp. 898–921. [Google Scholar]
- 2.Fishbein TM. Intestinal transplantation. N Engl J Med. 2009;361:998–1008. doi: 10.1056/NEJMra0804605. [DOI] [PubMed] [Google Scholar]
- 3.Binder HJ. Causes of chronic diarrhea. N Engl J Med. 2006;355:236–239. doi: 10.1056/NEJMp068124. [DOI] [PubMed] [Google Scholar]
- 4.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Cortina G, Wu SV, et al. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med. 2006;355:270–280. doi: 10.1056/NEJMoa054288. [DOI] [PubMed] [Google Scholar]
- 6.Martin MG, Turk E, Lostao MP, et al. Defects in Na+ glucose cotransporter (SGLT1) trafficking and function cause glucose-galactose malabsorption. Nat Genet. 1996;12:216–220. doi: 10.1038/ng0296-216. [DOI] [PubMed] [Google Scholar]
- 7.Sivagnanam M, Mueller JL, Lee H, et al. Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology. 2008;135:429–437. doi: 10.1053/j.gastro.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller T, Hess MW, Schiefermeier N, et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet. 2008;40:1163–1165. doi: 10.1038/ng.225. [DOI] [PubMed] [Google Scholar]
- 9.Hartley JL, Zachos NC, Dawood B, et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy) Gastroenterology. 2010;138:982388–982398. doi: 10.1053/j.gastro.2010.02.010. e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz-Erian P, Muller T, Krabichler B, et al. Mutations in SPINT2 cause a syndromic form of congenital sodium diarrhea. Am J Hum Genet. 2009;84:188–196. doi: 10.1016/j.ajhg.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaydon DC, Biancheri P, Di WL, et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N Engl J Med. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
- 12.Shendure J. Next-generation human genetics. Genome Biol. 2011;12:408. doi: 10.1186/gb-2011-12-9-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singleton AB. Exome sequencing: a transformative technology. Lancet Neurol. 2011;10:942–946. doi: 10.1016/S1474-4422(11)70196-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino A, Lindberg I. Peptide biosynthesis: prohormone convertases 1/3 and 2. In: Fricker LD, Devi L, editors. Colloquium Series on Neuropeptides. 1st ed. San Rafael, CA: Morgan & Claypool Life Sciences Publishers; 2004. [Google Scholar]
- 15.Martin MG, Lindberg I, Solorzano-Vargas RS, et al. Congenital proprotein convertase 1/3 deficiency causes malabsorptive diarrhea and other endocrinopathies in a pediatric cohort. Gastroenterology. 2013;145:138–148. doi: 10.1053/j.gastro.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Harrow J, Harte RA, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 18.Flicek P, Amode MR, Barrell D, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 26.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 28.Sunyaev S, Ramensky V, Koch I, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vindrola O, Lindberg I. Biosynthesis of the prohormone convertase mPC1 in AtT-20 cells. Mol Endocrinol. 1992;6:1088–1094. doi: 10.1210/mend.6.7.1508222. [DOI] [PubMed] [Google Scholar]
- 31.Cortina G, Smart CN, Farmer DG, et al. Enteroendocrine cell dysgenesis and malabsorption, a histopathologic and immunohistochemical characterization. Hum Pathol. 2007;38:570–580. doi: 10.1016/j.humpath.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Grant JR, Arantes AS, Liao X, et al. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics. 2011;27:2300–2301. doi: 10.1093/bioinformatics/btr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Center for Biotechnology Information NLoM. dbSNP134. Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine; Database of Single Nucleotide Polymorphisms (dbSNP Build ID: 134) [Google Scholar]
- 34.The 1000 Genomes Project ConsortiumA map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NHLBI Exome Sequencing Project. Exome Variant Server Volume September. Seattle, WA: NHLBI Exome Sequencing Project; 2011. [Google Scholar]
- 36.NIEHS Environmental Genome Project. NIEHS Exome Variant Server Volume September. Seattle, WA: NIEHS Environmental Genome Project; 2011. [Google Scholar]
- 37.Jackson RS. Small-intestinal dysfunction accompanies the complex endocrinopathy of human proprotein convertase 1 deficiency. J Clin Invest. 2003;112:1550–1560. doi: 10.1172/JCI18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farooqi IS, Volders K, Stanhope R, et al. Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J Clin Endocrinol Metab. 2007;92:3369–3373. doi: 10.1210/jc.2007-0687. [DOI] [PubMed] [Google Scholar]
- 39.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 41.Ueda K, Lipkind GM, Zhou A, et al. Mutational analysis of predicted interactions between the catalytic and P domains of prohormone convertase 3 (PC3/PC1) Proc Natl Acad Sci U S A. 2003;100:5622–5627. doi: 10.1073/pnas.0631617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou A, Martin S, Lipkind G, et al. Regulatory roles of the P domain of the subtilisin-like prohormone convertases. J Biol Chem. 1998;273:11107–11114. doi: 10.1074/jbc.273.18.11107. [DOI] [PubMed] [Google Scholar]
- 43.Maxmen A. Exome sequencing deciphers rare diseases. Cell. 2011;144:635–637. doi: 10.1016/j.cell.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 44.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lifton RP. Individual genomes on the horizon. N Engl J Med. 2010;362:1235–1236. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 46a.Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson RS, Creemers JW, Ohagi S, et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 48.O’Rahilly S, Gray H, Humphreys PJ, et al. Brief report: impaired processing of prohormones associated with abnormalities of glucose homeostasis and adrenal function. N Engl J Med. 1995;333:1386–1390. doi: 10.1056/NEJM199511233332104. [DOI] [PubMed] [Google Scholar]
- 49.Benzinou M, Creemers JW, Choquet H, et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 50.Strawbridge RJ, Dupuis J, Prokopenko I, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Creemers JWM, Choquet H, Stijnen P, et al. Heterozygous mutations causing partial prohormone convertase 1 deficiency contribute to human obesity. Diabetes. 2012;61:383–390. doi: 10.2337/db11-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gradwohl G, Dierich A, LeMeur M, et al. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.