Abstract
Infection of laboratory mice with murine noroviruses (MNV) is widely prevalent. MNV alters various mouse models of disease, including the Helicobacter bilis-induced mouse model of inflammatory bowel disease (IBD) in Mdr1a−/− mice. To further characterize the effect of MNV on IBD, we used mice deficient in the immunoregulatory cytokine IL10 (Il10−/− mice). In vitro infection of Il10−/− bone marrow-derived macrophages (BMDM) with MNV4 cocultured with H. bilis antigens increased the gene expression of the proinflammatory cytokines IL1β, IL6, and TNFα as compared with that of BMDM cultured with H. bilis antigens only. Therefore, to test the hypothesis that MNV4 infection increases inflammation and alters disease phenotype in H. bilis-infected Il10−/− mice, we compared the amount and extent of inflammation in Il10−/− mice coinfected with H. bilis and MNV4 with those of mice singly infected with H. bilis. IBD scores, incidence of IBD, or frequency of severe IBD did not differ between mice coinfected with H. bilis and MNV4 and those singly infected with H. bilis. Mice infected with MNV4 only had no appreciable IBD, comparable to uninfected mice. Our findings suggest that, unlike in Mdr1a−/− mice, the presence of MNV4 in Il10−/− mouse colonies is unlikely to affect the IBD phenotype in a Helicobacter-induced model. However, because MNV4 altered cytokine expression in vitro, our results highlight the importance of determining the potential influence of MNV on mouse models of inflammatory disease, given that MNV has a tropism for macrophages and dendritic cells and that infection is widely prevalent.
Abbreviations: BMDM, bone marrow-derived macrophages; IBD, inflammatory bowel disease; MLN, mesenteric lymph node; MNV, murine norovirus
Inflammatory bowel disease (IBD), which includes both ulcerative colitis and Crohn disease, is a chronic and relapsing inflammatory disorder of the gastrointestinal tract. In addition, patients with IBD may be at increased risk of developing colorectal cancer.15,46 Although the exact mechanisms of disease are still not understood fully, the pathogenesis of disease is likely multifactorial, with components of the innate and adaptive immune systems, host genetics, and environmental factors (for example, the commensal gut microflora) all playing a role.4,37,55
Animal models of IBD have been used to advance our knowledge and understanding of IBD pathogenesis and treatment.16,20,37,38,52 One such model that has been widely used to elucidate the mechanisms of IBD is the interleukin10–deficient (Il10−/−) mouse.3,5,6,20,21,29,33,57 The antiinflammatory cytokine IL10 modulates both innate and adaptive immune responses.41 Produced mainly by dendritic cells, monocytes, macrophages, and T regulatory cells, IL10 exerts its immunomodulatory effects by various mechanisms including decreasing secretion of proinflammatory cytokines (for example, interferon γ, IL1, IL2, IL6, IL12 and TNFα) and downregulating important components of innate immune responses and T-cell activation (for example, MHC class II, costimulatory molecules, and nitric oxide production) in antigen presenting cells.14,41 As a consequence, Il10−/− mice, which lack the suppressive effects of IL10, develop IBD in response to their commensal gut microflora or to certain microbial triggers such as Helicobacter infections.5,6,11,21,29,52,57
Antigen-presenting cells such as macrophages and dendritic cells play key roles in the inflammatory responses in IBD.32,47,50 In 2003, a newly discovered murine norovirus (MNV) in laboratory mice was shown to infect macrophages and dendritic cells.27,53 Subsequent studies indicated widespread MNV infection in laboratory mice used for biomedical research, with a serologic prevalence as high as 32%.25,43 Members of the genus Norovirus are regarded as gastrointestinal pathogens in humans and animals, eliciting both innate and adaptive immune responses.19 Therefore, in light of the cellular (macrophages and dendritic cells) and tissue (gastrointestinal) tropisms of MNV as well as the high prevalence of MNV infection in laboratory mice, we hypothesized that MNV infection could be a potential confounder in mouse models of inflammatory diseases including IBD. In support of this idea, our laboratory recently reported that MNV infection in Mdr1a−/− mice (FVB.129P2-Abcb1atm1Bor) accelerated weight loss and exacerbated IBD progression initiated by H. bilis infection.31 This effect potentially was mediated in part through modulating dendritic cell and cytokine responses. In addition, others have reported gastrointestinal abnormalities as a result of MNV infection in some strains of mice,7,26,36 whereas others have described the importance of both innate and adaptive immune responses during MNV infection.8,9,10,28,34,36,48 Collectively, these data indicate that MNV could alter inflammatory responses in laboratory mice.
Here we extended our studies of MNV beyond Mdr1a−/− mice to Il10−/− mice, another common animal model of IBD, to further examine the potential effect of MNV on IBD research. Disease was initiated in Il10−/− mice with H. bilis, and we determined whether coinfection with MNV altered disease development, incidence, and severity and the production of cytokines. We demonstrated that although MNV stimulates a Th1 skewing of cytokines in Il10−/− bone marrow-derived macrophages (BMDM) in vitro, MNV does not alter the development, incidence, or severity of disease in vivo. Therefore, although MNV may not affect disease in Il10−/− mouse models, the virus may influence in vitro cytokine phenotypes and thus complicate interpretation of such data. To our knowledge, this report is the first to describe the evaluation of MNV infection in the Helicobacter-induced Il10−/− mouse model of IBD.
Materials and Methods
Animals and husbandry.
Female Il10−/− mice (B6.129P2-Il10tm1Cgn/J; Stock 002251) were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were certified by the vendor to be free of specific rodent pathogens including ectoparasites, endoparasites, Pneumocystis murina, Helicobacter spp., known enteric and respiratory bacterial pathogens and antibodies to murine norovirus, mouse hepatitis virus, Sendai virus, pneumonia virus of mice, reovirus 3, Theiler murine encephalomyelitis virus, ectromelia virus, polyoma virus, lymphocytic choriomeningitis virus, mouse adenovirus, minute virus of mice, mouse parvovirus, mouse rotavirus, mouse cytomegalovirus, mouse thymic virus, Hantaan virus, K virus, Encephalitozoon cuniculi, CAR bacillus, Mycoplasma pulmonis, and Clostridium piliforme. Mice were maintained under SPF conditions (except for experimental infection with MNV and H. bilis) during the study according to sentinel surveillance of the animal housing room. Il10−/− mice were group housed by infection status in autoclaved, individually ventilated cages (Thoren, Hazleton, PA) with corncob bedding (The Andersons, Maumee, OH), fed standard irradiated rodent chow ad libitum (Purina Lab Diet 5053, Brentwood, MO), and provided acidified, reverse-osmosis–purified, autoclaved water in bottles. All manipulations were performed in a vertical flow animal transfer station (AniGard II, The Baker Company, Sanford, ME) disinfected with chlorine dioxide (dilution, 1:18:1; Clidox S, Pharmacal Research Laboratories, Naugatuck, CT). The University of Washington's animal facilities are AAALAC-accredited, and all animal studies were approved by the IACUC.
PCR for Helicobacter and RT-PCR for MNV.
Primers to detect Helicobacter spp., H. bilis, and MNV have been previously described.6,24 DNA was extracted from fecal pellets either by using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's recommended protocol or by mixing 1.2 mL PBS with a fecal sample, shaking for 20 min at room temperature, centrifuging the mixture at approximately 300 × g for 5 min, and then using 20 µL of the supernatant in the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to manufacturer's recommended protocol. Helicobacter-specific PCR was performed by using 2 µL DNA template and the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich, St. Louis, MO) with thermocycling parameters of 95 °C for 4 min, followed by 40 cycles of 95 °C for 2 s, 60 °C for 2 s, and 72 °C for 1 min. H. bilis-specific PCR was performed by using 2 µL of DNA template and the GoTaq Green Master Mix (Promega, Madison, WI), with thermocycling parameters of 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 60 °C for 20 s, and 72 °C for 1 min. For RNA extraction, feces were homogenized in RLT buffer for 40 s by using Lysing Matrix E Tubes and the FastPrep-24 (MP Biomedicals, Solon, OH), centrifuged at full speed for 3 min, and the resulting supernatant used with the RNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer's recommended protocol. RT-PCR for MNV was performed by using 2 µL of RNA template in a 25-µL reaction volume and the OneStep RT-PCR Kit (Qiagen, Valencia, CA), with thermocycling parameters as previously described.24
Experimental infections with H. bilis and MNV4.
H. bilis was grown as previously described,31 with the modification that organisms initially were streaked onto Brucella blood agar plates in a 1:1 mixture of brain heart infusion broth and Brucella broth. MNV4 was propagated in RAW 264.7 cells and plaque-assayed as previously described,25 with the modifications of using 1% penicillin–streptomycin instead of ciprofloxacin and no HEPES. Clarified supernatants of low-passage (passage 7) MNV4 were used for experimental inoculations, and clarified supernatants of uninfected RAW 264.7 cells were used for control inoculations.
Prior to any experimental manipulation, mice were acclimated for at least 1 wk and confirmed to be negative for Helicobacter spp. and MNV by fecal PCR or RT-PCR, respectively. At study termination, infection status was confirmed by fecal PCR or RT-PCR. Two independent animal experiments (studies 1 and 2) were performed to evaluate histopathology of the cecum, colon, mesenteric lymph nodes (MLN), and abdominal mesenteric lymphatics. Mice were divided into 4 groups: coinfected with MNV4 and H. bilis; singly infected with H. bilis; singly infected with MNV4; and uninfected controls. Mice were weighed and monitored for signs of illness and colitis. When experimental mice met preestablished endpoint criteria (for example, weight loss of 10% to 20% of baseline, signs of illness such as lethargy, decreased activity, and hunched posture), they were euthanized with an inhaled overdose of carbon dioxide gas, and samples were collected for analysis. When approximately 25% of the mice within a study had reached endpoint criteria, the remaining mice were euthanized for analysis.
In study 1, female, 7-wk-old Il10−/− mice were used. On day 0, coinfected mice (n = 20) were orally gavaged with 1 × 106 pfu MNV4 and then orally gavaged with 2 × 107 cfu H. bilis 5 d later. H. bilis singly infected mice (n = 20) were orally gavaged with uninfected RAW cell clarified lysate on day 0 and with H. bilis on day 5. Concurrently and to serve as controls, female, 6-wk-old, Il10−/− mice in the MNV4 singly infected group (n = 10) were orally gavaged with MNV4 on day 0 and then Brucella broth on day 5, whereas uninfected control mice (n = 10) were orally gavaged with uninfected RAW cell clarified lysate on day 0 and Brucella broth on day 5. All mice were monitored at least 3 times each week for the development of disease, and individual mice were necropsied when they met endpoint criteria. The remaining mice in the study were euthanized at 24 d after H. bilis inoculation.
In study 2, female Il10−/− mice were used as described in Study 1, except that some mice were slightly older (6 to 8 wk old) and age-matched across groups. In addition, larger groups (n = 35) than those in study 1 were used to compare IBD in mice coinfected with MNV4 and H. bilis compared with mice singly infected with H. bilis. All mice were monitored and necropsied as in study 1, and all remaining mice were euthanized 16 d after H. bilis inoculation.
Histopathology and scoring.
The cecum, MLN with mesentery, and a ‘Swiss roll’ of the colon were immersion-fixed in 10% phosphate-buffered formalin and prepared for routine histologic exam. Sections of the large bowel (cecum, proximal colon, mid-colon, and distal colon) stained with hematoxylin and eosin were scored for IBD by a pathologist, who was masked to experimental groups as previously described,49 with the modification that section scores were summed directly and were not size-weighted. Scores ranged between 0 and a theoretical maximum of 64. Lymphangitis scores were determined by evaluating the mesentery surrounding the MLN and attached to the colon, by using the grading criteria previously described.31 A theoretical maximum lymphangitis score of 12 was possible for each animal. MLN hyperplasia and inflammation was scored by using the grading criteria shown in Figure 1. The extent of histiocytes, plasma cells, and mucin-filled macrophages each were scored independently for both sinusoids and medullary cords of the MLN, whereas the germinal center formation score was based on number and size. A theoretical maximal MLN inflammation score of 28 was possible for each mouse.
Figure 1.
MLN hyperplasia and inflammation scoring system. a, The extent of histiocytes, plasma cells, and mucin-filled macrophages were each scored individually for the sinusoids and medullary cords.
BMDM and real-time RT–PCR gene expression.
Bone marrow was isolated from Il10−/− mice and macrophages differentiated in cell culture as previously described,30 with modifications including differentiating cells for 1 to 2 wk and then overnight-incubating 0.8 to 1.4 × 106 macrophages per well (6-well plates) in RPMI 1640 media supplemented with 10% FBS and 30% conditioned media. The next day, treatment was performed in RPMI 1640 media with 1% Nutridoma-SP (Roche, Indianapolis, IN), 5 ng/mL mouse recombinant macrophage colony-stimulating factor (PeproTech, Rocky Hill, NJ), 1% penicillin–streptomycin, and 20 µg/mL gentamicin. BMDM were infected with MNV4 (endotoxin-free, < 0.5 EU/mL) at a multiplicity of infection of 0.2, and then treated with H. bilis sonicates (final concentration, 0.2 or 2 µg/mL) 24 h later. After incubation for another 24 h, RNA extraction and gene expression analysis by real-time RT–PCR was performed in triplicate as previously described.30 Target gene expression was normalized to the housekeeping gene hypoxanthine–guanine phosphoribosyltransferase and calculated as the fold change relative to the average value obtained from the uninfected controls. Primers for PCR were: IL6 forward, 5′ AGA GTT GTG CAA TGG CAA TTC TGA 3′; IL6 reverse, 5′ TGG TAC TCC AGA AGA CCA GAG GAA 3′; TNFα forward, 5′ CTG AAC TTC GGG GTG ATC GG 3′; and TNFα reverse, 5′ GGC TTG TCA CTC GAA TTT TGA GA 3′. Primers for IL1β have been described previously.17
Serum cytokine expression.
Serum cytokine levels were analyzed by using the Milliplex MAP Mouse Cytokine/Chemokine Premixed 13-Plex (Millipore, Billerica, MA) according to the manufacturer's recommendations and run on a Bio-Plex 200 system with Bio-Plex Manager software version 5.0 (Bio-Rad, Hercules, CA). Cytokines evaluated included GM-CSF, IFNγ, IL1β, IL2, IL4, IL5, IL6, IL7, IL10, IL12 (p70), IL13, MCP1, and TNFα.
Statistical analysis.
Statistical analyses were performed by using Prism version 5 or 6 (GraphPad, San Diego, CA). An unpaired Student t test or Mann–Whitney test was used to compare histologic scores of IBD, lymphangitis, and MLN inflammation and serum cytokine levels between mice singly infected with H. bilis and those coinfected with MNV4 and H. bilis. The Fisher exact test was used to compare the incidence and severity of IBD between mice singly infected with H. bilis and those coinfected with MNV4 and H. bilis. Correlation between lymphangitis or MLN inflammation scores and IBD scores was assessed via Spearman nonparametric correlation. To evaluate effects of mouse age at inoculation on IBD development, a Kruskal–Wallis test with a Dunn multiple comparison test was performed to compare IBD scores and time to necropsy of all mice that received H. bilis (that is, regardless of MNV4 infection status) at 6, 7, or 8 wk of age.
Results
MNV4 alters gene expression in Il10−/− BMDM stimulated with H. bilis antigen.
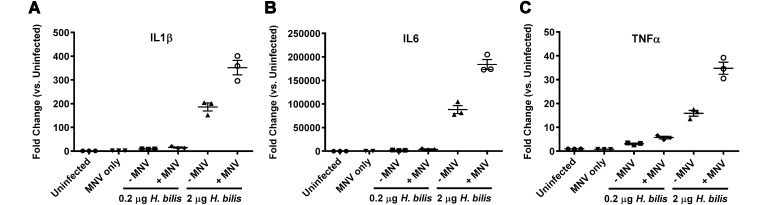
To determine whether MNV infection could augment development of H. bilis-induced IBD in Il10−/− mice, we first examined whether the cytokine profile of coinfected Il10−/− macrophages was altered in vitro. BMDM isolated from Il10−/− mice were incubated with MNV4 alone, H. bilis sonicate alone, or with MNV4 plus H. bilis sonicate and then evaluated for changes in gene expression of proinflammatory cytokines. Although H. bilis treatment alone increased expression of IL1β, IL6, and TNFα, coculture of H. bilis sonicate with MNV4 increased expression of these proinflammatory cytokines even further. The addition of MNV4 increased the expression of IL1β from 9-fold to 15-fold (Figure 2 A), IL6 from 1970-fold to 3903-fold (Figure 2 B), and TNFα from 3-fold to 6-fold (Figure 2 C) when cells were exposed to 0.2 µg of H. bilis sonicate. When a higher dose (2 µg) of H. bilis sonicate was used, the fold changes were even greater. The addition of MNV4 further elevated the expression of IL1β from 186-fold to 352-fold (Figure 2 A); IL6 from 88,133-fold to 184,000-fold (Figure 2 B); and TNFα from 16-fold to 35-fold (Figure 2 C) when cells were exposed to 2 µg of H. bilis sonicate. Notably, increasing the concentration of H. bilis 10-fold augmented the effects of MNV4 in a dose-dependent manner. MNV4 infection alone minimally increased the expression of IL1β and IL6 (1.5- and 3-fold, respectively) and did not alter TNFα expression when compared with those in uninfected controls.
Figure 2.
Inflammatory cytokine gene expression is increased in MNV4 infected Il10−/− BMDM stimulated with H. bilis antigen. Fold change of (A) IL1β, (B) IL6, and (C) TNFα compared with uninfected controls are shown. Data are given as mean ± SE. The results shown are representative of 1 of 2 independent experiments.
MNV4 does not alter incidence or severity of H. bilis-induced colitis in Il10−/− mice.
Because macrophages are an important component in innate responses in IBD,32,50 we reasoned that coinfection with both H. bilis and MNV4 would alter the development and disease phenotype in the Il10−/− mouse model. Therefore, we performed 2 independent in vivo experiments in which histologic scores, incidence of disease, and frequency of severe IBD were compared between mice inoculated with H. bilis alone and mice coinfected with both MNV4 and H. bilis.
In study 1, IBD scores (mean ± SEM) in mice coinfected with MNV4 and H. bilis (24.9 ± 4.7) tended to be greater (P = 0.07) than in mice infected with H. bilis alone (14.8 ± 4.6; Figure 3 A). According to liberal criteria in defining IBD (that is, an IBD score of greater than 0), 15 of 20 (75%) mice coinfected with MNV4 and H. bilis showed histologic evidence of IBD as compared with only 9 of 20 (45%) mice infected with H. bilis alone (Figure 3 B). Although an interesting trend, this difference in proportions (0.30; 95% CI, 0.011 to 0.589) was not statistically significant (P = 0.11). Defining severe IBD as any animal with an IBD score of greater than 30, a similar but statistically insignificant trend emerged. Whereas 9 of 20 (45%) mice coinfected with MNV4 and H. bilis had severe IBD, only 5 of 20 (25%) mice infected with H. bilis alone did (P = 0.32; Figure 3 C).
Figure 3.
MNV4 does not alter (A and D) histologic IBD scores, (B and E) incidence of IBD, or (C and F) frequency of severe IBD in H. bilis-infected Il10−/− mice. Results of 2 independent experiments are shown in panels A through C (study 1, n = 20 mice per group) and panels D through F (study 2, n = 35 mice per group). The incidence of IBD was calculated based on mice with IBD scores greater than 0, whereas mice with severe IBD were defined as those with IBD scores greater than 30 (dotted horizontal line). Data are given as mean ± SE.
Because the infection of mice with MNV4 in study 1 suggested a trend toward an altered phenotype in H. bilis-induced disease in Il10−/− mice, we used larger groups in study 2. Despite increasing animal numbers to give study 2 more statistical power, similar results to those in study 1 were seen, with no significant difference in IBD scores (P = 0.88) when comparing mice coinfected with MNV4 and H. bilis (23.6 ± 3.5) and those infected with H. bilis alone (21.7 ± 3.5; Figure 3 D). Likewise, no significant differences were noted in the incidence of IBD (P = 0.59; Figure 3 E) or frequency of severe IBD (P = 0.81; Figure 3 F) when mice coinfected with MNV4 and H. bilis were compared with mice infected with H. bilis alone. In both studies, uninfected Il10−/− mice and those infected with MNV4 alone had minimal to no disease (Figure 3 A and D).
Lymphangitis and MLN inflammation correlate with IBD scores.
We previously noted changes in lymphatic tissue associated with MNV infection in another bacterial-induced mouse model of IBD.31 Therefore, mice from study 1 also were evaluated for lymphangitis within the abdominal mesentery surrounding the MLN and attached to the colon and for hyperplasia and inflammation within the MLN itself. There was no difference in lymphangitis scores (mean ± SEM) between mice coinfected with MNV4 and H. bilis (2.8 ± 0.6) compared with mice infected with H. bilis alone (2.0 ± 0.7; P = 0.29). Likewise, inflammation scores in the MLN did not differ between mice coinfected with MNV4 and H. bilis (7.8 ± 0.7) compared with mice infected with H. bilis alone (6.2 ± 0.8; P = 0.16). Even though there was no differential effect with MNV4, lymphangitis and MLN inflammation scores positively correlated with IBD scores (lymphangitis scores: coinfected MNV4 and H. bilis group, P = 0.0027, Spearman r = 0.6484; H. bilis alone group, P < 0.0001, Spearman r = 0.8679; MLN inflammation scores: coinfected MNV4 and H. bilis group, P < 0.0001, Spearman r = 0.8890; H. bilis alone group, P < 0.0001, Spearman r = 0.8331).
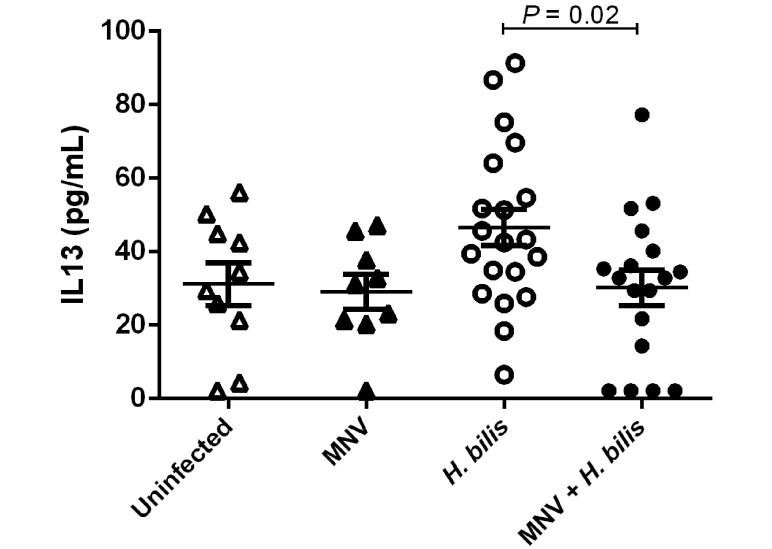
MNV4 abrogates H. bilis-induced increases in serum IL13 levels.
Serum samples collected at necropsy from study 1 mice were evaluated for cytokines and chemokines (GM-CSF, IFNγ, IL1β, IL2, IL4, IL5, IL6, IL7, IL10, IL12 (p70), IL13, MCP1, and TNFα). IL13 was increased in mice infected with H. bilis alone as compared with uninfected controls (Figure 4), although the difference was not statistically significant (P = 0.068). In mice coinfected with MNV4 and H. bilis, this increase in IL13 by H. bilis alone was abrogated (P = 0.02). No differences were found in any of the other serum analytes evaluated when mice coinfected with MNV4 and H. bilis were compared with those infected with H. bilis alone. Likewise, MNV4 infection alone (that is, without H. bilis) did not alter any serum cytokine or chemokine levels when compared with those in uninfected mice at study endpoint (IL13, Figure 4; remaining cytokines and chemokines, data not shown).
Figure 4.
Increased serum levels of IL13 induced by H. bilis are abrogated by MNV4. Serum cytokine levels of IL13 were measured from Il10−/− mice at the endpoint of study 1. Data are given as mean ± SE.
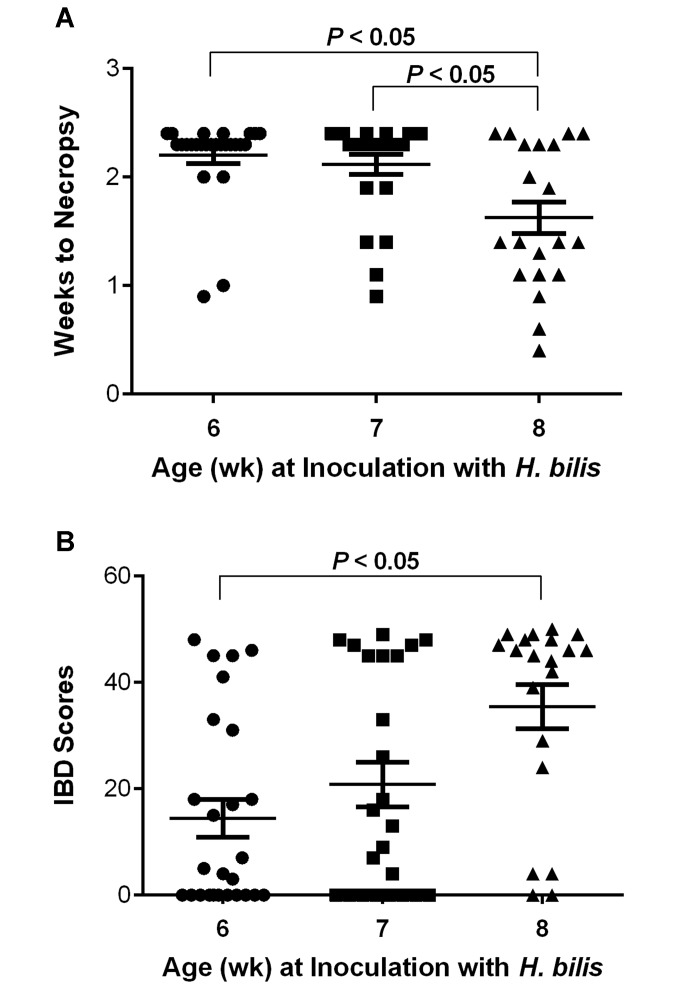
Older age at time of H. bilis infection in Il10−/− mice results in a more rapid progression of clinical disease and higher IBD scores, independent of MNV4 infection.
We noted that IBD phenotype was affected by age of mice at study initiation. In study 1, all mice that received H. bilis (either with or without MNV4) were 7 wk of age at time of infection. However, due to animal availability, study 2 used a mixed-age range of 6- to 8-wk-old Il10−/−mice. During the course of Study 2, a greater proportion of 8-wk-old mice reached endpoint criteria earlier and therefore had to be euthanized sooner than did younger, 6- to 7-wk-old mice. Given that MNV4 infection did not alter IBD scores, incidence of IBD, or frequency of severe IBD in study 2 (Figure 3 D through F), data were pooled for all Il10-/- mice inoculated with H. bilis, regardless of MNV4 infection status. The data then were stratified by animal age at time of H. bilis infection (6 wk, n = 26; 7 wk, n = 24; 8 wk, n = 20) to evaluate whether age at infection affected IBD scores or time to necropsy. Il10−/− mice that were 8 wk old at the time of infection with H. bilis reached euthanasia endpoints sooner as a result of their IBD (1.63 ± 0.15 wk, mean ± SEM; P < 0.05) than did those inoculated at 7 wk (2.12 ± 0.09 wk) or 6 wk (2.20 ± 0.07 wk) of age (Figure 5 A). Similarly, Il10−/− mice that were 8 wk old at the time of H. bilis infection had higher IBD scores (35.5 ± 4.1) when compared with those dosed at 7 wk (20.8 ± 4.2) or 6 wk (14.5 ± 3.5) of age, although statistical significance was reached only between mice infected at 8 wk of age compared with 6 wk (Figure 5 B; P < 0.05).
Figure 5.
Older age at time of H. bilis infection resulted in a more rapid progression of IBD and higher IBD scores. (A) Weeks to necropsy and (B) IBD scores are shown for all H. bilis-inoculated Il10−/− mice (that is, regardless of MNV4 infection status) from study 2 and are segregated according to animal age at time of inoculation with H. bilis. Data from mice coinfected with MNV4 and H. bilis were pooled with those of mice infected with H. bilis only because MNV4 did not affect the development of H. bilis-induced IBD. Data are given as mean ± SE.
Discussion
Infection of laboratory mice with a novel norovirus was first reported in 2003,27 and the virus has subsequently been shown to be highly prevalent and present worldwide.18,25,35,43,56 Results thus far regarding the effect of MNV infection in various mouse models of disease have been variable. Some studies have shown alteration to animal models with MNV infection,7,31,40 whereas others describe minimal to no effects.1,13,22,23,30,39 Our laboratory recently reported that MNV4 infection modulates progression of IBD in Mdr1a−/− mice induced with H. bilis infection.31 In these mice, coinfection with MNV4 and H. bilis resulted in greater weight loss and higher IBD scores than in mice infected with H. bilis alone.31 Other laboratories have reported alterations in intestinal injury as a result of MNV infection in a dextran sodium sulfate-induced model of colitis in mutant mice.7 These mice had a mutation in the autophagy gene Atg16L1, and variations of the ATG16L1 gene in people may increase the risk of Crohn disease.44 Therefore, given the widespread prevalence of MNV as well as the documented effects of MNV on some mouse models of IBD, our laboratory hypothesized that MNV would alter the disease phenotype in another common model of IBD, Il10−/− mice. In support of this hypothesis, MNV has a cellular tropism for macrophages and dendritic cells,53 which are key antigen-presenting cells and contribute to the cytokine milieu and development of inflammation in the gut. In the present study, mice lacking Il10 were infected with H. bilis to trigger the development of IBD6 and then evaluated for the progression of disease in the presence or absence of MNV infection.
We initially used in vitro studies to evaluate the effect of MNV infection on macrophages derived from Il10−/− mice and stimulated with H. bilis antigens. When stimulated with H. bilis sonicates, MNV4-infected BMDM had increased mRNA expression of IL1β, IL6, and TNFα when compared with that of H. bilis stimulated BMDM without MNV4. Similarly, MNV1 has previously been reported to enhance the expression of IL6 and TNFα in BMDM infected with either Escherichia coli or Pseudomonas aeruginosa when compared with cytokine expression after infection with bacteria alone.28 To test whether these in vitro findings translated to in vivo effects, we conducted 2 independent studies in which Il10−/− mice were infected first with MNV4 and then 5 d later with H. bilis. Despite the proinflammatory effect of MNV4 on BMDM, there were no statistically significant changes in disease development or histopathology. Therefore, MNV exacerbated the progression of IBD in Mdr1a−/− mice31 and altered responses to dextran sodium sulfate-induced intestinal injury in Atg16L1 mutant mice7 but had no effect on disease development in Il10−/− mice. Although both the Mdr1a−/− and Il10−/− mice can be triggered with the same bacteria to induce IBD, the different mechanisms by which each strain develops IBD may account for the differences seen. Mdr1a−/− mice have a gut epithelial barrier defect, which has been speculated to result in altered antigen presentation and innate immune responses due to inappropriate exposure to commensals in the intestine.16,37,45 Similarly, the toxic effects of dextran sodium sulfate7 damage the gut epithelial barrier.52 In comparison, Il10−/− mice develop IBD as a result of the lack of the immunoregulatory cytokine IL10, impacting both innate and adaptive immune responses and impairing regulatory T-cell function.16,37,45 In addition, the background mouse strain may have contributed to the different results regarding MNV4’s ability to modulate IBD, in that Mdr1a−/− mice have an FVB background, Atg16L1 mutant mice a mixed background, and Il10−/− mice are on a C57BL/6 background.
MNV4 infection did not alter levels of most of the serum cytokines and chemokines measured at the endpoint of study 1. The one exception was IL13, in which MNV4 abrogated increases in serum IL13 induced by H. bilis infection in Il10−/− mice. A recent study23 reported decreased levels of IL13 mRNA expression (as measured by quantitative RT-PCR analysis) in the cecum of MNV4- (as well as MNV1-) infected C57BL/6 mice when compared with uninfected mice. This result contrasts with our finding, in which MNV4 infection alone did not decrease serum IL13 compared with that in uninfected mice. In addition, infection with Salmonella typhimurium alone decreased expression of IL13, and coinfection with MNV did not significantly alter this response.23 This finding also contrasts with our results, in which MNV4 coinfection appeared to modulate IL13 responses in mice infected with H. bilis. IL13 is associated with a Th2 immune response, is functionally related to IL4,54 and has recently been reported to have a protective role in the piroxicam- or helminth-induced colitis model in Il10-/- mice by suppressing Th1/Th17-mediated inflammation.12,51 Determining whether the abrogation of increased IL13 by MNV4 in the context of H. bilis infection that we noted in the current study is biologically significant or just an inconsequential finding requires additional studies.
During the course of study 2, we noted that a subset of mice had to be euthanized sooner after H. bilis infection than did other mice in the study, and this observation stratified with age of the mouse at time of H. bilis infection. Why the age at the time of H. bilis infection affected survival times and IBD scores in our study is unclear. Similarly, other laboratories have reported differences in inflammatory disease phenotype as a result of mouse age at the time of infection with other Helicobacter spp.11
Many strains of MNV have been reported since the virus was first identified in 2003.2,18,24,42 MNV4 was originally derived from a colony of naturally infected laboratory mice24 and efforts were made to passage the virus minimally in vitro before use in vivo. Therefore, MNV4 is thought to closely resemble other field isolates of MNV circulating in laboratory mice, most of which have been shown to cause persistent infections. We elected to study MNV4 since our laboratory had previously published a number of studies with MNV4, including the report in which MNV4 altered disease progression in the Mdr1a−/− IBD mouse model.30,31,39,40 It is unknown, however, whether other strains of MNV may impact the H. bilis-induced Il10−/− mouse model of IBD.
Although a previous study in our laboratory indicated that MNV4 infection is an intercurrent variable in the H. bilis-induced Mdr1a−/− IBD mouse model,31 we did not detect any differences in IBD after MNV4 infection in the Il10−/− H. bilis-induced IBD model. However, MNV4 did alter in vitro cytokine gene expression in BMDM derived from Il10−/− mice, even though this finding did not translate to differential effects on IBD in vivo. These results are consistent with other reports in the literature, in which MNV infection had minimal or no effects on other mouse models of disease, including colon cancer, obesity and insulin resistance, Salmonella typhimurium infection, and various viral infections.1,13,22,23,30,39 However, given MNV's ability to influence some mouse models of disease,7,31,36,40 its cellular tropism for macrophages and dendritic cells,53 and its ability to influence cytokine gene expression in vitro, reporting of these negative findings adds to the knowledge and characterization of this highly prevalent virus.
Acknowledgments
We thank Lela Riley, Bob Livingston, and Greg Purdy for providing MNV4 and H. bilis. We thank Gary Knowels and Jennifer Finley for their technical assistance. This work was supported by NIH grant R01-OD011149.
References
- 1.Ammann CG, Messer RJ, Varvel K, Debuysscher BL, Lacasse RA, Pinto AK, Hasenkrug KJ. 2009. Effects of acute and chronic murine norovirus infections on immune responses and recovery from Friend retrovirus infection. J Virol 83:13037–13041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barron EL, Sosnovtsev SV, Bok K, Prikhodko V, Sandoval-Jaime C, Rhodes CR, Hasenkrug K, Carmody AB, Ward JM, Perdue K, Green KY. 2011. Diversity of murine norovirus strains isolated from asymptomatic mice of different genetic backgrounds within a single US research institute. PLoS ONE 6:e21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, Thompson-Snipes L, Leach MW, Rennick D. 1996. Enterocolitis and colon cancer in interleukin10-deficient mice are associated with aberrant cytokine production and CD4+ Th1-like responses. J Clin Invest 98:1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouma G, Strober W. 2003. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol 3:521–533 [DOI] [PubMed] [Google Scholar]
- 5.Buchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, Hedrich HJ, Bleich A. 2012. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota–host interactions. Inflamm Bowel Dis 18:943–954 [DOI] [PubMed] [Google Scholar]
- 6.Burich A, Hershberg R, Waggie K, Zeng W, Brabb T, Westrich G, Viney JL, Maggio-Price L. 2001. Helicobacter-induced inflammatory bowel disease in IL10- and T-cell–deficient mice. Am J Physiol Gastrointest Liver Physiol 281:G764–G778 [DOI] [PubMed] [Google Scholar]
- 7.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus plus susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. 2008. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog 4:e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chachu KA, Strong DW, LoBue AD, Wobus CE, Baric RS, Virgin HW.4th. 2008. Antibody is critical for the clearance of murine norovirus infection. J Virol 82:6610–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Leach D, Hunter DA, Sanfelippo D, Buell EJ, Zemple SJ, Grayson MH. 2011. Characterization of intestinal dendritic cells in murine norovirus infection. Open Immunol J 4:22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chichlowski M, Sharp JM, Vanderford DA, Myles MH, Hale LP. 2008. Helicobacter typhlonius and Helicobacter rodentium differentially affect the severity of colon inflammation and inflammation-associated neoplasia in IL10-deficient mice. Comp Med 58:534–541 [PMC free article] [PubMed] [Google Scholar]
- 12.Danese S. 2011. IBD: of mice and men—shedding new light on IL13 activity in IBD. Nat Rev Gastroenterol Hepatol 8:128–129 [DOI] [PubMed] [Google Scholar]
- 13.Doom CM, Turula HM, Hill AB. 2009. Investigation of the impact of the common animal facility contaminant murine norovirus on experimental murine cytomegalovirus infection. Virology 392:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duell BL, Tan CK, Carey AJ, Wu F, Cripps AW, Ulett GC. 2012. Recent insights into microbial triggers of interleukin 10 production in the host and the impact on infectious disease pathogenesis. FEMS Immunol Med Microbiol 64:295–313 [DOI] [PubMed] [Google Scholar]
- 15.Dyson JK, Rutter MD. 2012. Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk? World J Gastroenterol 18:3839–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. 2005. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev 206:260–276 [DOI] [PubMed] [Google Scholar]
- 17.Ericsson AC, Myles M, Davis W, Ma L, Lewis M, Maggio-Price L, Franklin C. 2010. Noninvasive detection of inflammation-associated colon cancer in a mouse model. Neoplasia 12:1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farkas T, Fey B, Keller G, Martella V, Egyed L. 2012. Molecular detection of murine noroviruses in laboratory and wild mice. Vet Microbiol 160:463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green KY. 2007. Caliciviridae: the noroviruses, p 949–979. In: Fields BN, Knipe DM, Howley PM, editors. Fields’ virology, 5th ed. Philadelphia (PA): Lippincott, Williams, and Wilkins. [Google Scholar]
- 20.Hale LP, Greer PK. 2012. A novel murine model of inflammatory bowel disease and inflammation-associated colon cancer with ulcerative colitis-like features. PLoS ONE 7:e41797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale LP, Perera D, Gottfried MR, Maggio-Price L, Srinivasan S, Marchuk D. 2007. Neonatal co-infection with Helicobacter species markedly accelerates the development of inflammation-associated colonic neoplasia in Il10−/− mice. Helicobacter 12:598–604 [DOI] [PubMed] [Google Scholar]
- 22.Hensley SE, Pinto AK, Hickman HD, Kastenmayer RJ, Bennink JR, Virgin HW, Yewdell JW. 2009. Murine norovirus infection has no significant effect on adaptive immunity to vaccinia virus or influenza A virus. J Virol 83:7357–7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins PD, Johnson LA, Sauder K, Moons D, Blanco L, Taube S, Wobus CE. 2011. Transient or persistent norovirus infection does not alter the pathology of Salmonella typhimurium-induced intestinal inflammation and fibrosis in mice. Comp Immunol Microbiol Infect Dis 34:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu CC, Riley LK, Wills HM, Livingston RS. 2006. Persistent infection and serologic crossreactivity of 3 novel murine noroviruses. Comp Med 56:247–251 [PubMed] [Google Scholar]
- 25.Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. 2005. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol 12:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kahan SM, Liu G, Reinhard MK, Hsu CC, Livingston RS, Karst SM. 2011. Comparative murine norovirus studies reveal a lack of correlation between intestinal virus titers and enteric pathology. Virology 421:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 28.Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H, Akira S, Wobus C, Nunez G. 2011. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe 9:496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. 1993. Interleukin10-deficient mice develop chronic enterocolitis. Cell 75:263–274 [DOI] [PubMed] [Google Scholar]
- 30.Lencioni KC, Drivdahl R, Seamons A, Treuting PM, Brabb T, Maggio-Price L. 2011. Lack of effect of murine norovirus infection on a mouse model of bacteria-induced colon cancer. Comp Med 61:219–226 [PMC free article] [PubMed] [Google Scholar]
- 31.Lencioni KC, Seamons A, Treuting PM, Maggio-Price L, Brabb T. 2008. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp Med 58:522–533 [PMC free article] [PubMed] [Google Scholar]
- 32.Mahida YR. 2000. The key role of macrophages in the immunopathogenesis of inflammatory bowel disease. Inflamm Bowel Dis 6:21–33 [DOI] [PubMed] [Google Scholar]
- 33.Mahler M, Leiter EH. 2002. Genetic and environmental context determines the course of colitis developing in IL10-deficient mice. Inflamm Bowel Dis 8:347–355 [DOI] [PubMed] [Google Scholar]
- 34.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. 2008. MDA5 recognition of a murine norovirus. PLoS Pathog 4:e1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller B, Klemm U, Mas Marques A, Schreier E. 2007. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch Virol 152:1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mumphrey SM, Changotra H, Moore TN, Heimann-Nichols ER, Wobus CE, Reilly MJ, Moghadamfalahi M, Shukla D, Karst SM. 2007. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J Virol 81:3251–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nell S, Suerbaum S, Josenhans C. 2010. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol 8:564–577 [DOI] [PubMed] [Google Scholar]
- 38.Ostanin DV, Bao J, Koboziev I, Gray L, Robinson-Jackson SA, Kosloski-Davidson M, Price VH, Grisham MB. 2009. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 296:G135–G146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paik J, Fierce Y, Drivdahl R, Treuting PM, Seamons A, Brabb T, Maggio-Price L. 2010. Effects of murine norovirus infection on a mouse model of diet-induced obesity and insulin resistance. Comp Med 60:189–195 [PMC free article] [PubMed] [Google Scholar]
- 40.Paik J, Fierce Y, Mai PO, Phelps SR, McDonald T, Treuting P, Drivdahl R, Brabb T, LeBoeuf R, O'Brien KD, Maggio-Price L. 2011. Murine norovirus increases atherosclerotic lesion size and macrophages in Ldlr−/− mice. Comp Med 61:330–338 [PMC free article] [PubMed] [Google Scholar]
- 41.Paul G, Khare V, Gasche C. 2012. Inflamed gut mucosa: downstream of interleukin10. Eur J Clin Invest 42:95–109 [DOI] [PubMed] [Google Scholar]
- 42.Perdue KA, Green KY, Copeland M, Barron E, Mandel M, Faucette LJ, Williams EM, Sosnovtsev SV, Elkins WR, Ward JM. 2007. Naturally occurring murine norovirus infection in a large research institution. J Am Assoc Lab Anim Sci 46:39–45 [PubMed] [Google Scholar]
- 43.Pritchett-Corning KR, Cosentino J, Clifford CB. 2009. Contemporary prevalence of infectious agents in laboratory mice and rats. Lab Anim 43:165–173 [DOI] [PubMed] [Google Scholar]
- 44.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39:596–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenstiel P, Sina C, Franke A, Schreiber S. 2009. Towards a molecular risk map—recent advances on the etiology of inflammatory bowel disease. Semin Immunol 21:334–345 [DOI] [PubMed] [Google Scholar]
- 46.Rubin DC, Shaker A, Levin MS. 2012. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol 3:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutella S, Locatelli F. 2011. Intestinal dendritic cells in the pathogenesis of inflammatory bowel disease. World J Gastroenterol 17:3761–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thackray LB, Duan E, Lazear HM, Kambal A, Schreiber RD, Diamond MS, Virgin HW. 2012. Critical role for interferon regulatory factor 3 (IRF3) and IRF7 in type I interferon-mediated control of murine norovirus replication. J Virol 86:13515–13523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrence AE, Brabb T, Viney JL, Bielefeldt-Ohmann H, Treuting P, Seamons A, Drivdahl R, Zeng W, Maggio-Price L. 2008. Serum biomarkers in a mouse model of bacterial-induced inflammatory bowel disease. Inflamm Bowel Dis 14:480–490 [DOI] [PubMed] [Google Scholar]
- 50.Watanabe N, Ikuta K, Okazaki K, Nakase H, Tabata Y, Matsuura M, Tamaki H, Kawanami C, Honjo T, Chiba T. 2003. Elimination of local macrophages in intestine prevents chronic colitis in interleukin10-deficient mice. Dig Dis Sci 48:408–414 [DOI] [PubMed] [Google Scholar]
- 51.Wilson MS, Ramalingam TR, Rivollier A, Shenderov K, Mentink-Kane MM, Madala SK, Cheever AW, Artis D, Kelsall BL, Wynn TA. 2011. Colitis and intestinal inflammation in Il10−/− mice results from IL13Rα2-mediated attenuation of IL13 activity. Gastroenterology 140:254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wirtz S, Neurath MF. 2007. Mouse models of inflammatory bowel disease. Adv Drug Deliv Rev 59:1073–1083 [DOI] [PubMed] [Google Scholar]
- 53.Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wynn TA. 2003. IL13 effector functions. Annu Rev Immunol 21:425–456 [DOI] [PubMed] [Google Scholar]
- 55.Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434 [DOI] [PubMed] [Google Scholar]
- 56.Yeom SC, Yu SA, Choi EY, Lee BC, Lee WJ. 2009. Prevalence of Helicobacter hepaticus, murine norovirus, and Pneumocystis carinii and eradication efficacy of cross-fostering in genetically engineered mice. Exp Anim 58:497–504 [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Danon SJ, Grehan M, Chan V, Lee A, Mitchell H. 2005. Natural colonization with Helicobacter species and the development of inflammatory bowel disease in interleukin-10–deficient mice. Helicobacter 10:223–230 [DOI] [PubMed] [Google Scholar]