SUMMARY
Tumor suppressors safeguard the fidelity of the mitotic checkpoint by transcriptional regulation of genes that encode components of the mitotic checkpoint complex (MCC). Here we report a new role for the tumor suppressor and transcription factor, WT1, in the mitotic checkpoint. We show that WT1 regulates the MCC by directly interacting with the spindle assembly checkpoint protein, MAD2. WT1 colocalizes with MAD2 during mitosis and preferentially binds to the functionally active, closed-conformer, C-MAD2. Furthermore, WT1 associates with the MCC containing MAD2, BUBR1 and CDC20, resulting in prolonged inhibition of the anaphase promoting complex/cyclosome (APC/C), and delayed degradation of its substrates SECURIN and CYCLIN B1. Strikingly, RNAi-mediated depletion of WT1 leads to enhanced turnover of SECURIN, decreased lag time to anaphase, and defects in chromosome-segregation. Our findings identify WT1 as a regulator of the mitotic checkpoint and chromosomal stability.
INTRODUCTION
WT1 is a zinc finger transcription factor that can activate or repress target genes that control cell growth and development1–3. WT1 is subject to alternative splicing in two regions; a 17 amino acid insertion within the central region of the protein (17AA) and the insertion of three amino acids (Lys-Thr-Ser) within the zinc finger region (KTS). WT1 is expressed in several organs and tissues of the embryo and is particularly important for the development of the urogenital system where it functions as a tumor suppressor4,5. Recent findings suggest that WT1 is a key regulator of mesenchyme to epithelial balance during development, and is also required for the maintenance of several adult tissues6. A considerable body of evidence suggests that WT1 can also act as an oncogene7,8. WT1 is overexpressed in several cancers including leukemia, breast, ovary, bone, lung and brain, and is a promising therapeutic target9,10.
The accuracy of cell division is monitored at several steps and is initiated at the beginning of mitosis by the spindle assembly checkpoint (SAC). The SAC components, which include MAD1, MAD2, BUBR1 and BUB3, play key roles to ensure correct attachment of chromosomes to the mitotic spindle and fidelity of chromosomal segregation during cell division. Impaired SAC function promotes aneuploidy and contributes to genomic instabilities and tumorigenesis11–13. Indeed, a majority of human tumors accumulate mutations that deregulate the expression of proteins essential for mitotic checkpoint function14–16. This results in mis-segregation of chromosomes during mitosis and contributes to chromosome instability (CIN).
MAD2 adopts two native conformations, open (O-MAD2) and closed (C-MAD2), and has the ability to self-dimerize. C-MAD2 is the functionally active form of MAD2 which engages in the formation and maintenance of the checkpoint signal cascade17–20. The presence of un/mis-attached kinetochores activates the SAC signal in which the MAD2-MAD1 complex generates a diffusible anaphase wait signal. This mitotic checkpoint complex (MCC) composed of MAD2, BUBR1, CDC20 and BUB3 then inhibits the anaphase-promoting complex/cyclosome (APC/C). The ubiquitin ligase activity of APC/C is critical for the degradation of SECURIN and CYCLIN B1 and eventual anaphase entry12,21–25. Several lines of evidence suggest that human tumors with CIN have misregulated expression of MAD2. Studies in mice heterozygous for MAD2 showed increased frequency towards aneuploidy26–28.
Here we show that WT1 associates with C-MAD2 during mitosis and regulates the mitotic checkpoint function. We demonstrate that, through interaction with MAD2, WT1 inhibits APC/C-mediated degradation of SECURIN and CYCLIN B1 and that ablation of WT1 protein in cells that normally express WT1 leads to chromosomal-segregation defects and early anaphase entry. Our results reveal a previously unknown role of WT1 in the direct regulation of mitotic checkpoint function and genomic stability via its interaction with MAD2.
RESULTS
WT1 interacts with MAD2 during mitosis
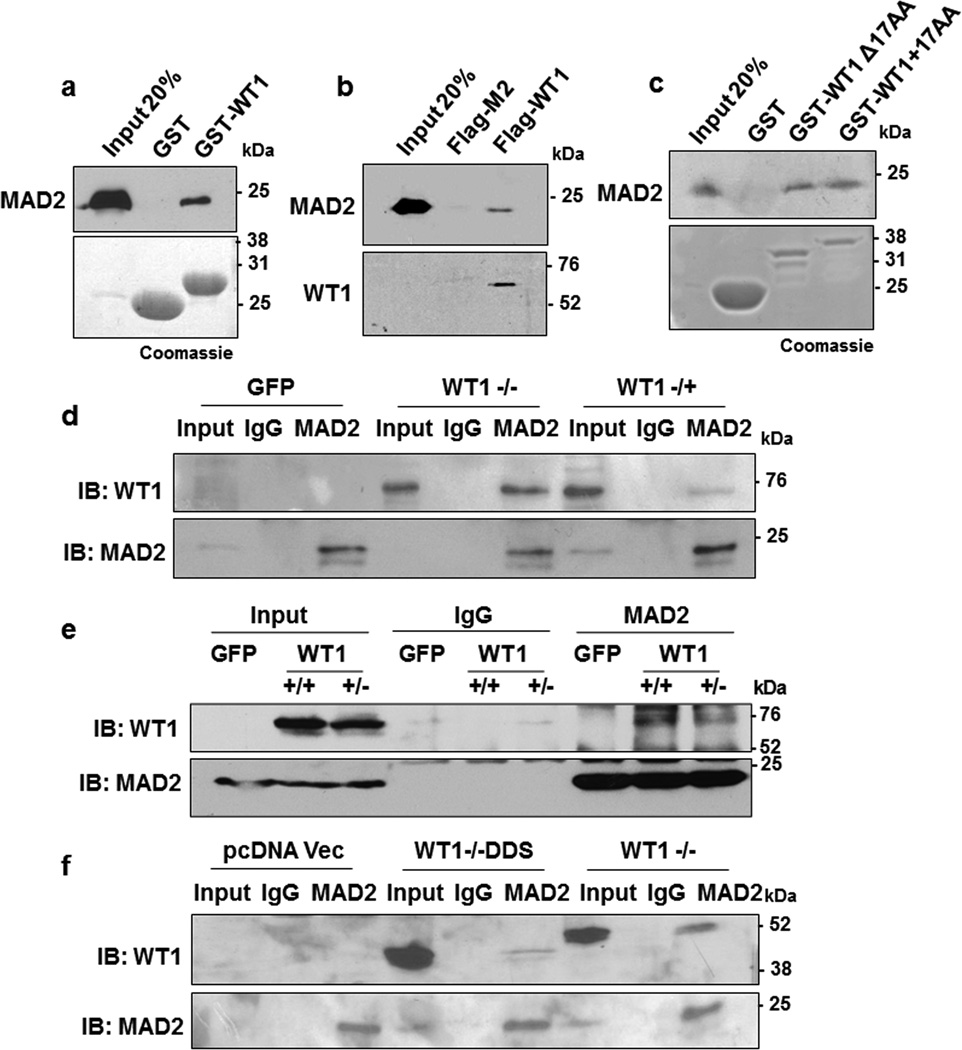
A yeast two-hybrid screen performed with a discrete region of mouse WT1 protein (residues 245–297; that contains the 17AA) using a HeLa cDNA library revealed MAD2 as a potential interaction partner. A direct interaction between WT1 and MAD2 was confirmed in vitro by GST-pulldown assay where recombinant full length (FL) His-tagged MAD2 protein associated with GST-WT1 (245–297) but not with the control GST-beads (Fig.1a). Furthermore, Flag-tagged full length human WT1 protein was also found to interact with MAD2 in vitro (Fig. 1b). Binding assays comparing GST-WT1+17AA (residues 180–297) and GST-WT1 Δ17AA (lacking the 17AA) revealed that the 17AA region of WT1 was dispensable for the MAD2 interaction (Fig. 1c).
Figure 1. WT1 interacts with MAD2.
(a) In vitro interaction assay was performed with either GST-WT1 (residues 245–297) or GST in the presence of full length His-MAD2. The interaction was analyzed by immunoblotting with anti-MAD2 antibody. Bound proteins were also resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. (b) In vitro pulldown assay was also performed with either Flag-M2 magnetic beads alone or incubated with full length Flag-tagged WT1 protein in presence of His-MAD2. The interaction was analyzed by immunoblotting with anti-WT1 and anti-MAD2 antibodies (c) GST-interaction assay was carried out with WT1 containing the 17 amino acid insertion (residues 180-297, +17AA) or lacking it (residues 180-297, Δ17 AA) with His-MAD2. The interaction was analyzed by immunoblotting with anti-MAD2 antibody. Bound proteins were also resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. (d) HeLa cells were transfected with GFP Vector, full length GFP-WT1 (−/−) or GFP-WT1 (−/+) isoforms and 48 hours later whole cell extracts were prepared followed by immunoprecipitation with anti-MAD2 antibodies. The immunoprecipitates were probed with anti-WT1 antibody. Blotting with anti-MAD2 antibody was performed as a control. (e) The MAD2 interaction with ectopically expressed GFP tagged full length WT1 (+/+) and WT1 (+/−) isoforms was analyzed as in part d (f) HeLa cells were transfected with pcDNA vector, pcDNA driving expression of WT1−/−DDS (R394X) or wild type WT1−/− followed by immunoprecipitation with anti-MAD2 antibodies. The immunoprecipitates were probed with anti-WT1 antibodies. Blotting with anti-MAD2 antibodies was performed as a control.
We next sought to determine if MAD2 interacts with all the four major isoforms of human WT1 in the cells. HeLa cells (which do not express WT1) were transfected with a plasmid driving expression of either GFP, GFP linked to full length WT1 (−/− isoform, which does not contain either the 17AA or KTS insertions) or full length GFP-tagged WT1 (−/+ isoform, which lacks the 17AA but contains the KTS insertion). Whole cell extracts were prepared and MAD2-immunoprecipitates from the transfected cells were analysed by immunoblotting with anti-WT1 antibodies, which confirmed that both the −/− and −/+ WT1 isoforms interact with endogenous MAD2 (Fig. 1d). Similar interaction assays were carried out with the other full length GFP-tagged WT1 isoforms, the WT1 (+/+) which contains both the 17AA and the KTS regions and WT1 (+/−) which contains the 17AA but not the KTS domain (Fig. 1e). The results showed that endogenous MAD2 could interact with all four major isoforms of WT1. We also analyzed a WT1 mutant derivative (R394X) that mimics those found in Denys Drash syndrome (WT1 −/−DDS) and lacks an intact DNA-binding domain. The results revealed a weaker MAD2-binding efficiency of WT1 −/−DDS with respect to full length WT1 −/− (Fig. 1f) suggesting that DNA-binding might play a role in bringing WT1 in close proximity to MAD2.
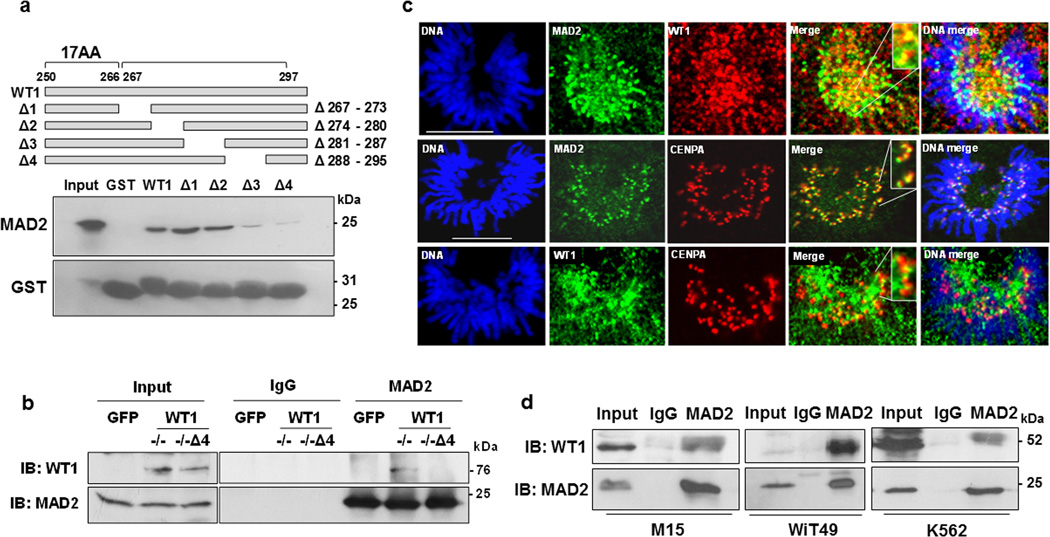
To determine the MAD2-binding domain of WT1, several internal deletion mutants of GST-WT1 were generated (Fig. 2a, schematic) and tested for their ability to interact with MAD2. Deletion of the region between residues 280 and 295 (Δ3 and Δ4) led to a significant reduction in the WT1-MAD2 interaction (Fig. 2a). We therefore deleted the (Δ4) region spanning amino acids 288–295 in the full length human GFP-WT1−/− isoform and tested whether this region of WT1 is crucial for MAD2 interaction in cells. The results showed significant reduction in GFP-WT1−/−Δ4 and MAD2 interaction when compared to GFP-WT1−/− (Fig. 2b).
Figure 2. Endogenous WT1 interacts with MAD2.
(a) GST-interaction assays were performed with the internal deletion mutants of GST-WT1 (schematic) with full length His-MAD2 and immunoblotted with anti-MAD2 and anti-GST antibodies. (b) Co-immunoprecipitation assays were carried out with whole cell extracts derived from HeLa cells that had been transfected with either GFP vector, GFP WT1 (−/−) or GFP-WT1 (−/−Δ4) using anti-MAD2 or anti-WT1 antibodies. (c) Co-immunofluorescence analysis was carried out in WiT49 cells with anti-MAD2 and anti-WT1 antibodies at the pro-metaphase stage of mitosis. DNA was stained with Hoechst. The scale bar is 10 microns. (d) M15, WiT49 and K562 cells were treated with 60 ng/ml nocodazole for 24 hours, whole cell extracts prepared and immunoprecipitation performed with anti-MAD2 antibodies followed by immunoblotting with WT1 antibody. Input is 10% of the total whole cell extract.
We next performed co-immunofluorescence analysis in WiT49 cells (a Wilms’ tumor-derived cell line that expresses endogenous wild type WT1) with anti-MAD2 and anti-WT1 antibodies, which revealed a significant co-localization pattern during the pro-metaphase of mitosis (Fig. 2c). The centromeric localization of MAD2 and WT1 was also studied using anti-CENPA antibodies in our colocalization assays. MAD2 and CENPA were juxtaposed close to the centromeres while WT1 was localized at the surrounding regions of the centromeres. To study the association between endogenous WT1 and MAD2 during activation of the checkpoint, co-immunoprecipitation analysis was performed with whole-cell extracts prepared from nocodazole-treated M15 cells (mouse mesonephric cell line), WiT49 cells (human Wilms' tumor cell line) and K562 cells (derived from human chronic myelogenous leukemia). In each case the endogenous WT1 was present in MAD2-immunoprecipitates (Fig. 2d). These results raised the possibility that the association of WT1 with MAD2 might play a role in the regulation of spindle assembly checkpoint function.
WT1 interacts with the active, closed MAD2 conformer
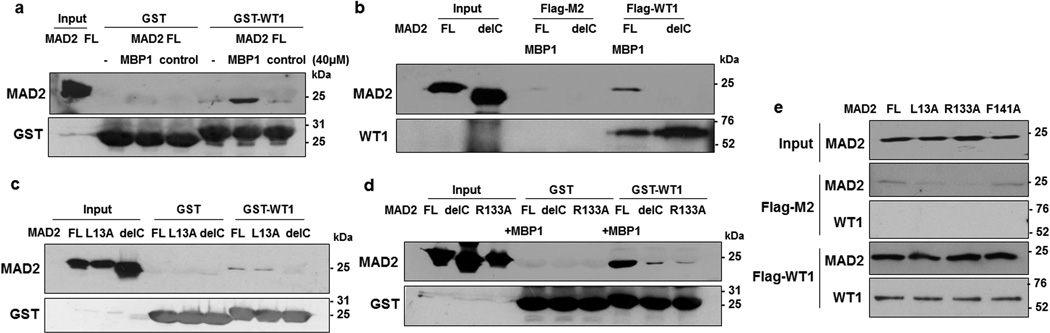
MAD2 protein can adopt two native conformations; the closed (C-MAD2) or the open (O-MAD2) conformers17–20. At the onset of mitosis, O-MAD2 is recruited to the unattached kinetochores of the pro-metaphase chromosomes with the help of its partner MAD1 which catalyses the conversion of O-MAD2 to C-MAD2 conformer. The MAD1-C-MAD2 tetramer acts as a template for additional O- to C-MAD2 conversion and initiates active SAC signaling. CDC20 is a critical C-MAD2-binding partner which occupies the same binding site on MAD2 as MAD1. Binding of either MAD1 or CDC20 facilitates the formation of the C-MAD2 conformation29,30. A 12-amino acid MAD2-binding peptide (MBP1) has been generated that competes for the same site on MAD2 that is bound by MAD1 or CDC20. Comparative NMR and co-crystal studies have revealed that the MAD2-MBP1 complex adopts the C-MAD2 conformation, mimicking MAD2 structure in either the MAD2-MAD1 or MAD2-CDC20 complexes18,29. We therefore employed MBP1 in our interaction studies to test the binding preference of WT1 with the MAD2 conformers. Recombinant wild type MAD2 protein equilibrates between O- and C-MAD2 conformers and incubation of MAD2 with MBP1 shifts the equilibrium towards C-MAD2 conformer. Pulldown assays were carried out with GST-WT1 (245–297) and MAD2 that had been pre-incubated with either MBP1, or a control peptide. The data revealed that GST-WT1 (245–297) has a higher affinity for MAD2 that was pre-bound to MBP1 and therefore for C-MAD2 (Fig. 3a).
Figure 3. WT1 has higher affinity for the active closed MAD2 conformer.
(a) GST-interaction assays were carried out with WT1 (residues 245–297) and full length (FL) His-MAD2 alone and also with MAD2 pre-incubated with either 40µM of MAD2-binding peptide 1 (MBP1), or a control peptide. The pulldown complexes were resolved by SDS-PAGE and immunoblotted with anti-MAD2 antibodies. (b) In vitro pulldown assays were performed with either Flag-M2 magnetic beads alone or full length Flag-tagged WT1 protein in the presence of Full length His-MAD2 (pre-incubated with MBP1) or the MAD2 delC mutant. The interaction was analyzed by immunoblotting with anti-WT1 and anti-MAD2 antibodies. (c) GST-interaction assays were carried out with His-MAD2, MAD2 L13A and MAD2-C-terminal deletion mutant (Del-C). Immunoblotting was performed using anti-MAD2 and anti- GST antibodies. (d) GST-interaction assays were carried out with His-MAD2 + MBP1, MAD2-Del-C, and MAD2 dimerization mutant, R133A. Immunoblotting was performed using anti-MAD2 and anti-GST antibodies. (e) Pull down assays were performed with full length Flag-WT1 protein and the different MAD2 point mutant derivatives. Immunoblotting was performed using anti-MAD2 and anti-WT1 antibodies. Input is 20% of the different MAD2 proteins used for interaction analysis.
Studies with several deletion and point mutants of MAD2 have identified residues that are important for its dimerization and also for acquiring the C/O-MAD2 state18,31. Deletion of 10 amino acids at the C-terminus results in a MAD2 derivative that is locked in the O-conformation and cannot bind MAD1, CDC20 or MBP131. Similarly, when MAD2 lysine residue 13 is substituted to alanine (L13A), it results in a MAD2 derivative locked in C-conformation, while substitution of arginine residue 133 with alanine (R133A), generates the monomeric form of MAD2 which cannot self-dimerize. We generated a C-terminal 10 aa deletion mutant of MAD2 (Del-C), C-MAD2 mutant (L13A) and MAD2 dimerization mutant, R133A and F141A and then compared their ability to interact with WT1 with respect to MBP1-bound C-MAD2.
We performed pull down assays with full length MAD2 (pre-incubated with MBP1 to generate C-MAD2) and MAD2 Del-C mutant (O-MAD2) in presence of either Flag-M2 beads or Flag-tagged full length WT1 protein. Interestingly, full length WT1 showed a greater level of binding to MAD2 that was pre-incubated with MBP1 suggesting that WT1 has a higher affinity to interact with C-MAD2 (Fig. 3b). Furthermore, GST-pulldown assays showed that WT1 (residues 245–297) has higher affinity to interact with C-MAD2 (L13A) when compared to MAD2 Del-C (Fig. 3c). WT1 can also bind to O-MAD2 (Del-C), but at a lower affinity than that observed for C-MAD2 that is induced by MBP1 (Fig. 3d). The different structural mutants of MAD2 (L13A, R133A and F141A) were also found to efficiently interact with full length Flag-WT1 (Fig. 3e). Taken together, these results suggest that WT1 binds preferentially to C-MAD2 and that WT1 does not bind to the same site in MAD2 as either MAD1 or CDC20.
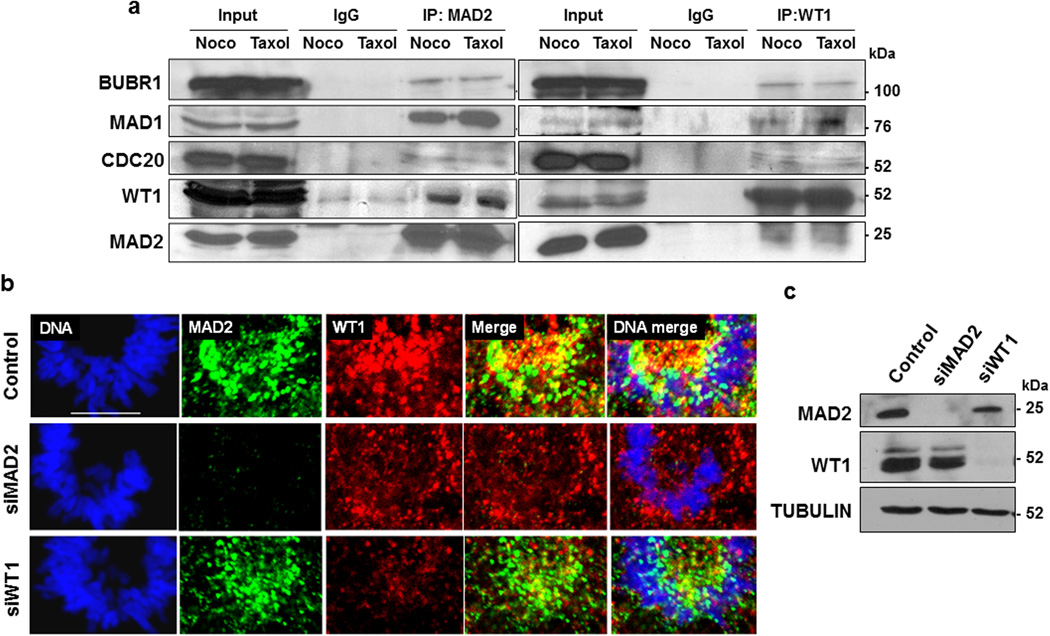
In order to test whether WT1 can form a complex with MAD2 along with other components of the MCC, we induced the spindle checkpoint in K562 cells with nocodazole or taxol, prepared whole cell extracts and immunoprecipitated either MAD2 or WT1 followed by immunoblotting with anti-BUBR1, MAD1, CDC20, WT1 and MAD2 antibodies (Fig. 4a). Both anti-WT1 and anti-MAD2 antibodies co-immunoprecipitated MAD1, CDC20 and BUBR1, suggesting that WT1 can associate with activated spindle/mitotic checkpoint complexes. The association of WT1 with the MAD2-MAD1 complex is consistent with its localization during the pro-metaphase stage (as seen in Fig. 2c). We therefore determined the effect of MAD2 depletion on the sub-cellular localization of WT1 during pro-metaphase. Co-immunofluorescence analysis of WT1 and MAD2 was performed in WiT49 cells that had been transfected with MAD2 siRNA, which resulted in the loss of WT1 localization pattern at the chromosomes when compared to cells transfected with control siRNA (Fig.4b and 4c). Comparable effects were seen when we performed a similar experiment in M15 cells (Supplementary Fig. 1). However, RNAi-mediated depletion of WT1 did not adversely affect the localization of MAD2 (Fig. 4b and Supplementary Fig. 1). Taken together, these results show that WT1 associates with the active C-MAD2 complex during mitosis suggesting a potential role in MAD2-dependent checkpoint functions.
Figure 4. WT1 interacts with the mitotic checkpoint complex.
(a) K562 cells were treated with 60 ng/ml of either nocodazole or taxol. MAD2 and WT1-immunoprecipitates were analyzed by immunoblotting with the indicated antibodies. Input is 10% of the total whole cell extract used for immunoprecipitations. (b) WiT49 cells were transfected with control, MAD2 siRNA or WT1 siRNA for 48 hours followed by immunofluorescence analysis with anti-MAD2 and anti-WT1 antibodies in the pro-metaphase stage of mitosis. DNA was stained with Hoechst. The scale bar is 10 microns. (c) Immunoblotting was performed with anti-MAD2, anti-WT1 and anti-TUBULIN antibodies for control and MAD2 or WT1 siRNA transfected cells.
WT1 inhibits APC/C function
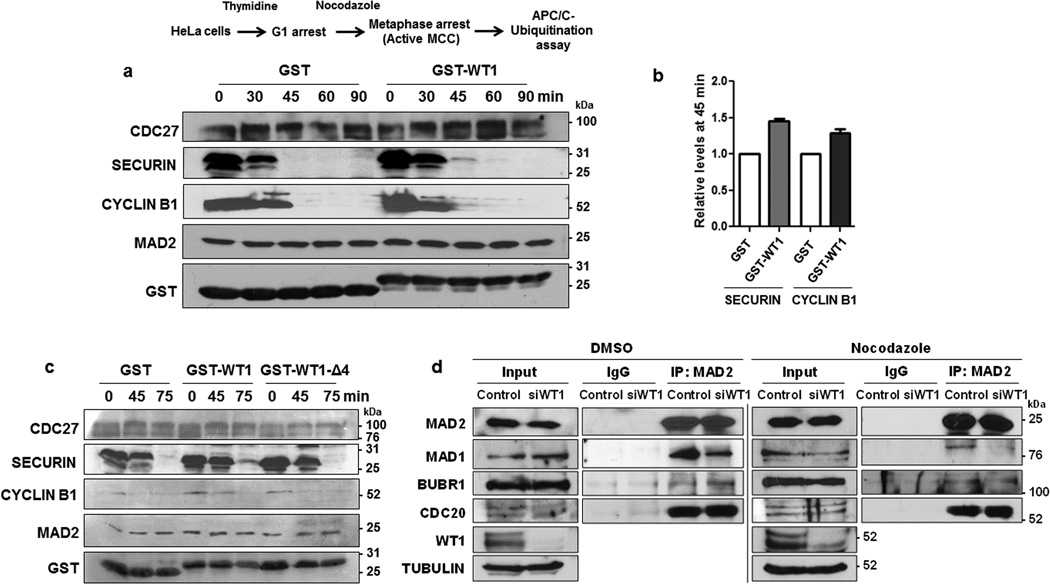
Our data so far suggest that WT1 associates with C-MAD2 that forms part of the MCC. An essential function of the MCC is to inhibit APC/C, an E3-ubiquitin ligase that marks SECURIN and CYCLIN B1 for degradation leading to chromosomal separation during anaphase. In order to determine if WT1 plays a role in the regulation of MAD2-dependent APC/C inhibition, a ubiquitination/degradation assay was performed with mitotic extracts prepared from nocodazole-arrested HeLa cells (which do not express WT1), to which we added either GST or GST-WT1 and incubated for different time periods. The results showed that GST-WT1 significantly delayed the degradation of SECURIN and CYCLIN B1 by APC/C compared to GST (Fig. 5a). Immunoblotting with anti-MAD2, GST and CDC27 (a subunit of APC/C) antibodies was performed as controls. Quantitation of the levels of SECURIN and CYCLIN B1 at the 45 min time point are plotted as graphs (Fig. 5b). The GST-WT1-Δ4 mutant (MAD2-interaction defective mutant lacking residues 288–295; see Fig. 2a) did not delay SECURIN and CYCLIN B1 degradation, confirming that direct association with MAD2 is essential for WT1-mediated APC/C inhibition (Fig. 5c).
Figure 5. WT1 inhibits APC/C function.
(a) APC/C ubiquitination assay was performed with mitotic extracts derived from nocodazole-arrested HeLa cells in the presence of GST control or GST-WT1 (residues 245–297) for different time periods. The reaction was stopped at 0, 30, 45, 60, and 90 minutes and immunoblotting was performed with the indicated antibodies. The degradation of SECURIN and CYCLIN B1 was monitored for the indicated time points. (b) The band intensity at 45 min time point for SECURIN and CYCLIN B1 degradation in the presence of GST control or GST-WT1 from three independent experiments was quantified using Image J software and plotted graphically. Error bars are standard deviation from the mean. (c) APC/C ubiquitination assay was performed as in part A, with GST, GST-WT1 (residues 245–297) and GST-WT1Δ4 (Δ 288–295) at different time points and immunoblotted with the antibodies indicated. (d) WiT49 cells were transfected with either control or WT1 siRNA for 48 hours followed by treating the cells with either DMSO or nocodazole (60 ng/ml) for a further 12 hours. Whole cell extracts were prepared and MAD2 was immunoprecipitated then immunoblotted with anti-MAD1, anti-BUBR1, anti-CDC20 and anti-MAD2 antibodies. Input is 10% of the total whole cell extract used for immunoprecipitation.
To study the possible mechanism by which WT1 inhibits APC/C activity, co-immunoprecipitation analysis was carried out with whole cell extracts derived from WiT49 cells that had been transfected with either control or WT1 siRNA followed by treatment with DMSO or nocodazole (60 ng/ml). The results showed that upon WT1 silencing the association of MAD2 with MAD1 was significantly reduced in both DMSO as well as nocodazole-treated cells (Fig. 5d), suggesting a potential role of WT1 in promoting MAD2-MAD1 interaction during the initiation of active checkpoint signaling. Thus, WT1 integrates with and enhances the formation of MAD2-containing spindle/mitotic checkpoint complexes leading to a sustained inhibition of APC/C activity. The results also show that WT1 does not compete with MAD1 for MAD2 binding. Presumably, WT1 may promote the stability of MAD1-MAD2 tetramer or recruit more O-MAD2 for amplifying the SAC signal. Alternatively, WT1 could also induce or stabilize C-MAD2 conformation. Structural insights from MAD2-WT1 association would reveal more information regarding the mode of action of WT1 during checkpoint activation.
Depletion of WT1 promotes mitotic segregation defects
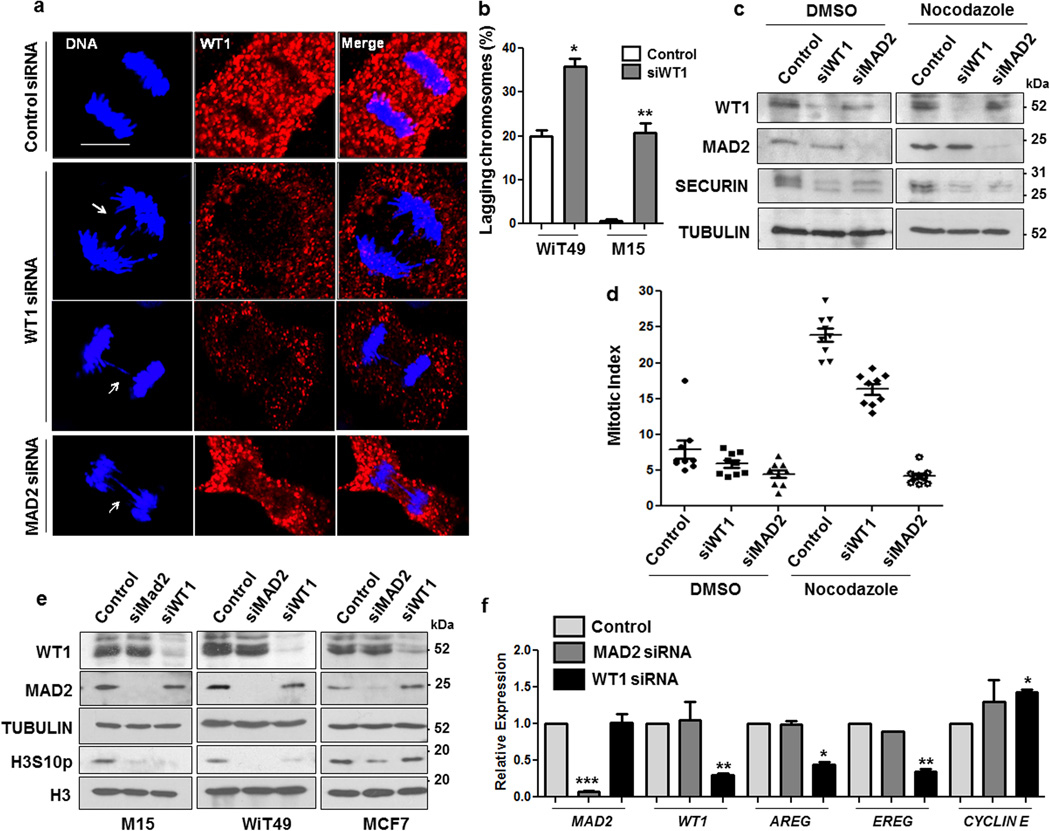
Our results show that the interaction between WT1 and MAD2 enhances MCC function and leads to prolonged inhibition of APC/C activity. This suggests that stimulation of SAC function by WT1 could potentially affect chromosomal stability and genomic integrity. To test this possibility, WiT49 cells were treated with control siRNA, WT1 siRNA or MAD2 siRNA for 48 hours, followed by immunofluorescence analysis using anti-WT1 antibodies (Fig. 6a). Depletion of WT1 protein resulted in an enhanced rate of chromosome-lagging and -bridges during anaphase which was strikingly similar to the defects observed upon MAD2 silencing in WiT49 cells (Fig. 6a, compare central two panels with lower panel). Quantitation of lagging chromosomes in control and WT1 siRNA-transfected WiT49 cells is shown in Fig. 6b. Transfection of M15 cells with WT1 siRNA also caused a significant increase in lagging chromosomes (Fig. 6b). Consistent with these observations, siRNA-mediated knockdown of either WT1 or MAD2 resulted in faster degradation of SECURIN when compared to control siRNA in WiT49 cells treated with either DMSO or nocodazole (Fig. 6c).
Figure 6. WT1 knockdown increases the rate of mitotic abnormalities.
(a) WiT49 cells were transfected with control, WT1, or MAD2 siRNA and 48 hours later immunofluorescence analysis was performed with anti-WT1 antibodies to study the anaphase stage of mitosis. DNA was stained with Hoechst. The scale bar is 10 microns. (b) The percentage of WiT49 and M15 cells exhibiting mitotic defects (chromosome lagging/bridges) was scored over total number of cells that were in anaphase for >100 optical fields and plotted graphically. Error bars are standard deviation from the mean for three independent experiments and statistical significance was determined by Student’s t test (P < 0.05). (c) WiT49 cells were transfected with control, WT1 or MAD2 siRNAs for 48 hours followed by treatment with either DMSO or nocodazole (60 ng/ml) for another 24 hours. Whole cell extracts were prepared and immunoblotted with anti-WT1, MAD2, SECURIN and TUBULIN antibodies (d) Mitotic indices were calculated for WiT49 cells transfected with control, WT1 or MAD2 siRNA followed by DMSO or nocodazole (60 ng/ml) treatment. The results were plotted as graphs after analyzing >100 optical fields for at least three independent experiments. Error bars are standard deviation from the mean. (e) M15, WiT49 and MCF7 cells were transfected with control, MAD2 or WT1 siRNAs for 48 hours followed by immunoblotting with anti-WT1, anti-MAD2, anti-TUBULIN, anti-phosphorylated Histone H3 serine10 (H3S10p) and anti-Histone H3 antibodies. (f) Quantitative RT-PCR analysis was performed to analyze the expression of MAD2, WT1, AREG, EREG and CYCLIN E genes in WiT49 cells transfected with control, MAD2 siRNA or WT1 siRNA for 48 hours. Error bars denote standard deviation from the mean of three independent experiments and statistical significance was analyzed by Student’s t test (P < 0.05).
Ablation of MAD2 protein results in defective SAC signaling. As a consequence, cells exit mitosis without stringent monitoring of kinetochore-spindle attachment errors. We scored the relative mitotic indices for WiT49 cells transfected with control, MAD2 or WT1 siRNAs (Fig. 6d). The mitotic index was drastically reduced upon MAD2 silencing in nocodazole treated cells. Knockdown of WT1 also showed significant reduction in the number of metaphase cells in nocodazole treated cells when compared to control. These results suggest that the absence of either MAD2 or WT1 abrogates SAC function leading to faster exit from mitosis and accumulation of chromosome-segregation defects in the cells. We also determined if chromatin condensation during mitosis was affected upon MAD2 or WT1 silencing. Interestingly, we observed a large reduction in the level of Histone H3 serine10 phosphorylation (H3S10p) in WiT49 and M15 cells transfected with either MAD2 or WT1 siRNAs (Fig. 6e). However, the effect of WT1 knockdown was not significant in MCF7 cells, suggesting the possible role of other cell-type specific components in the regulation of WT1 functions during mitosis. Taken together, the data in Fig. 6a–e suggests that loss of WT1 results in a hypo-active SAC signaling and early anaphase entry, leading to an increase in erroneous or incomplete spindle attachments to the kinetochores and chromosome-lagging during anaphase.
DISCUSSION
WT1 is a transcription factor with diverse functions in the cell ranging from organogenesis and tumor suppression to oncogenesis6–10, 32. In this study we have uncovered a unique role for WT1 as a mitotic checkpoint regulator. WT1 performs this function without significantly affecting the expression of genes crucial for SAC/MCC regulation. We also found that depletion of MAD2 by siRNA does not influence transcription of WT1-target genes (Fig. 6f). Several transcription factors are known to regulate the mitotic checkpoint through control of the expression of SAC and MCC components33. Our finding of a direct role for WT1 in the MCC has demonstrated a novel mechanism of action for a tumor suppressor/oncogene in the control of mitosis.
WT1 is aberrantly expressed in tumors of several different origins7,9,10. Thus a role in the regulation of APC/C activity could potentially contribute to the mis-regulation of the mitotic checkpoint in tumorigenesis. Conversely, loss of WT1 protein could compromise the SAC function and thereby contribute to increased mitotic checkpoint failure and eventually chromosome instability. Indeed, loss of WT1 has also been associated with abnormal chromosome number34,35. It would be of interest to study whether aberrant WT1 expression contributes to aneuploidy in different cancers as an outcome of its role in the regulation of cell cycle checkpoint.
There are several factors that modulate MAD2-dependent checkpoint functions. p31 comet is a critical regulator of MAD2 which interacts with MAD1- or CDC20-bound MAD2 and negatively regulates the SAC function. p31 comet binds to the dimerization interface of MAD2 and promotes dissociation of CDC20-MAD2 containing MCC36,37. Interestingly, WT1 influences MAD2 function in an opposite manner to p31 comet and instead stabilizes MCC-mediated inhibition of APC/C and positively regulates the SAC function. WT1 preferentially binds to MBP1-bound MAD2, suggesting it favors the C-MAD2 conformer and does not compete with either MAD1 or CDC20 binding. Furthermore, knockdown of WT1 reduces MAD1-MAD2 interaction in the cells which indicate that WT1 is not a competitive inhibitor of MAD1. Rather it augments MAD1-MAD2 association crucial for active SAC signaling. This property of WT1 is also different from the role played by Shugoshin (human Sgo2), which competes with MAD1 and CDC20 for binding to MAD2 during meiosis38. WT1 neither competes with MAD2-dimerization interface nor the MAD1/CDC20 binding site, yet significantly influences MAD2-dependent MCC functions. The interplay among these cellular regulators of MAD2 and their relative expression could therefore influence the final timing of anaphase entry and stringency of chromosome segregation. Our findings also raise the likelihood that other tumor suppressors might also act on the MCC directly and not only through the regulation of the abundance of MCC components.
METHODS
Cell culture and transfection
WiT49 (derived from human anaplastic Wilms’ tumor), HeLa (derived from human cervical carcinoma), MCF7 (derived from human breast ductal carcinoma) and M15 (derived from developing mouse kidney) cells were grown in DMEM and K562 cells (derived from chronic myelogenous leukemia) were grown in RPMI, supplemented with 10% (vol/vol) FBS at 37°C. Transfection of plasmids was performed using Effectene reagent (Qiagen). Cells were harvested 48 hours after transfection and processed for different assays. The human and mouse MAD2 siRNAs and mouse WT1 siRNA were obtained from Qiagen. Human WT1 siRNA was obtained from Ambion. siRNAs were transfected using Hiperfect reagent (Qiagen) for 48 hours.
Plasmid constructs and protein purification
Full length human MAD2 was cloned from HeLa cDNA library in PRSET-A His-tagged vector. Point and deletion mutants were generated using a site directed mutagenesis kit (Agilent). GST, GST-WT1 (and deletion mutants) were expressed in BL21-DE3 competent cells, and His-tagged human MAD2 (and mutant derivatives) were expressed in BL21-DE3(pLysS) competent cells followed by affinity purification as described before39. Flag-tagged full length WT1 protein (residue 1–449, including exon 5/17AA and KTS) was purchased from Creative Biomart. The MAD2-binding peptide 1 (MBP1) was purchased from Peptide 2.0 (sequence: SWYSYPPPQRAV); Control peptide sequence: CKATKDPSRVGDS. GFP-tagged full length isoforms WT1 (−/−) and WT1 (−/+) were kind gifts from Prof. Brigitte Royer-Pokora. GFP-tagged full length isoform WT1 (+/+) was purchased from GeneCopoeia Inc., and GFP WT1 (+/−) was generated by deleting the KTS region using site directed mutagenesis kit (Agilent).
RNA analysis
Total RNA was prepared using the QIAGEN RNeasy kit and cDNA was prepared using the BioRad iScript cDNA synthesis kit. Real-time PCR was performed using a BioRad MiniOpticon System and BioRAd SYBR Green assay reagents. Primers used in the analysis were: MAD2, Fwd: ACGGTGACATTTCTGCCACT, Rev: TGGTCCCGACTCTTCCCATT; WT1, Fwd: CCGCTATTCGCAATCAGGGT, Rev: ACTTGTTTTACCTGTATGAGTCCTG; AREG, Fwd: TGGATTGGACCTCAATGACA, Rev: ACTGTGGTCCCCAGAAAATG; EREG, Fwd: GCTCTGCCTGGGTTTCCATC, Rev: CCACACGTGGATTGTCTTCTGTC,; CYCLIN E, Fwd: CTCCAGGAAGAGGAAGGCAA, Rev: TCGATTTTGGCCATTTCTTCA; GAPDH, Fwd: ACAGTCAGCCGCATCTTCTT, Rev: ACGACCAAATCCGTTGACTC
Western blotting and immunofluorescence analysis
Western blotting analysis was performed as described before40. In brief, cells were incubated with non-denaturing lysis buffer (20 mMTris HCl pH 8, 150 mM NaCl, 1% (v/v) NP-40, 2 mM EDTA) for 30 minutes and prepared for Western blotting. Whole cell lysates were electrophoresed and transferred to PVDF membrane (Millipore). The membranes were blocked in Phosphate Buffered Saline (PBS), 0.1% (v/v) Tween 20 and 5% (w/v) milk for 1 hour at room temperature followed by incubation with primary antibodies (1:1000 dilution) prepared in PBS, 0.1% (v/v) Tween 20 and 2.5% milk for 12 hours at 4C. Membranes were then washed three times with PBS and incubated for 3 hours with a horseradish peroxidase-conjugated anti-rabbit, anti-mouse or anti-goat IgG secondary antibody (1:5000 dilution, Jackson ImmunoResearch). Membranes were washed three times with PBS and proteins were detected using ECL Western blotting detection kit (Amersham). Uncropped scans of western blots are provided in Supplementary Figures 2–7.
Immunofluorescence analysis was performed as previously described41. Briefly, cells were grown on coverslips and were washed with PBS followed by crosslinking in 4% (v/v) paraformaldehyde (PFA) in PBS for 15 minutes at room temperature. The cells were washed once in PBS and incubated in blocking solution for 30 minutes at room temperature. The cells were then incubated in PBS containing primary antibody (1:100 dilution) for 1 hour and washed twice with PBS followed by incubation in DyLight 488- or DyLight 549-conjugated goat anti-rabbit/anti-mouse IgG secondary antibodies (1:500 dilution, Thermo Scientific) for 1 hour. DNA was stained with Hoechst dye and the cells were mounted using Fluoromount-G (Southern Biotech). The immunofluorescence signal was visualized using Zeiss LSM 710 Confocal Microscope.
The following antibodies were used for Western blotting and immunofluorescence analysis: MAD2 (C-19-sc-6329, FL-205-sc-28261), CDC20 (sc-1907), CYCLIN B1 (sc-245), WT1-F6 (sc-7385), WT1-C-19 obtained from Santa Cruz Biotechnology. MAD2 (A300-301A), MAD1 (A300-339A), BUBR1 (A300-386A), and CDC27 (A301-184A) antibodies were obtained from Bethyl laboratories. SECURIN (ab3305), H3S10p (ab5176), H3 (ab1791), CENPA (ab13939) and β-TUBULIN (ab6046) antibodies were obtained from Abcam.
APC/C-Ubiquitination activity assay
The APC/C-ubiquitination activity assay was performed as described before20. HeLa cells were blocked in prometaphase stage by treatment with 2.5 mM thymidine for 24 hours, followed by treatment with 60 ng/ml nocodazole for 12 hours. Nocodazole-arrested HeLa cells were washed with ice-cold PBS and resuspended in 75% of pellet volume of hypotonic buffer (20 mM Hepes-NaOH, pH 7.6, 5 mM KCl, 1 mM DTT) containing protease inhibitor cocktail (Roche). After repeated freeze-thawing on dry ice, the cell lysates were centrifuged at 13000 rpm for 1 hour. The supernatant was supplemented with glycerol to 10% (v/v) and used in APC/C activity assay. 20-µl reaction mixtures contained 10 µl of concentrated mitotic extract (~ 10 mg/ml), 2 µl of 10X degradation mixture (100 mM Tris-HCl, pH 7.6, 50 mM MgCl2, 10 mM DTT, 10 mg/ml ubiquitin, 100 mM phosphocreatine, 5 mM ATP, 0.1 mg/ml UbcH10), and 1 µl of 20° creatine phosphokinase (1 mg/ml) (Sigma). Reactions were incubated at 30°C, in presence of different recombinant proteins and 3-µl samples were withdrawn at indicated time points in SDS-PAGE sample buffer. Degradation of CYCLIN B and SECURIN was followed by Western blotting.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Keith Brown, Chris Paraskeva and Lindsey Marsh for comments on the manuscript. We thank Alan Siegel and UB North Campus Imaging Facility funded by NSF-MRI Grant DBI 0923133, for the confocal images. This work was funded by the National Institute of General Medical Sciences [1R01GM098609] (to KFM and SGER).
Footnotes
AUTHOR CONTRIBUTIONS
J.S. designed and performed research, analyzed data and prepared the manuscript. E.T. performed research. D.J.R. performed the yeast-two hybrid experiment. K.F.M. provided reagents and edited the manuscript. S.G.E.R. designed research, analyzed data and prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Discenza MT, Pelletier J. Insights into the physiological role of WT1 from studies of genetically modified mice. Physiol. genomics. 2004;16:287–300. doi: 10.1152/physiolgenomics.00164.2003. [DOI] [PubMed] [Google Scholar]
- 2.Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum. Mol. Genet. 2006;15(Spec No 2):R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 3.Roberts SG. Transcriptional regulation by WT1 in development. Curr. Opin. Genet. Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Kreidberg JA. WT1 and kidney progenitor cells. Organogenesis. 2010;6:61–70. doi: 10.4161/org.6.2.11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholz H, Kirschner KM. A role for the Wilms' tumor protein WT1 in organ development. Physiology. 2005;20:54–59. doi: 10.1152/physiol.00048.2004. [DOI] [PubMed] [Google Scholar]
- 6.Chau YY, Hastie ND. The role of Wt1 in regulating mesenchyme in cancer, development, and tissue homeostasis. Trends Genet. 2012;28:515–524. doi: 10.1016/j.tig.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat. Rev. Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicent S, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J. Clin. Invest. 2010;120:3940–3952. doi: 10.1172/JCI44165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiyama H. WT1 (Wilms' tumor gene 1): biology and cancer immunotherapy. Jpn. J. Clin. Oncol. 2010;40:377–387. doi: 10.1093/jjco/hyp194. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia. 2007;21:868–876. doi: 10.1038/sj.leu.2404624. [DOI] [PubMed] [Google Scholar]
- 11.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr. Biol. 2012;22:966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Schuyler SC, Wu YF, Kuan VJ. The Mad1–Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. J. Cell Sci. 2012;125:4197–4206. doi: 10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- 13.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest. 2012;122:1138–1143. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr B, Compton DA. A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front. Oncol. 2013;3:164. doi: 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 19.Skinner JJ, Wood S, Shorter J, Englander SW, Black BE. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. J. Cell Biol. 2008;183:761–768. doi: 10.1083/jcb.200808122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tipton AR. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J. Biol. Chem. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 2011;30:3322–3336. doi: 10.1038/emboj.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han JS. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol. Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol. Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Lau DT, Murray AW. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Curr Biol. 2012;22:180–190. doi: 10.1016/j.cub.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl Acad. Sci. USA. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30:1469–1474. doi: 10.1093/carcin/bgp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo X, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 31.Yang M, et al. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toska E, Roberts SG. Mechanisms of transcriptional regulation by WT1 (Wilms' tumour 1) Biochem J. 2014;461:15–32. doi: 10.1042/BJ20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 34.Nakadate H, et al. Correlation of chromosome abnormalities with presence or absence of WT1 deletions/mutations in Wilms tumor. Genes Chromosomes Cancer. 1999;25:26–32. doi: 10.1002/(sici)1098-2264(199905)25:1<26::aid-gcc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 35.Traub F, et al. Nephroblastomatosis and loss of WT1 expression associated with trisomy 13. Virchows Arch. 2006;448:214–217. doi: 10.1007/s00428-005-0067-3. [DOI] [PubMed] [Google Scholar]
- 36.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci. 2011;124:3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, et al. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orth M, et al. Shugoshin is a Mad1/Cdc20-like interactor of Mad2. EMBO J. 2011;30:2868–2880. doi: 10.1038/emboj.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard DJ, Schumacher V, Royer-Pokora B, Roberts SG. Par4 is a coactivator for a splice isoform-specific transcriptional activation domain in WT1. Genes Dev. 2001;15:328–339. doi: 10.1101/gad.185901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shandilya J, Wang Y, Roberts SG. TFIIB dephosphorylation links transcription inhibition with the p53-dependent DNA damage response. Proc. Natl Acad. Sci. USA. 2012;109:18797–18802. doi: 10.1073/pnas.1207483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toska E, et al. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2012;2:462–469. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.