Abstract
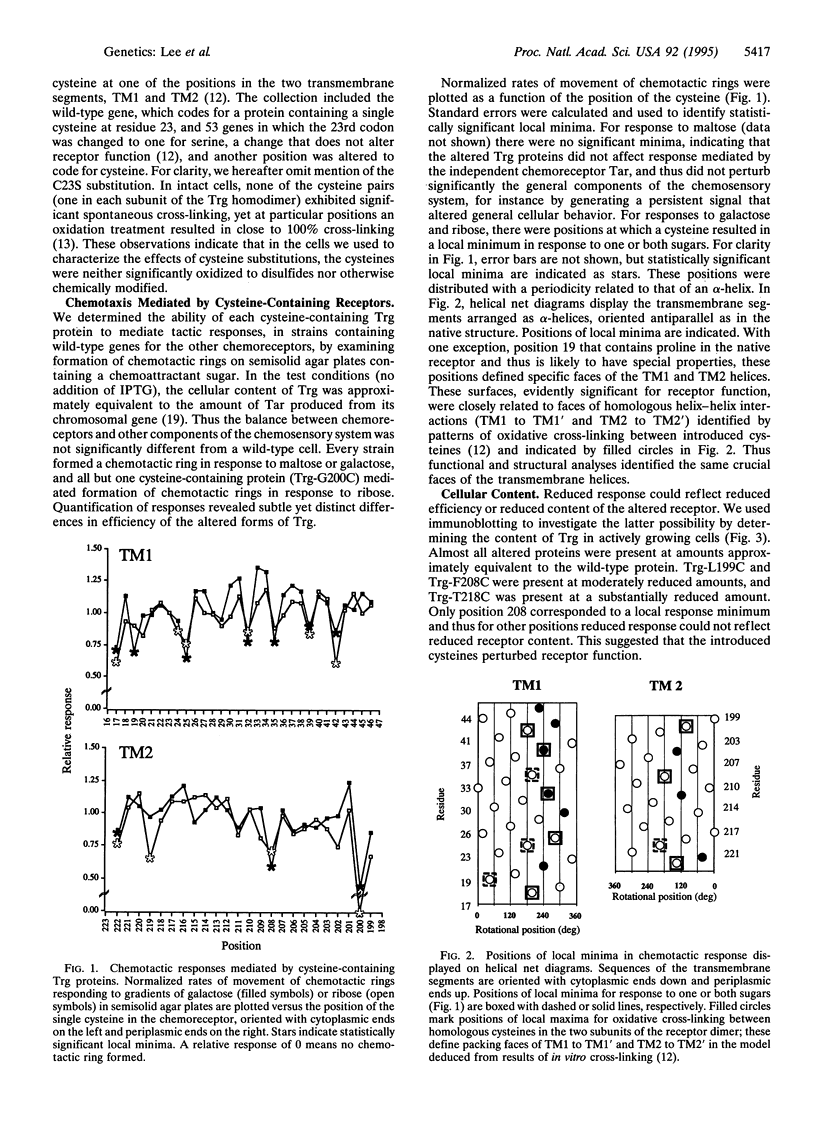
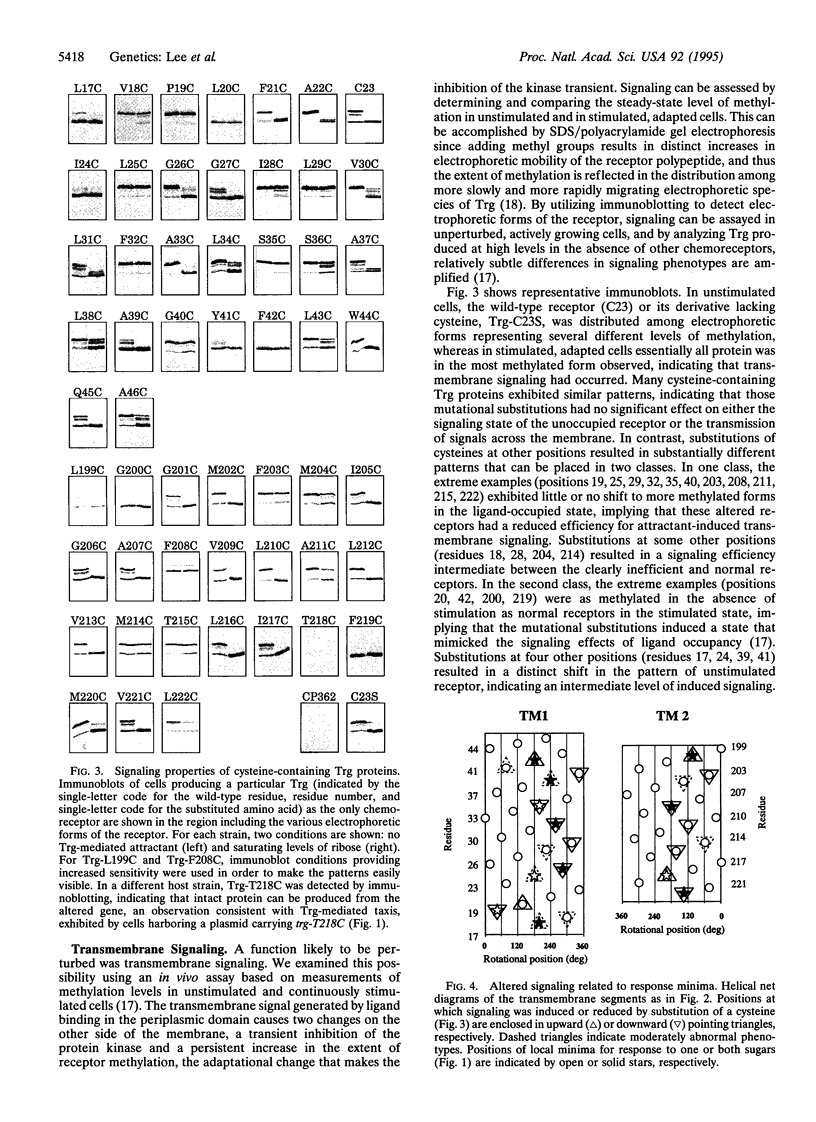
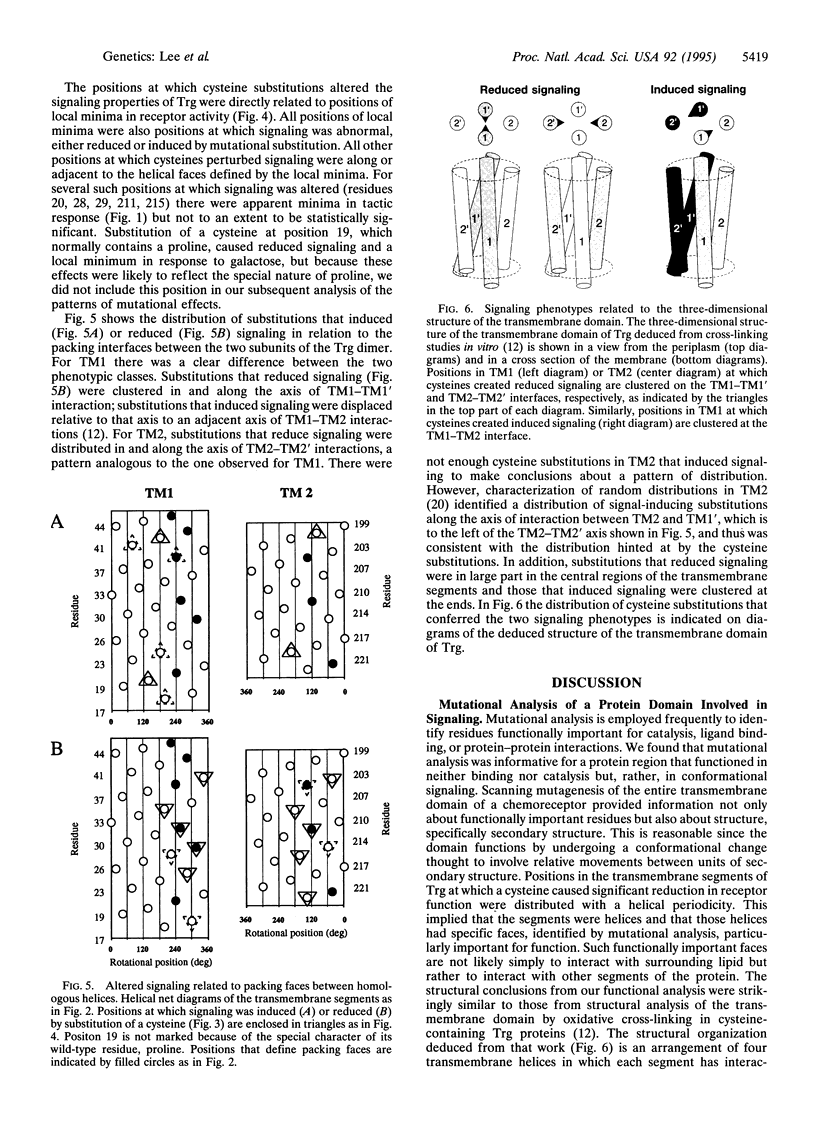
We applied mutational analysis to a protein domain that functions in neither catalysis nor binding but, rather, in transmembrane signaling. The domain is part of chemoreceptor Trg from Escherichia coli. It contains four transmembrane segments, two from each subunit of the homodimer. We used cysteine scanning to investigate the functional importance of each of 54 residues in the two transmembrane segments. Cysteines at some positions resulted in subtle but significant reductions in tactic response. Those positions defined a specific helical face on each segment, implying that the segments function as helices. The functionally important faces corresponded to structural, helical packing faces identified independently by biochemical studies. All functionally impaired receptors exhibited altered signaling properties, either reduced signaling upon stimulation or induced signaling in the absence of stimulation. The distribution of substitutions creating these two phenotypes implied that conformational signaling involves movement between the two transmembrane helices within a subunit and that signaling is optimal when stable interactions are maintained across the interface between subunits.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alex L. A., Simon M. I. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 1994 Apr;10(4):133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Burrows G. G., Newcomer M. E., Hazelbauer G. L. Purification of receptor protein Trg by exploiting a property common to chemotactic transducers of Escherichia coli. J Biol Chem. 1989 Oct 15;264(29):17309–17315. [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989 Jun 2;244(4908):1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Park C., Nowlin D. M. Adaptational "crosstalk" and the crucial role of methylation in chemotactic migration by Escherichia coli. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. F., Burrows G. G., Lebert M. R., Dutton D. P., Hazelbauer G. L. Deducing the organization of a transmembrane domain by disulfide cross-linking. The bacterial chemoreceptor Trg. J Biol Chem. 1994 Nov 25;269(47):29920–29927. [PubMed] [Google Scholar]
- Lee G. F., Lebert M. R., Lilly A. A., Hazelbauer G. L. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl cross-linking in vivo. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch B. A., Koshland D. E., Jr Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Intrasubunit signal transduction by the aspartate chemoreceptor. Science. 1991 Dec 13;254(5038):1651–1654. doi: 10.1126/science.1661030. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988 May 5;263(13):6268–6275. [PubMed] [Google Scholar]
- Morgan D. G., Baumgartner J. W., Hazelbauer G. L. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993 Jan;175(1):133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin D. M., Bollinger J., Hazelbauer G. L. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1987 May 5;262(13):6039–6045. [PubMed] [Google Scholar]
- Pakula A. A., Simon M. I. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Dutton D. P., Hazelbauer G. L. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J Bacteriol. 1990 Dec;172(12):7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Signal transduction schemes of bacteria. Cell. 1993 Jun 4;73(5):857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Yaghmai R., Hazelbauer G. L. Ligand occupancy mimicked by single residue substitutions in a receptor: transmembrane signaling induced by mutation. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):7890–7894. doi: 10.1073/pnas.89.17.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J. I., Biemann H. P., Pandit J., Koshland D. E., Kim S. H. The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J Biol Chem. 1993 May 5;268(13):9787–9792. [PubMed] [Google Scholar]





