SUMMARY
Most colorectal cancers (CRCs) containing activated BRAF (BRAF[V600E]) have a CpG island methylator phenotype (CIMP) characterized by aberrant hypermethylation of many genes, including the mismatch repair gene MLH1. MLH1 silencing results in microsatellite instability and a hypermutable phenotype. Through an RNA interference screen, here we identify the transcriptional repressor MAFG as the pivotal factor required for MLH1 silencing and CIMP in CRCs containing BRAF(V600E). In BRAF-positive human CRC cell lines and tumors, MAFG is bound at the promoters of MLH1 and other CIMP genes, and recruits a corepressor complex that includes its heterodimeric partner BACH1, the chromatin remodeling factor CHD8, and the DNA methyltransferase DNMT3B, resulting in hypermethylation and transcriptional silencing. BRAF(V600E) increases BRAF/MEK/ERK signaling resulting in phosphorylation and elevated levels of MAFG, which drives DNA binding. Analysis of transcriptionally silenced CIMP genes in KRAS-positive CRCs indicates that different oncoproteins direct the assembly of distinct repressor complexes on common promoters.
INTRODUCTION
A hallmark of human cancer genomes is aberrant DNA methylation, which is typified by both global DNA hypomethylation and site-specific DNA hypermethylation (reviewed in Baylin and Jones, 2011; Esteller, 2008; Hassler and Egger, 2012; Sharma et al., 2010). Site-specific DNA hypermethylation of promoter-associated CpG islands of tumor suppressor and DNA repair genes results in transcriptional silencing (commonly referred to as epigenetic silencing), thereby facilitating the initiation and progression of cancer (Baylin and Jones, 2011; Esteller, 2008; Hassler and Egger, 2012; Sharma et al., 2010).
Widespread CpG island promoter hypermethylation, referred to as the CpG island methylator phenotype (CIMP), was first identified in colorectal cancers (CRCs) (Toyota et al., 1999) and has since been extensively studied in this tumor type (reviewed in Lao and Grady, 2011). In fact, CRCs can be categorized into three subclasses based on aberrant CpG island methylation: CIMP-1 (also called CIMP-high), CIMP-2 (also called CIMP-low), and CIMP-negative (Kaneda and Yagi, 2011; Yagi et al., 2010). CIMP-1 CRCs, the focus of this study, are associated with an activating mutation in the BRAF oncoprotein (typically BRAF[V600E]), a serine/threonine kinase that stimulates cellular proliferation by signaling through the mitogen activated protein kinase pathway (BRAF/MEK/ERK) (reviewed in Dhomen and Marais, 2007).
The majority of CIMP-1 CRCs are characterized by promoter hypermethylation of the DNA mismatch repair gene MLH1, resulting in its transcriptional inactivation (Weisenberger et al., 2006). Loss of MLH1 expression results in microsatellite instability, a form of genetic instability characterized by length alterations within simple repeated microsatellite sequences of DNA (reviewed in Boland and Goel, 2010). Clinically, there is evidence to suggest that CIMP is associated with disease prognosis (Dahlin et al., 2010; Ogino et al., 2009) and it is also being investigated as a predictive marker for response to chemotherapy (Iacopetta et al., 2008; Jover et al., 2011; Van Rijnsoever et al., 2003).
How abnormal DNA methylation patterns develop in CRCs remains to be determined. To understand the basis of aberrant promoter hypermethylation, here we use MLH1 as a prototypical gene that is silenced in CIMP-1 CRCs and perform an RNA interference (RNAi) screen to identify factors required for MLH1 hypermethylation and silencing. Our results reveal a BRAF(V600E)-directed pathway that mediates silencing of MLH1 and, more generally, is responsible for CIMP.
RESULTS
An RNAi Screen to Identify Mediators of MLH1 Transcriptional Silencing
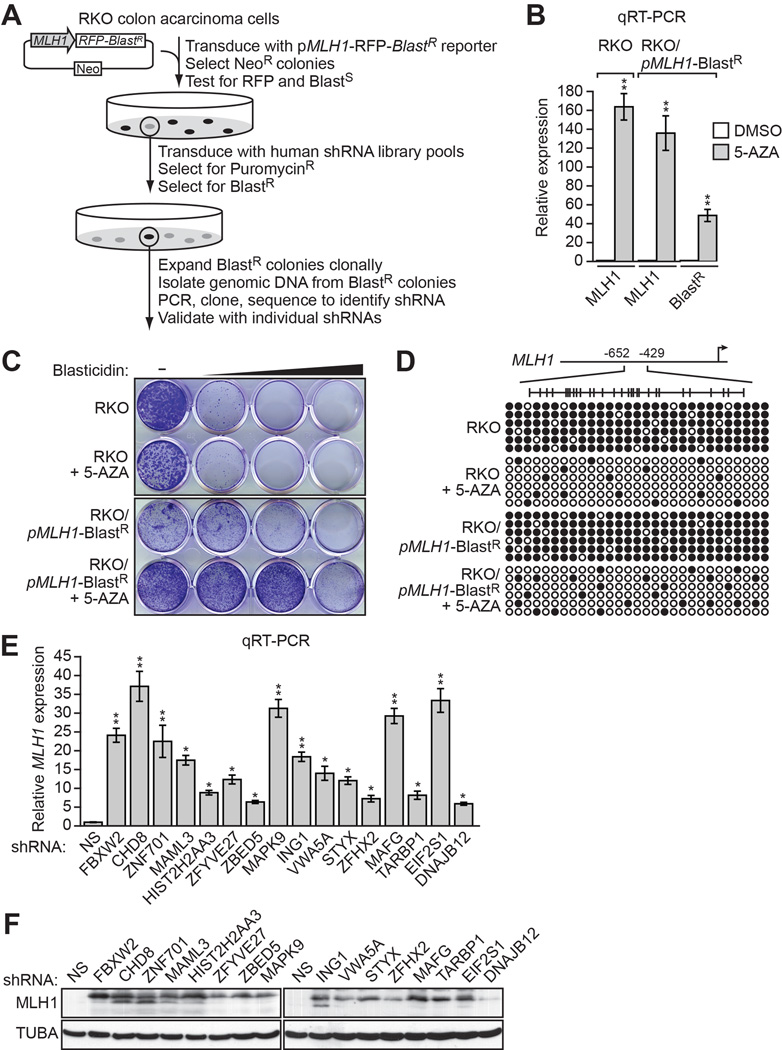
To screen for factors involved in transcriptional silencing of MLH1, we generated a reporter construct in which the MLH1 promoter was used to direct expression of the blasticidin-resistance (BlastR) gene (Figure 1A). This pMLH1-BlastR reporter construct was stably transduced into RKO cells, a human CRC cell line in which endogenous MLH1 is transcriptionally silenced (Veigl et al., 1998)and Figure 1B). We selected cells in which the reporter gene had been silenced, as evidenced by acquisition of blasticidin resistance (Figure 1C), transcriptional derepression (Figure 1B) and decreased promoter hypermethylation (Figure 1D) following treatment with the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine (5-AZA).
Figure 1. An RNAi Screen to Identify Mediators of MLH1 Transcriptional Silencing.
(A) Schematic of the shRNA screen.
(B) qRT-PCR analysis monitoring MLH1 expression in parental RKO cells, or MLH1 and BlastRexpression in RKO/pMLH1-BlastR cells, following treatment with either DMSO or 5-aza-2’-deoxycytidine (5-AZA). The results were normalized to that observed upon DMSO treatment, which was set to 1.
(C) Viability of RKO or RKO/pMLH1-BlastR cells treated with DMSO or 5-AZA for 3 days and then 0, 1, 2 or 5 µM blasticidin for 6 days. Cells were stained with crystal violet.
(D) Bisulfite sequencing analysis of the endogenous MLH1 promoter in parental RKO cells or the pMLH1-BlastR reporter in RKO/pMLH1-BlastR cells treated in the absence or presence of 5-AZA. (Top) Schematic of the MLH1 promoter; positions of CpGs are shown to scale by vertical lines. (Bottom) Each circle represents a methylated (black) or unmethylated (white) CpG dinucleotide. Each row represents a single clone.
(E) qRT-PCR analysis monitoring MLH1 expression in RKO cells expressing a non-silencing (NS) shRNA or an shRNA against one of the 16 candidates. The results were normalized to that observed with the NS shRNA, which was set to 1. Data are represented as mean ± SD. *P ≤0.05, **P≤0.01.
(F) Immunoblot analysis monitoring MLH1 levels in RKO cells upon knockdown of each of the 16 candidates. α-tubulin (TUBA) was monitored as a loading control. See also Figure S1.
A genome-wide human small hairpin (shRNA) library was divided into pools, which were packaged into lentivirus particles and used to stably transduce the RKO/pMLH1-BlastR reporter cell line. Blasticidin-resistant colonies, indicative of derepression of the reporter gene, were selected and the shRNAs identified by sequence analysis (see Figure 1A).
Positive candidates identified in the primary screen were validated by stably transducing parental RKO cells with an individual shRNA corresponding to that isolated from the primary screen, as well as a second, unrelated shRNA targeting the same gene, followed by analysis of endogenous MLH1 expression by quantitative RT-PCR (qRT-PCR). Only candidates that scored positively with two shRNAs were considered validated. Using this approach, we identified 16 genes that, following shRNA-mediated knockdown, resulted in derepression of endogenous MLH1 (Figures 1E and S1A; Table S1). qRT-PCR analysis confirmed that each shRNA reduced target gene expression (Figure S1B and S1C). As expected, the immunoblot of Figure 1F shows that shRNA-mediated knockdown of the 16 genes also resulted in derepression of MLH1 at the protein level.
A MAFG-Directed Corepressor Complex Mediates MLH1 Transcriptional Silencing
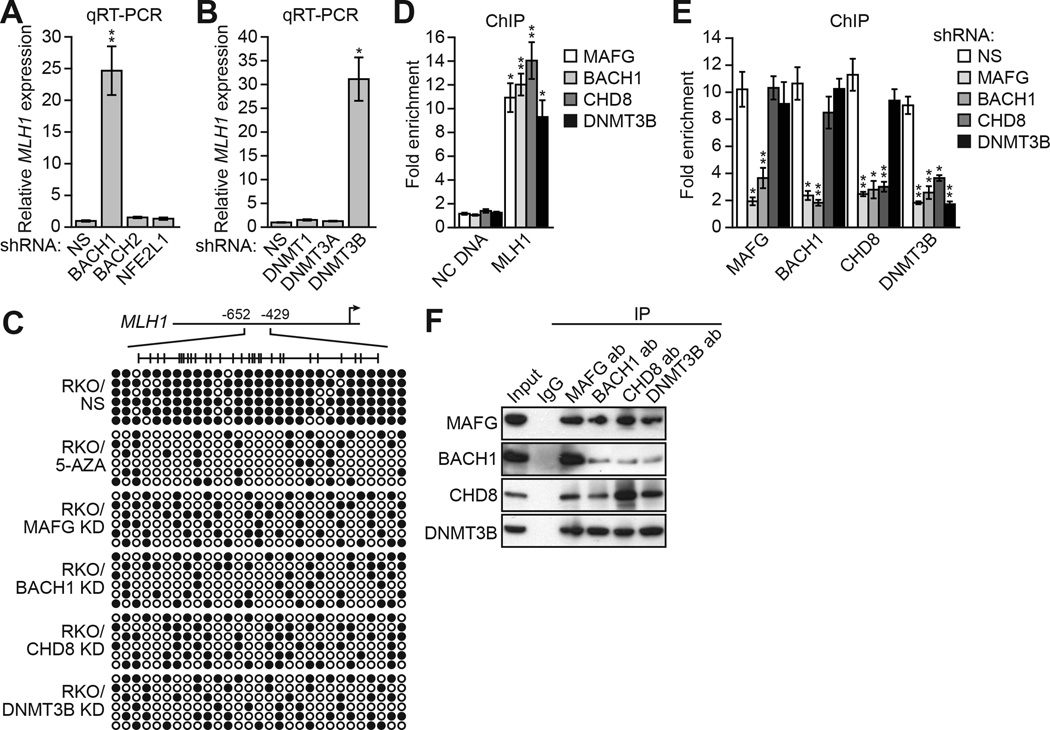
Several of the 16 candidates, including MAFG, CHD8, ING1 and ZNF701, have known or predicted transcriptional repression activity (Feng et al., 2002; Motohashi et al., 1997; Ronan et al., 2013; Urrutia, 2003). In a chromatin immunoprecipitation (ChIP) assay we could detect binding of MAFG and CHD8, but not ING1 and ZNF701, on the transcriptionally silenced MLH1 promoter (Figure S2A). We therefore elected to focus on MAFG and CHD8. MAFG is a sequence-specific DNA-binding protein (Motohashi et al., 1997), and expression profiling studies have found that MAFG is over-expressed in CRCs (Figure S2B). CHD8 is a chromatin remodeling factor that has also been linked to transcriptional repression (see, for examples, Sakamoto et al., 2000; Thompson et al., 2008).
Large-scale RNAi screens typically do not achieve saturation (reviewed in Mullenders and Bernards, 2009). We therefore performed several directed experiments to identify other factors involved in transcriptional silencing of MLH1. MAFG binds DNA as a heterodimer in conjunction with one of several proteins including BACH1, BACH2 or NFE2L1 (Fujiwara et al., 1993; Kataoka et al., 1995; Motohashi et al., 1997). Figure 2A shows that knockdown of BACH1, but not BACH2 or NFE2L1, derepressed MLH1 expression (see also Figure S2C and S2D). Our previous studies have shown that a specific DNA methyltransferase is involved in transcriptional silencing of a tumor suppressor gene (Gazin et al., 2007; Palakurthy et al., 2009; Serra et al., 2014). Figure 2B shows that MLH1 was also derepressed following knockdown of DNMT3B but not the other DNA methyltransferases, DNMT1 or DNMT3A (see also Figure S2C and S2D).
Figure 2. A MAFG-Directed Corepressor Complex Mediates MLH1 Transcriptional Silencing.
(A) qRT-PCR analysis monitoring MLH1 expression in RKO cells expressing a NS, BACH1, BACH2 or NFE2L1 shRNA.
(B) qRT-PCR analysis monitoring MLH1 expression in RKO cells expressing a NS, DNMT1, DNMT3A or DNMT3B shRNA.
(C) Bisulfite sequencing analysis of the MLH1 promoter in RKO cells expressing a NS, MAFG, BACH1, CHD8 or DNMT3B shRNA. Decreased promoter methylation in the presence of 5-AZA is shown as a control.
(D) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter or, as a control, an irrelevant DNA region (negative control [NC] DNA) in RKO cells. The results were normalized to that obtained with IgG, which was set to 1.
(E) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter in RKO cells expressing a NS, MAFG, BACH1, CHD8 or DNMT3B shRNA. Data are represented as mean ± SD. *P≤0.05, **P≤0.01.
(F) Co-immunoprecipitation analysis. RKO cell extracts were immunoprecipitated with a MAFG, BACH1, CHD8, DNMT3B or control (IgG) antibody, and the immunoprecipitate was analyzed for MAFG, BACH1, CHD8 or DNMT3B by immunoblotting. See also Figure S2.
The bisulfite sequencing experiment of Figure 2C shows that in RKO cells the MLH1 promoter is, as expected, hypermethylated. Treatment with 5-AZA or shRNA-mediated knockdown of MAFG, BACH1, CHD8 or DNMT3B resulted in a large reduction in MLH1 promoter methylation, consistent with the transcriptional derepression. By contrast, methylation of MINT6, which is hypermethylated in both normal and cancer cells (Toyota et al., 1999), was not dependent upon MAFG, BACH1, CHD8 or DNMT3B (Figure S2E).
To determine whether like MAFG and CHD8, BACH1 and DNMT3B also functioned by binding directly to the MLH1 promoter, we carried out a series of ChIP experiments. Figure 2D shows that MAFG, BACH1, CHD8 and DNMT3B were all enriched on the MLH1 promoter (see also Figure S2F). To ask whether binding of these factors to the MLH1 promoter was ordered, we determined the consequence of single gene knockdowns on promoter occupancy of all factors. The ChIP results of Figure 2E shows that knockdown of MAFG or BACH1 substantially decreased binding of all factors. Knockdown of CHD8 led to loss of DNMT3B binding but binding of MAFG or BACH1 was unaffected. Finally, knockdown of DNMT3B did not eliminate binding of MAFG, BACH1 or CHD8 to the MLH1 promoter. These results indicate that MAFG, along with its heterodimeric partner BACH1, initiates binding on the MLH1 promoter and directs recruitment of the other corepressors culminating in binding of DNMT3B.
The ordered recruitment of MAFG and its corepressors raised the possibility of physical interactions between the proteins. Consistent with this idea, the co-immunoprecipitation experiment of Figure 2F shows that MAFG, BACH1, CHD8 and DNMT3B were stably associated with one another. By contrast, there was no detectable association between MAFG and DNMT1 (Figure S2G). Interactions between MAFG and BACH1, CHD8 and DNMT3B were confirmed by liquid chromatography tandem mass spectrometry (Figure S2H and Table S2).
BRAF(V600E)-Mediated Upregulation of MAFG is Required for Transcriptional Silencing of MLH1
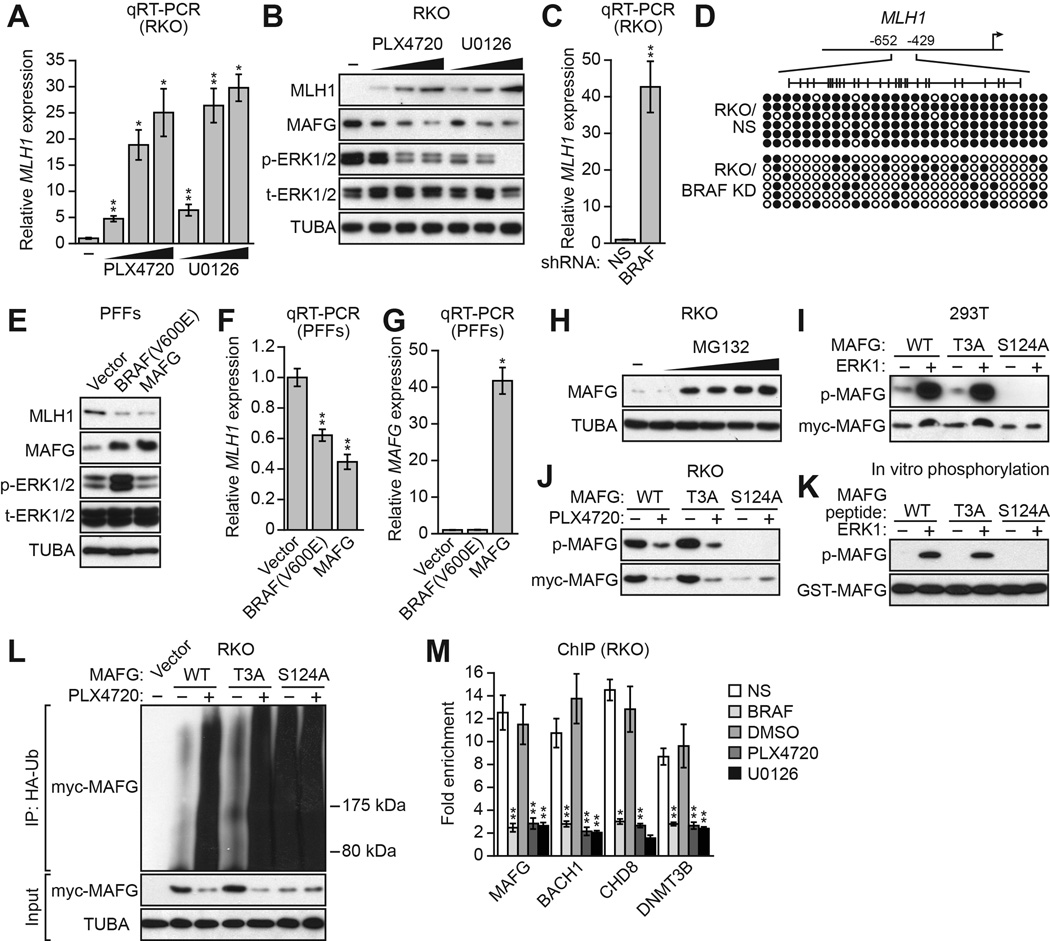
RKO cells contain an activated BRAF mutation, BRAF(V600E) (Yun et al., 2009), and we therefore investigated the relationship between BRAF(V600E), MAFG and its corepressors, and silencing of MLH1. The qRT-PCR results of Figure 3A show that inhibition of BRAF/MEK/ERK signaling in RKO cells using either PLX4720, a BRAF(V600E) inhibitor (Tsai et al., 2008), or U0126, a MEK inhibitor (Favata et al., 1998), led to derepression of MLH1. Derepression of MLH1 following treatment with PLX4720 or U0126 was also evident at the protein level (Figure 3B). Moreover, inhibition of BRAF/MEK/ERK signaling resulted in decreased levels of MAFG protein. As expected, shRNA-mediated knockdown of BRAF(V600E) (Figure S3A) also derepressed MLH1 expression (Figure 3C and S3B) and resulted in decreased methylation of the MLH1 promoter (Figure 3D). By contrast, inhibition of BRAF/MEK/ERK signaling did not affect the levels of the MAFG corepressors BACH1, CHD8 or DNMT3B (Figure S3C).
Figure 3. BRAF(V600E)-Mediated Upregulation of MAFG is Required for Transcriptional Silencing of MLH1.
(A) qRT-PCR analysis monitoring MLH1 expression in RKO cells following treatment with PLX4720 or U0126.
(B) Immunoblot analysis monitoring MLH1, MAFG, phosphorylated ERK1/2 (p-ERK1/2) and total ERK1/2 (t-ERK1/2) levels in RKO cells treated with DMSO, PLX4720 or U0126.
(C) qRT-PCR analysis monitoring MLH1 expression in RKO cells expressing a NS or BRAF shRNA.
(D) Bisulfite sequencing analysis of the MLH1 promoter in RKO cells expressing a NS or BRAF shRNA.
(E) Immunoblot analysis monitoring MLH1, MAFG, p-ERK1/2 and t-ERK1/2 levels in PFFs expressing BRAF(V600E) or MAFG.
(F) qRT-PCR analysis monitoring MLH1 expression in PFFs expressing BRAF(V600E) or MAFG. The results were normalized to that obtained with a vector control, which was set to 1.
(G) qRT-PCR analysis monitoring MAFG expression in PFFs expressing BRAF(V600E) or MAFG.
(H) Immunoblot analysis showing MAFG levels in RKO cells treated with 0–8 µM MG132 for 4 hours.
(I) In vivo phosphorylation assay. 293T cells were transfected with myc-tagged MAFG-wild-type (WT), -T3A or -S124A in the presence or absence of an ERK1-expression plasmid. Cell lysate was immunoprecipitated with a myc antibody and immunoprecipitates were analyzed by immunoblotting with a phosphorylated-(S/T)P antibody.
(J) RKO cells were transfected with myc-tagged MAFG-WT, -T3A or –S124A and treated in the presence or absence of PLX4720, and analyzed as described in (I).
(K) In vitro kinase assay. GST-tagged MAFG-WT, -T3A or -S124A peptides were incubated in the presence or absence of ERK1 and γ-ATP and analyzed for incorporation of the radiolabel by autoradiography.
(L) HA-ubiquitination pull-down assay. Extracts from RKO cells expressing HA-tagged ubiquitin and myc-tagged MAFG-WT, -T3A or -S124A and treated in the presence or absence of PLX4720 were immunoprecipitated using an HA antibody, and the immunoprecipitate was analyzed by immunoblotting using a myc antibody.
(M) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter in RKO cells expressing a NS or BRAF shRNA or treated with DMSO, PLX4720 or U0126. Data are represented as mean ± SD. *P≤0.05, **P≤0.01. See also Figure S3.
To further investigate the relationship between BRAF(V600E) and MAFG levels we stably expressed BRAF(V600E) or, as a control, MAFG in primary foreskin fibroblasts (PFFs). As expected, MAFG levels were substantially lower in PFFs compared to RKO cells (Figure S3D), and the MAFG-BACH1-CHD8-DNMT3B complex was not detected in PFFs (Figure S3E). Moreover, in PFFs, MAFG and its corepressors were not bound to the MLH1 promoter (Figure S3F) and RNAi-mediated knockdown of MAFG did not affect MLH1 expression (Figure S3G). Stable expression of BRAF(V600E) in PFFs led to increased phosphorylated ERK1/2 levels (Figure 3E), elevated MAFG levels (Figure 3E), and decreased MLH1 expression (Figures 3E and 3F). Stable expression of MAFG in PFFs also resulted in decreased MLH1 expression (Figures 3E and 3F). Notably, BRAF(V600E) did not increase MAFG mRNA levels (Figure 3G), indicating that the increased MAFG protein levels occurred by a predominantly post-transcriptional mechanism. Consistent with this idea, treatment of RKO cells with the proteasome inhibitor MG132 substantially increased MAFG levels (Figure 3H).
We next investigated whether MAFG was a direct substrate of BRAF/MEK/ERK signaling. We noticed that MAFG has two potential ERK phosphorylation sites ([S/T]P; Davis, 1993) at T3 and S124, of which the S124 site is more highly conserved (Figure S3H). We analyzed ERK-directed phosphorylation of wild-type MAFG and two MAFG mutants bearing mutations in each phosphorylation site, MAFG-T3A and MAFG-S124A. Each MAFG derivative was C-terminally myc tagged and transfected into 293T cells in the presence or absence of an ERK1-expression plasmid. Cell lysate was immunoprecipitated with a myc antibody and immunoprecipitates were analyzed by immunoblotting with an antibody that recognizes a phosphorylated ERK consensus site. Figure 3I shows that wild-type MAFG and MAFG-T3A were phosphorylated in an ERK1-dependent fashion. By contrast, MAFG-S124A was not phosphorylated, indicating that ERK1 phosphorylates MAFG at S124. Notably, cotransfection of ERK1 increased total levels of wild-type MAFG and MAFG-T3A but not MAFG-S124A, indicating that phosphorylation increased MAFG stability.
We also analyzed phosphorylation of MAFG in RKO cells, which contain BRAFV600E, in the presence or absence of PLX4720. Figure 3J shows that ectopically expressed wild-type MAFG and MAFG-T3A were phosphorylated in untreated RKO cells, and the levels of the phosphorylated proteins were greatly reduced following PLX4720 addition. MAFG-S124A was again not phosphorylated and was expressed at low levels. Finally, the in vitro kinase assay of Figure 3K shows that ERK1 could phosphorylate wild-type MAFG and MAFG-T3A, but not MAFG-S124A. Our results are consistent with a previous phosphoproteomics study reporting phosphorylation of MAFG at S124 (Olsen et al., 2010).
Protein stability is frequently regulated by polyubiquitination (Desterro et al., 2000). To gain insight into how ERK-directed phosphorylation increased MAFG stability, we analyzed MAFG polyubiquitination in RKO cells. Figure 3L shows that in untreated RKO cells there was relatively little ubiquitination of wild-type MAFG and MAFG-T3A. However, ubiquitination of wild-type MAFG and MAFG-T3A increased greatly following PLX4720 treatment. Notably, in untreated RKO cells MAFG-S124A, which cannot be phosphorylated by ERK, had a high level of ubiquitination, which was unaffected by PLX4720 treatment. Collectively, these results indicate that phosphorylation of MAFG at S124 by ERK increases MAFG levels, at least in part, by preventing polyubiquitination and subsequent proteasomal-mediated degradation. FBXW2, an F-box protein involved in protein degradation, was one of the 16 candidates isolated in the RNAi screen (Figure 1E and Table S1). As expected, knockdown of FBXW2 had no effect on MAFG levels (Figure S3I).
Finally, we investigated how BRAF/MEK/ERK signaling affected binding of MAFG and its corepressors to the MLH1 promoter. The ChIP results of Figure 3M show that shRNA-mediated knockdown of BRAF(V600E), or inhibition of BRAF/MEK/ERK signaling by treatment with PLX4720 or U0126, resulted in loss of binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter.
Validation of the Role of MAFG and its Corepressors in MLH1 Silencing in Other CRC Cell Lines and BRAF-Positive Human Tumor Samples
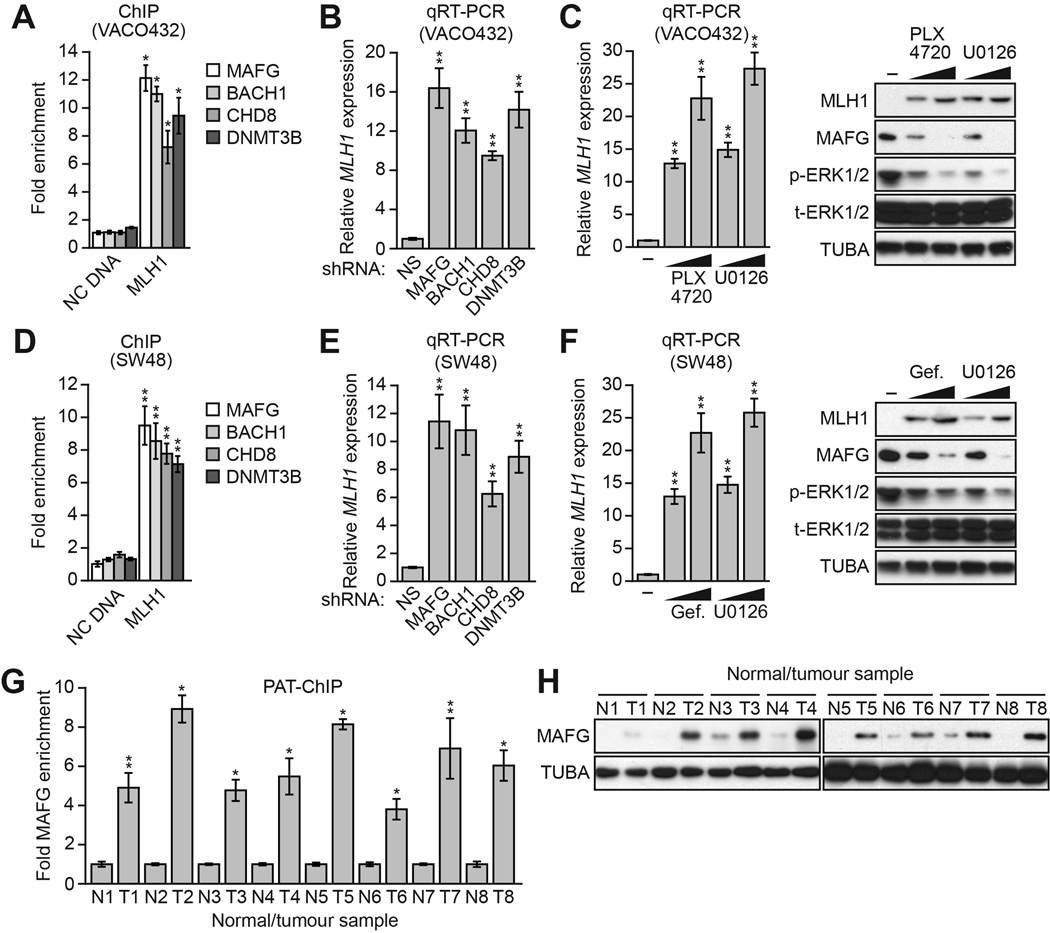
To determine the generality and clinical relevance of these results, we analyzed other human CRC cell lines and tumor samples in which BRAF/MEK/ERK signaling was increased. We first analyzed the CRC cell line VACO432, which contains BRAF(V600E) (Yun et al., 2009) and transcriptionally silenced MLH1 (Veigl et al., 1998). We found that in VACO432 cells, MAFG and its corepressors were associated with the MLH1 promoter (Figure 4A), and their knockdown derepressed the transcriptionally silenced MLH1 (Figure 4B). Moreover, treatment of VACO432 cells with PLX4720 or U0126 resulted in reduced MAFG levels and depreression of MLH1 (Figure 4C). We also analyzed the CRC cell line SW48, which is wild-type for BRAF but contains an activating EGFR mutation (EGFR[G719S]; Yeh et al., 2009) and transcriptionally silenced MLH1 (Deng et al., 1999). Like BRAF(V600E), EGFR(G719S) increases RAF/MEK/ERK signaling (Hodoglugil et al., 2013). In SW48 cells, MAFG and its corepressors were associated with the MLH1 promoter (Figure 4D), and their knockdown derepressed the transcriptionally silenced MLH1 (Figure 4E). Furthermore, treatment of SW48 cells with the EGFR inhibitor gefitinib (Barker et al., 2001; Ward et al., 1994) or U0126 resulted in reduced MAFG levels and derepression of MLH1 (Figure 4F). These results with chemical inhibitors confirm the role of EGFR(G719S)-directed increased RAF/MEK/ERK signaling in MLH1 silencing.
Figure 4. Validation of the Role of MAFG and its Corepressors in MLH1 Silencing in Other CRC cell lines and BRAF-Positive Human Tumor Samples.
(A) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter or, as a control, an irrelevant DNA region (NC DNA) in VACO432 cells.
(B) qRT-PCR analysis monitoring MLH1 expression in VACO432 cells expressing an NS, MAFG, BACH1, CHD8 or DNMT3B shRNA.
(C) qRT-PCR analysis monitoring MLH1 expression (left) or immunoblot analysis monitoring MLH1 levels (right) in VACO432 cells treated with DMSO or 1 or 5 µM PLX4720 or U0126.
(D) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to the MLH1 promoter or, as a control, an irrelevant DNA region (NC DNA) in SW48 cells.
(E) qRT-PCR analysis monitoring MLH1 expression in SW48 cells expressing an NS, MAFG, BACH1, CHD8 or DNMT3B shRNA.
(F) qRT-PCR analysis monitoring MLH1 expression (left) or immunoblot analysis monitoring MLH1 levels (right) in SW48 cells treated with DMSO or 1 or 5 µM gefitinib (Selleck) or U0126 for 24 hours.
(G) PAT-ChIP analysis monitoring binding of MAFG to the MLH1 promoter in matched adjacent normal (N) and BRAF-positive CRC human tumor (T) samples. Data are represented as mean ± SD. *P≤0.05, **P≤0.01.
(H) Immunoblot analysis monitoring MAFG levels in samples described in (G). See also Figure S4.
We next used a pathology tissue ChIP (PAT-ChIP) assay (Fanelli et al., 2011) to measure association of MAFG with the MLH1 promoter in BRAF-positive human CRC tumor samples. Figure 4G shows that MAFG was substantially enriched at the MLH1 promoter in CRC tumor samples relative to adjacent normal colon. Consistent with these PAT-ChIP results, the immunoblot analysis of Figure 4H shows that MAFG levels were higher in CRC tumor samples relative to adjacent normal colon. Bisulfite sequencing confirmed MLH1 promoter hypermethylation in the BRAF-positive human CRC tumor samples but not in the matched normal controls (Figure S4).
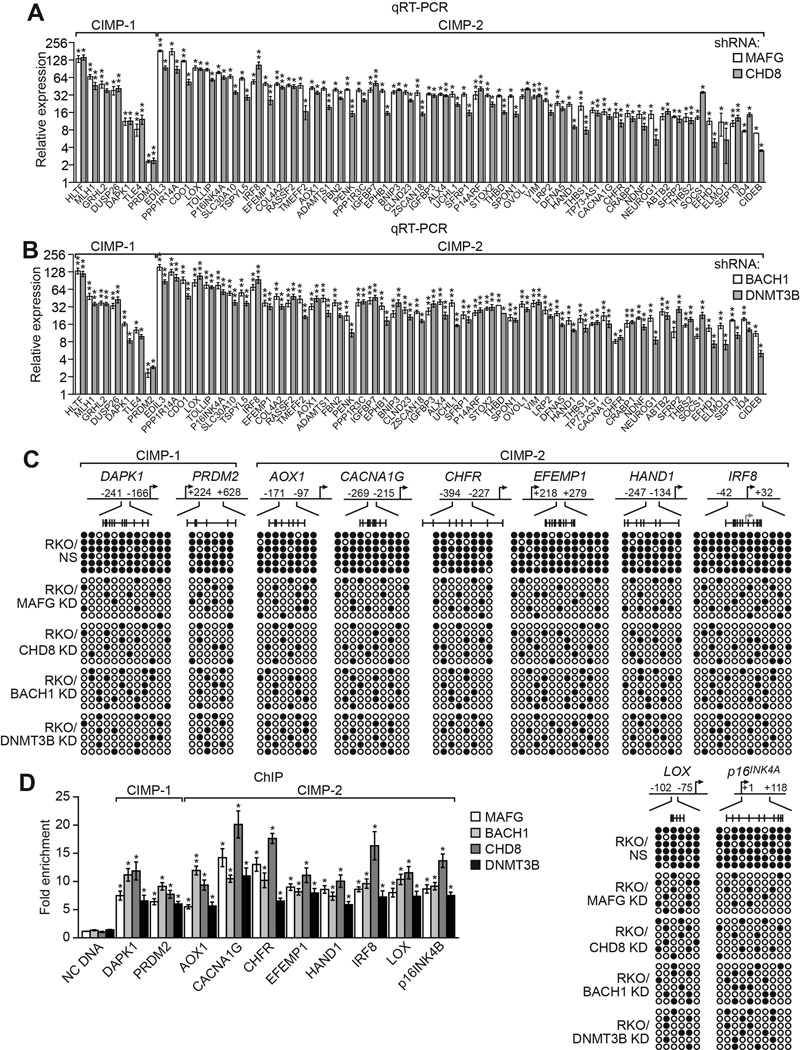
MAFG and its Corepressors Mediate CIMP in BRAF-Positive RKO Cells
Most CRCs containing BRAF(V600E) can be classified as CIMP-1, as defined by aberrant promoter hypermethylation of ~60 so-called CIMP-1 and CIMP-2 marker genes, which include MLH1 and multiple tumor suppressors (Kaneda and Yagi, 2011; Toyota et al., 1999; Toyota et al., 2000; Yagi et al., 2010). Notably, MLH1 is a CIMP-1 marker gene, which prompted to ask whether MAFG and its corepressors have a general role in the aberrant promoter hypermethylation and transcriptional silencing characteristic of CIMP genes in CRCs containing BRAF(V600E). Remarkably, knockdown of MAFG, CHD8, BACH1 or DNMT3B in CIMP-positive RKO cells (Ahmed et al., 2013 and Figure S5A) derepressed expression of both CIMP-1 and CIMP-2 marker genes (Figure 5A and 5B). Interestingly, knockdown of MAFG or its corepressors also derepressed two non-CIMP genes, VIM and SEPT9 (Figure S5B), whose promoter hypermethylation is used to diagnose CRC (Gyparaki et al., 2013) as well as several hypermethylated non-CIMP genes that flank MLH1 (Figure S5C) (Hitchins et al., 2007). Analysis of a representative subset of 10 other CIMP genes revealed that, like MLH1, knockdown of MAFG or its corepressors also decreased promoter hypermethylation (Figure 5C). Furthermore, as with MLH1, ChIP analysis showed significant enrichment of MAFG, BACH1, CHD8, and DNMT3B on the promoters of the CIMP genes (Figure 5D), whose expression was also derepressed by knockdown of the MAFG corepressors (Figure S5D).
Figure 5. MAFG and its Corepressors Mediate CIMP in BRAF-Positive RKO Cells.
(A and B) qRT-PCR analysis monitoring expression of CIMP genes in RKO cells expressing a MAFG or CHD8 shRNA (A) or a BACH1 or DNMT3B shRNA (B). The results were normalized to that obtained with the NS control, which was set to 1.
(C) Bisulfite sequencing analysis of representative CIMP genes in RKO cells expressing a NS, MAFG, CHD8, BACH1 or DNMT3B shRNA.
(D) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B on representative CIMP gene promoters in RKO cells. Data are represented as mean ± SD. *P≤0.05, **P≤0.01. See also Figure S5.
MAFG is known to bind to a consensus site (Kataoka et al., 1994) (see Table S3 legend). We could identify one or more MAFG consensus binding sites in the promoter regions of 50 of the 57 CIMP genes (Table S3). The absence a MAFG consensus site in some CIMP genes is not unexpected and may be explained by the high concentrations of MAFG in BRAF-positive CRC cells resulting in binding to non-consensus, weak sites.
MAFG and its Corepressors Mediate CIMP in Other CRC Cell Lines and BRAF-Positive Human Tumor Samples
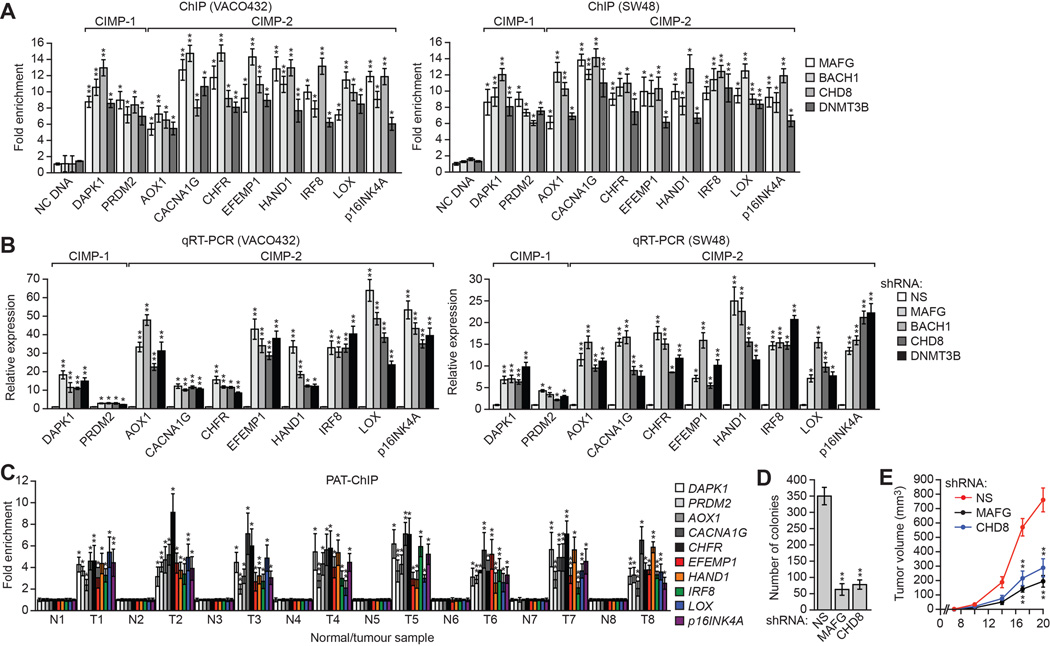
We next asked whether MAFG and its corepressors silenced CIMP genes in other CIMP-positive human CRC cell lines and tumor samples. In CIMP-positive VACO432 (BRAF[V600E]) and SW48 (EGFR[G719S]) CRC cells (Ahmed et al., 2013) (Figure S6A), MAFG, BACH1, CHD8, and DNMT3B were associated with the promoters of the 10 representative CIMP genes (Figure 6A), whose expression was derepressed by knockdown of MAFG or its corepressors (Figure 6B), similar to what we had observed for MLH1.
Figure 6. MAFG and its Corepressors Mediate CIMP in Other CRC Cell Lines and BRAF-Positive Human Tumor Samples.
(A) ChIP analysis monitoring binding of MAFG, BACH1, CHD8 and DNMT3B to representative CIMP gene promoters in VACO432 (left) and SW48 (right) cells.
(B) qRT-PCR analysis monitoring expression of representative CIMP genes in VACO432 (left) or SW48 (right) cells expressing a NS, MAFG, BACH1, CHD8 or DNMT3B shRNA.
(C) PAT-ChIP analysis monitoring binding of MAFG to representative CIMP gene promoters in matched adjacent normal (N) and BRAF-positive CRC human tumor (T) samples.
(D) Soft agar assay measuring colony formation of RKO cells expressing a NS, MAFG or CHD8 shRNA.
(E) Tumor formation assay. RKO cells expressing a NS, MAFG or CHD8 shRNA were subcutaneously injected into the flanks of nude mice (n=3), and tumor formation was measured. Data are represented as mean ± SD. *P≤0.05, **P≤0.01. See also Figure S6.
The PAT-ChIP results of Figure 6C show that, as with MLH1, MAFG was substantially enriched at the promoters of the 10 CIMP genes in BRAF-positive human CRC tumors relative to matched normal controls. Analysis of representative CIMP genes confirmed promoter hypermethylation in all BRAF-positive CRCs (Figure S6B).
Finally, we assessed the role of the activated oncoprotein and RAF/MEK/ERK signaling on transcriptional silencing of CIMP genes. shRNA-mediated knockdown of BRAF(V600E) or treatment with PLX4720 derepressed all ~60 CIMP genes in RKO cells (Figure S6C). Likewise, treatment of VACO432 (BRAF[V600E]) cells with PLX4720 (Figure S6D) and treatment of SW48 cells (EGFR[G719S]) with gefitinib (Figure S6E) derepressed all representative CIMP genes tested. Furthermore, inhibition of RAF/MEK/ERK signaling by U0126 also derepressed all CIMP genes tested in the three cell lines (Figure S6D–S6F).
We predicted that loss of MAFG or CHD8, which results in derepression of multiple tumor suppressor genes, would reduce tumorigenicity. Consistent with this prediction, knockdown of MAFG or CHD8 substantially reduced the ability of RKO cells to grow in soft agar (Figure 6D) or form tumors in mice (Figure 6E).
Oncogenic BRAF and KRAS Direct the Assembly of Distinct Repressor Complexes on Common CIMP Gene Promoters
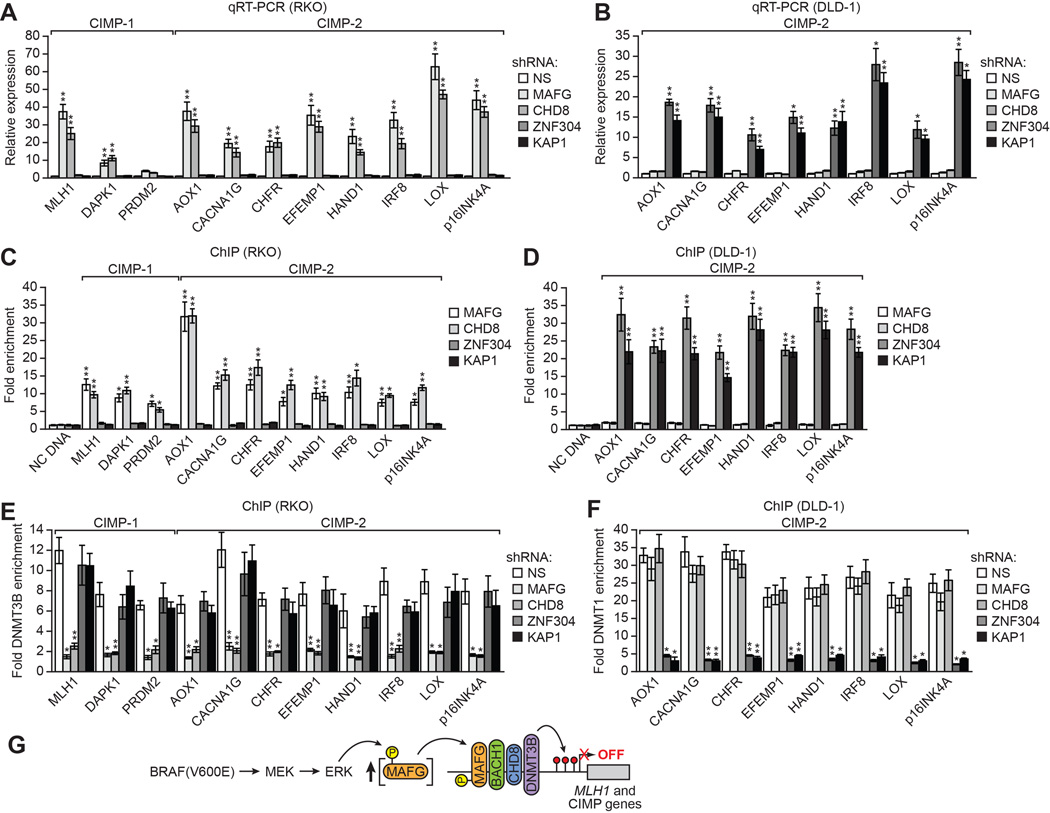
Approximately 70% of CRCs containing activated KRAS are classified as CIMP-2 and have a DNA hypermethylation pattern that is related to but less severe than that of CIMP-1 CRCs, which typically contain BRAF(V600E) (Ogino et al., 2009; Weisenberger et al., 2006). Many of the same CIMP genes are aberrantly hypermethylated in both BRAF- and KRAS-positive CRCs (CIMP-2 marker genes), whereas a small set of genes are hypermethylated and silenced only in BRAF-positive CRCs (CIMP-1 marker genes). Recently, we identified a silencing pathway directed by the transcriptional repressor ZNF304 that mediates CIMP in KRAS-positive CRCs, which involves a completely different set of corepressors from that described above (Serra et al., 2014). We were therefore interested in assessing the relative contributions of these two silencing pathways in repressing transcription of common CIMP genes in BRAF- and KRAS-positive CRC cell lines.
Knockdown of ZNF304, KAP1, SETDB1 or DNMT1 (Figure S7A), which are required for silencing of CIMP genes in KRAS-positive CRC cell lines (Serra et al., 2014), did not significantly derepress CIMP gene expression in BRAF-positive RKO cells (Figure 7A and S7B). Conversely, knockdown of MAFG or its corepressors (Figure S7C), which are required for silencing of CIMP genes in BRAF-positive CRC cell lines, did not significantly derepress CIMP gene expression in KRAS-positive DLD-1 cells (Figures 7B and S7D). Consistent with these results, in BRAF-positive RKO cells there was not significant association of ZNF304 and its corepressors with CIMP gene promoters (Figure 7C and S7E) and, conversely, in KRAS-positive DLD-1 cells there was not significant association of MAFG and its corepressors with CIMP gene promoters (Figure 7D and S7F). Finally, in BRAF-positive RKO cells association of DNMT3B with CIMP gene promoters was not affected by knockdown of either ZNF304 or its corepressors (Figures 7E and S7G) and, conversely, in KRAS-positive DLD-1 cells association of DNMT1 with CIMP gene promoters was not affected by knockdown of MAFG or its corepressors (Figures 7F and S7H). Collectively, these results show that different oncoproteins can selectively direct the assembly of distinct repressor complexes on common promoters.
Figure 7. Oncogenic BRAF and KRAS Direct the Assembly of Distinct Repressor Complexes on Common CIMP Gene Promoters.
(A and B) qRT-PCR analysis monitoring CIMP gene expression in RKO (A) and DLD-1 (B) cells expressing a NS, MAFG, CHD8, ZNF304 or KAP1 shRNA. Comparable analysis of BACH1, DNMT3B, SETDB1 and DNMT1 shRNAs is shown in Figures S7B and S7D.
(C and D) ChIP analysis monitoring binding of MAFG, CHD8, ZNF304 and KAP1 on CIMP gene promoters in RKO (C) and DLD-1 (D) cells. Comparable analysis of BACH1, DNMT3B, SETDB1 and DNMT1 is shown in Figures S7E and S7F.
(E and F) ChIP analysis monitoring binding of DNMT3B to CIMP gene promoters in RKO cells (E) and binding of DNMT1 to CIMP gene promoters in DLD-1 cells (F) expressing a NS, MAFG, CHD8, ZNF304 or KAP1 shRNA. Comparable analysis of BACH1, DNMT3B, SETDB1 and DNMT1 is shown in Figures S7G and S7H. Data are represented as mean ± SD. *P≤0.05, **P≤0.01.
(G) Model for BRAF(V600E)-directed recruitment of MAFG and its corepressors to MLH1 and CIMP gene promoters. See also Figure S7.
DISCUSSION
It is well established that in many cancers specific genes affecting cellular growth control are hypermethylated and transcriptionally silenced (Baylin, 2005; Esteller, 2006). However, in general, the factors involved in and the mechanistic basis of promoter hypermethylation and transcriptional silencing in cancer is not understood. In addition, the relationship between the initiating genetic events responsible for tumorigenesis (e.g., acquisition of activating mutations in oncogenes) and the subsequent promoter hypermethylation and transcriptional silencing remain to be determined.
In this study, we have identified a specific pathway that mediates CIMP in BRAF-positive CRCs (Figure 7G). The pathway is initiated on DNA by binding of the transcriptional repressor MAFG, which recruits a corepressor complex that includes BACH1, CHD8 and DNMT3B, leading to promoter hypermethylation and transcriptional silencing. Notably, a previous study analyzing a large collection of CRCs implicated a role for DNMT3B in CIMP-1 (Nosho et al., 2009). As discussed above, like other large-scale RNAi screens (Mullenders and Bernards, 2009), our RNAi screen was not saturating. Thus, it is likely that there are additional corepressors involved in MAFG-directed silencing of MLH1 and other CIMP genes. In this regard, our mass spectrometry analysis identified several MAFG interacting proteins with known roles in transcriptional repression (Figure S2H and Table S2).
BRAF(V600E) promotes transcriptional silencing through increased BRAF/MEK/ERK signaling, resulting in ERK-directed phosphorylation of MAFG at S124. Phosphorylation of MAFG at S124 prevents polyubiquitination and subsequent proteasomal-mediated degradation leading to increased MAFG levels, which drives DNA binding and transcriptional silencing. Notably, this mechanism explains why MLH1 and other CIMP genes are repressed in BRAF(V600E)-containing CRCs but not in normal cells. It is well known that a high percentage of BRAF-positive CRCs are also CIMP-positive (Ogino et al., 2009; Weisenberger et al., 2006). However, our results provide the first demonstration that BRAF, as well as EGFR, is not merely associated with but rather directly responsible for CIMP.
The results presented here, in conjunction with previous studies (Gazin et al., 2007; Palakurthy et al., 2009; Serra et al., 2014; Wajapeyee et al., 2013), suggest a general model by which tumor suppressor genes become transcriptionally silenced during cancer development. Activated oncoproteins alter cell signaling, protein stability or transcriptional regulatory pathways leading to elevated concentrations of a specific transcriptional repressor or corepressor, such as a DNA methyltransferase (see, for example, Palakurthy et al., 2009). The increased levels of repressors or corepressors results in their association with promoters that are not bound by these factors in normal cells, leading to promoter hypermethylation and transcriptional silencing. Moreover, here we have shown that through this mechanism different oncoproteins can direct the assembly of distinct repressor complexes on the same promoter. These latter results provide particularly strong support for the so-called instructive model of tumor suppressor gene silencing (Struhl, 2014), which invokes a specific pathway, comprising a defined set of components, initiated by an oncoprotein.
EXPERIMENTAL PROCEDURES
Cell Line Culture and Drug Treatment
RKO, SW48, VACO432 and DLD-1 cells were obtained from ATCC and grown as recommended by the supplier. For drug treatments in Figures 3A and 3B, RKO cells were treated with 0.1, 1 or 5 µM PLX4720 (Selleckchem), or 0.2, 2 or 10 µM U0126 (Cell Signaling Technology) for 24 hr. For Figure 3H, RKO cells were treated with 0.01, 1, 2, 4 or 8 µM MG132 (Cayman Chemical) for 4 hr. For Figure 3M, RKO cells were treated with 1 µM PLX4720 or 1 µM U0126 for 24 hr.
PFFs (ATCC) stably expressing BRAF(V600E) or MAFG were derived as follows. BRAF(V600E) cDNA (from plasmid pBabe-Puro-BRAF-V600E; plasmid 15269, Addgene) and MAFG cDNA (pMT2-MAFG (Blank et al., 1997); kindly provided by Volker Blank, McGill University/Lady Davis Institute for Medical Research) were cloned into pGIPZ-CMV (Open Biosystems), and the lentiviruses were transduced into PFFs and selected with puromycin for 24 hr.
Reporter Construct Cloning and Validation
The pMLH1-BlastR reporter was constructed as previously described (Serra et al., 2014), except 5 kb of the MLH1 promoter was PCR amplified from a BAC using primers engineered with NheI and XhoI restriction sites. Linearized plasmid was stably transfected into RKO cells using Effectene reagent (QIAGEN), cells were selected, and surviving colonies isolated and expanded as previously described. Clones were treated with 10 µM 5-aza-2’-deoxycytodine (Calbiochem) every 24 hr for 72 hr. After 24 hr treatment, 0, 5 or 10 µM blasticidin (Sigma-Aldrich) was added for 6 days, and cells were fixed and stained with 0.1% crystal violet to assess viability. Treatment with 5-aza-2’-deoxycytodine and subsequent challenge with blasticidin was used to identify a clone with robust survival when treated with both drugs.
RNAi Screen
The RNAi Consortium (TRC) lentiviral human shRNA library (Open Biosystems/Thermo Scientific) was obtained through the University of Massachusetts RNAi Core facility. Twenty-two lentivirus pools, each comprising ~5000 shRNA clones, were generated with titers of ~2×107 cfu/ml, as previously described (Gazin et al., 2007). 2×106 RKO cells were transduced at a multiplicity of infection of 0.2 with the lentiviral stocks in 10 cm dishes, and 2 days later puromycin selected (5 µg/ml) for 5 days. Cells were challenged with blasticidin (10 µg/ml) for 14 days. Cells that bypassed the basticidin challenge formed colonies that were isolated and individually expanded, and shRNAs were identified by sequence analysis as previously described (Gazin et al., 2007). Individual knockdown cell lines were generated by stable transduction of 1×105 cells with a single shRNA (listed in Table S4) followed by puromycin selection.
qRT-PCR
Total RNA was isolated and reverse transcription was performed as described (Gazin et al., 2007), followed by qRT-PCR using Power SYBR Green PCR Master Mix (Applied Biosystems). GAPDH was used as an internal reference gene for normalization. See Table S5 for primer sequences.
Bisulfite Sequencing
Bisulfite modification was carried out using an EpiTect Bisulfite Kit (QIAGEN) followed by assay kits from EpigenDX for AOX1 (ADS2444), CACNA1G (ADS2300), CHFR (ADS1462), DAPK1 (ADS037), EFEMP1 (ADS043), HAND1 (ADS1690), IRF8 (ADS1254), LOX (ADS852), PRDM2 (ADS297), p14ARF (ADS2130) and p16INK4A (ADS1067), or PCR primers for MLH1 and MINT6 (see Table S5). Multiple independent clones were sequenced from each PCR product within each cell line, of which six representative clones are displayed.
Immunoblot Analysis
Cell extracts were prepared by lysis in Laemmli buffer in the presence of protease inhibitor cocktail (Roche). The following commercial antibodies were used: MLH1 (Cell Signaling Technology), MAFG (Santa Cruz), BACH1 (Santa Cruz), CHD8 (Bethyl Laboratories), DNMT3B (Abcam), phospho-ERK1/2 and total ERK1/2 (both from Cell Signaling Technology), myc (Roche) and α-tubulin (TUBA; Sigma).
ChIP Assays
ChIP assays were performed as previously described (Gazin et al., 2007) using the following antibodies: MAFG (Santa Cruz), BACH1 (Santa Cruz), CHD8 (Bethyl Laboratories), DNMT3B (Abcam), ZNF304 (Serra et al., 2014), KAP1 (Bethyl Laboratories), SETDB1 (Millipore), DNMT1 (Imgenex), ING (Abcam), MAML3 (Santa Cruz), VWA5A (Antibody Online), ZBED5 (Abnova), ZFHX2 (Santa Cruz), ZFYVE27 (Santa Cruz) and ZNF701 (Sigma). ChIP products were analyzed by qRT-PCR (see Table S5 for primers). Samples were quantified as percentage of input, and then normalized to an irrelevant region in the genome (~3.2 kb upstream from the transcription start site of GCLC). Fold enrichment was calculated by setting the IgG control IP sample to a value of 1.
Co-immunoprecipitation Assays
RKO or PFF cell lysate was immunopreciptated with a MAFG, BACH1, CHD8, DNMT1, DNMT3B or control antibody (IgG; Millipore), and the immunoprecipitate was analyzed for MAFG, BACH1, CHD8 or DNMT3B by immunoblotting. Input lanes represent 5% of immunoprecipitated lanes.
In Vivo Kinase Assay
Wild-type MAFG was PCR amplified using primers that introduced a myc epitope tag at the C-terminus (Table S5) and cloned into pGEM-T Easy Vector (Promega) to generate pT-MAFG-myc. MAFG-T3A and MAFG-S124A were generated by inverse PCR using pT-MAFG-myc as a template (see Table S5 for primer sequences). The PCR fragments were agarose gel purified, treated with T4 polynucleotide kinase and self-ligated. Multiple clones of MAFG-wild-type (WT), MAFG-T3A and MAFG-S124A were confirmed by Sanger sequencing and cloned into pGIPZ-CMV to generate pGIPZ-CMV-MAFG-WT, pGIPZ-CMV-MAFG-T3A and pGIPZ-CMV-MAFG-S124A.
For assays in 293T cells (Figure 3I), each plasmid was transfected into 293T cells in the presence or absence of an ERK1-expression plasmid (pCMV-FLAG-ERK1; kindly provided by Roger Davis, University of Massachusetts Medical School). For assays in RKO cells (Figure 3J), plasmids were transfected into RKO cells and 24 hr later cells were treated with 1 µM PLX4720. Cell lysate was immunoprecipitated with a myc antibody and immunoprecipitates were analyzed by immunoblotting with a phospho-(S/T)P antibody (Abcam).
In Vitro Kinase Assay
A peptide of 161 amino acids (from 2 to 162) was amplified from plasmid pGIPZ-CMV-MAFG-WT, pGIPZ-CMV-MAFG-T3A or pGIPZ-CMV-MAFG-S124A (see Table S5 for primer sequences), and cloned into pGEX-4T1 (GE Healthcare). The GST-MAFG proteins were purified from E. coli. In vitro ERK1 kinase assays were performed as described previously (Canman et al., 1998). Briefly, 293T cells were transfected with plasmid expressing FLAG-tagged ERK1 (pCMV-FLAG-ERK1). ERK1 was immunoprecipitated using anti-Flag M2 beads (Sigma), incubated with 10 µCi [γ-32P]ATP and 1 µg of GST fusion substrates for 20 min at 30°C, and the reaction was stopped by the addition of SDS–PAGE protein sample buffer. Proteins were separated by SDS–PAGE and detected by autoradiography.
HA-Ubiquitin Pull-Down Assays
RKO cells (8×106) were plated on 10 cm dishes and transfected with 5 µg pGIPZ-CMV-MAFG-WT, pGIPZ-CMV-MAFG-T3A or pGIPZ-CMV-MAFG-S124A, 5 µg pcDNA3.1-HA-Ubiquitin (Addgene), and 0.5 µg pEGFP-N1 (Clontech) using Effectene reagent. To ensure equivalent transfection efficiency, EGFP expression was monitored 48 hr later. PLX4720 (1 µM) was added to cells 24 hr post transfection, and cells were incubated for another 24 hr. Cells were harvested in NETN-150 buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.05% NP-40) plus 1X protease inhibitor cocktail (Roche). Pull-downs were performed using an HA antibody (Cell Signaling Technology) and anti-rabbit Trublot beads (eBioscience). Beads were incubated with lysate for 18 hr, washed three times using NETN-150 buffer, and eluted in 2X sample buffer. Input samples were probed with a myc or TUBA antibody, and immunprecpitated samples were probed with a myc antibody.
PAT-ChIP Assay
This study was approved by the institutional review board at the University of Massachusetts Medical School (UMMS). Archived specimens (2010–2012) were obtained from the Department of Pathology at UMMS, and the CRC diagnosis was made by a UMMS pathologist. BRAF mutational analysis was performed by the UMass Memorial Laboratory of Diagnostic Molecular Oncology. Formalin-fixed paraffin-embedded tissue sections of matched adjacent normal colon and tumor samples isolated from individuals with invasive or metastatic BRAF-positive CRC were deparaffinized, rehydrated and processed as previously described (Serra et al., 2014).
Tumor Formation Assays
RKO cells (2×106) expressing either a NS, MAFG or CHD8 shRNA were suspended in 100 µl of serum-free RPMI and injected subcutaneously into the right flank of athymic BALB/c (nu/nu) mice (Taconic) (n=3 mice per shRNA). Tumor dimensions were measured every 3 days for 3 weeks and tumor volume was calculated using the formula π/6 x (length) x (width)2. All experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines.
Statistics
All quantitative data were collected from experiments performed in at least triplicate, and expressed as mean ± standard deviation. Differences between groups were assayed using two-tailed student t-test using Microsoft Excel. Significant differences were considered when P<0.05; *P≤0.05 and **P≤0.01.
Supplementary Material
HIGHLIGHTS.
MAFG mediates MLH1 silencing and CIMP in BRAF-positive colorectal cancers
BRAF(V600E) promotes silencing by increasing levels of MAFG to drive DNA binding
MAFG binds to CIMP genes and recruits a corepressor complex that includes DNMT3B
BRAF and KRAS direct assembly of distinct repressor complexes on common promoters
ACKNOWLEDGEMENTS
We thank Volker Blank and Roger Davis for providing reagents; the UMMS RNAi Core Facility for providing shRNA clones and libraries; the UMMS Proteomics and Mass Spectroscopy Facility for mass spectrometry analysis; and S. Deibler for editorial assistance. This work was supported by a grant from the NIH (R01GM033977) to M.R.G., who is also an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.F. and M.R.G. conceived and designed the experiments. M.F. performed the experiments. J.O. performed the bioinformatic analysis for MAFG binding sites, and L.H. provided the human CRC samples, performed the genotyping and characterized the samples. M.F. and M.R.G. analyzed the data and wrote the paper. All authors reviewed the paper and provided comments.
REFERENCES
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker AJ, Gibson KH, Grundy W, Godfrey AA, Barlow JJ, Healy MP, Woodburn JR, Ashton SE, Curry BJ, Scarlett L, et al. Studies leading to the identification of ZD1839 (IRESSA): an orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg. Med. Chem. Lett. 2001;11:1911–1914. doi: 10.1016/s0960-894x(01)00344-4. [DOI] [PubMed] [Google Scholar]
- Baylin SB. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Kim MJ, Andrews NC. Human MafG is a functional partner for p45 NF-E2 in activating globin gene expression. Blood. 1997;89:3925–3935. [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. e2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklof V, Rutegard J, Oberg A, Van Guelpen BR. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin. Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. Regulation of transcription factors by protein degradation. Cell Mol. Life Sci. 2000;57:1207–1219. doi: 10.1007/PL00000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Marais R. New insight into BRAF mutations in cancer. Curr. Opin. Genet. Dev. 2007;17:31–39. doi: 10.1016/j.gde.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br. J. Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fanelli M, Amatori S, Barozzi I, Minucci S. Chromatin immunoprecipitation and high-throughput sequencing from paraffin-embedded pathology tissue. Nat. Protoc. 2011;6:1905–1919. doi: 10.1038/nprot.2011.406. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–538. doi: 10.1016/s0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyparaki MT, Basdra EK, Papavassiliou AG. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J. Mol. Med. (Berl) 2013;91:1249–1256. doi: 10.1007/s00109-013-1088-z. [DOI] [PubMed] [Google Scholar]
- Hassler MR, Egger G. Epigenomics of cancer - emerging new concepts. Biochimie. 2012;94:2219–2230. doi: 10.1016/j.biochi.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Lin VA, Buckle A, Cheong K, Halani N, Ku S, Kwok CT, Packham D, Suter CM, Meagher A, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- Hodoglugil U, Carrillo MW, Hebert JM, Karachaliou N, Rosell RC, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for the epidermal growth factor receptor. Pharmacogenet. Genomics. 2013;23:636–642. doi: 10.1097/FPC.0b013e3283655091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int. J. Clin. Oncol. 2008;13:498–503. doi: 10.1007/s10147-008-0854-3. [DOI] [PubMed] [Google Scholar]
- Jover R, Nguyen TP, Perez-Carbonell L, Zapater P, Paya A, Alenda C, Rojas E, Cubiella J, Balaguer F, Morillas JD, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda A, Yagi K. Two groups of DNA methylation markers to classify colorectal cancer into three epigenotypes. Cancer Sci. 2011;102:18–24. doi: 10.1111/j.1349-7006.2010.01712.x. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Igarashi K, Itoh K, Fujiwara KT, Noda M, Yamamoto M, Nishizawa M. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25:2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28:4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- Nosho K, Shima K, Irahara N, Kure S, Baba Y, Kirkner GJ, Chen L, Gokhale S, Hazra A, Spiegelman D, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin. Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, Green MR. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol. Cell. 2009;36:219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M, Kikuchi A. A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J. Biol. Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
- Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Is DNA methylation of tumour suppressor genes epigenetic? Elife. 2014;3:e02475. doi: 10.7554/eLife.02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell. Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc. Natl. Acad. Sci. USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. USA. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin. Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl. Acad. Sci. USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N, Malonia SK, Palakurthy RK, Green MR. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27:2221–2226. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WH, Cook PN, Slater AM, Davies DH, Holdgate GA, Green LR. Epidermal growth factor receptor tyrosine kinase. Investigation of catalytic mechanism, structure-based searching and discovery of a potent inhibitor. Biochem. Pharmacol. 1994;48:659–666. doi: 10.1016/0006-2952(94)90042-6. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin. Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- Yeh JJ, Routh ED, Rubinas T, Peacock J, Martin TD, Shen XJ, Sandler RS, Kim HJ, Keku TO, Der CJ. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol. Cancer Ther. 2009;8:834–843. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.