Abstract
Purpose
Sporadic colorectal cancers with high-frequency microsatellite instability (MSI-H) are related to hypermethylation of mismatch repair (MMR) genes and a higher frequency of BRAF mutations than Lynch syndrome. We estimated the feasibility of hereditary colorectal cancer based on hMLH1 methylation and BRAF mutations.
Methods
Between May 2005 and June 2011, we enrolled all 33 analyzed patients with MSI-H cancer (male:female, 23:10; mean age, 65.5 ± 9.4 years) from a prospectively maintained database that didn't match Bethesda guidelines and who had results of hMLH1 methylation and BRAF mutations.
Results
Among the 33 patients, hMLH1 promoter methylation was observed in 36.4% (n = 12), and was not significantly related with clinicopathologic variables, including MLH1 expression. BRAF mutations were observed in 33.3% of the patients (n = 11). Four of 11 and five of 22 patients with MSI-H colon cancers were BRAF mutation (+)/hMLH1 promoter methylation (-) or BRAF mutation (-)/hMLH1 promoter methylation (+). Of the 33 patients, 21.2% were BRAF mutation (+)/hMLH1 promoter methylation (+), indicating sporadic cancers. Seventeen patients (51.5%) were BRAF mutation (-)/hMLH1 promoter methylation (-), and suggested Lynch syndrome.
Conclusion
Patients with MSI-H colorectal cancers not fulfilling the Bethesda guidelines possibly have hereditary colorectal cancers. Adding tests of hMLH1 promoter methylation and BRAF mutations can be useful to distinguish them from sporadic colorectal cancers.
Keywords: Colorectal neoplasms, hMLH1, BRAF, Hereditary colorectal cancer
INTRODUCTION
Microsatellite instability (MSI) is one of the main colorectal carcinogenic mechanisms. MSI is associated with germ line mutations of mismatch repair (MMR) genes in patients with Lynch syndrome and promoter hypermethylation of MMR in patients with sporadic colorectal cancer [1]. These high-frequency MSI (MSI-H) tumors share the same distinctive clinicopathologic characteristics and have different molecular profiles. Therefore, treatment and surveillance approaches need to be specific. Lynch syndrome occurs in approximately 5% of patients with colorectal cancer [2]. Lynch syndrome has the following characteristics: 80% lifetime risk of colorectal cancer; early-onset; family history of cancers; and multiple tumors and multiorgan involvement, including the endometrium, stomach, small intestine, hepatobiliary and genitourinary tracts, and ovary [3]. Mutations in the hMLH1 or hMSH2 genes are the most common defects in these families, and comprise approximately 94% of germ line mutations in equal proportions [4,5].
The revised Bethesda guidelines (BGs) [6] are based on clinicopathologic features, rather than molecular changes, as a screening tool to select patients who need to undergo MSI analysis for detecting Lynch syndrome. Current laboratory algorithms detecting Lynch syndrome include MSI testing, immunochemistry (IHC) of MMR proteins, and germ line testing for mutations in MMR genes [7]. The sensitivity and specificity of BGs have been reported to be 94% and 25%, respectively [8]. As a result, a number of published studies [7,9] have approached colorectal carcinomas in terms of molecular biology and an effective strategy to detect Lynch syndrome.
BRAF encodes a cytoplasmic serine/threonine kinase, which is an essential component of the mitogen-activated protein kinase-signaling pathway [10]. Mutations in BRAF occur in 15% of colorectal cancers, are frequently present in sporadic colorectal cancers with methylated hMLH1 promoters, and are rare in the Lynch syndrome [10,11]. This discrepancy may be useful to distinguish MSI-H sporadic colorectal cancers from Lynch syndrome.
In the current study, we estimated the feasibility of hereditary colorectal cancer based on hMLH1 promoter methylation and BRAF mutations in MSI-H colorectal cancers not fulfilling revised BGs.
METHODS
Patients
Between May 2005 and June 2011, 1,867 patients who had available clinicopathologic data, as well as MSI status, were selected from a prospectively maintained database, analyzed, and categorized based on the revised BGs [6]. We asked about their family and operation histories through individual interviews. BGs (-) means patients do not have any BGs components (n = 958, 51.3%), and 47 patients (4.9%) had MSI-H. Among the 47 patients with BGs (-) and MSI-H colorectal cancers, 33 who had available tissue blocks were analyzed with respect to hMLH1 promoter methylation and BRAF mutations. We excluded families with polyposis syndrome (familial adenomatous polyposis, Peutz-Jeghers syndrome, juvenile polyposis syndrome, and Cowden disease), hereditary nonpolyposis colorectal syndrome fulfilling the Amsterdam criteria, and inflammatory bowel disease-related cancer. The mean age was 65.5 ± 9.4 years (range, 51-84 years).
MSI-H tumors are defined as >2 mutations of the 5 microsatellite sequences in the tumor DNA [12,13]. Tumors are classified as right-sided (proximal of the splenic flexure) and left-sided (distal of the splenic flexure and rectum). This study was approved by the Institutional Review Board of Samsung Medical Center.
MSI analysis
DNA was extracted from formalin-fixed, paraffin-embedded tissues of tumor mucosa and corresponding normal mucosa by a standard procedure [14]. Areas containing >50% tumor cells were selected by microscopic evaluation on a reference H&E-stained slide. Slides (50 µm thick) were made, and if necessary, tumor cells were prepared using a scalpel. MSI status was determined by polymerase chain reaction (PCR) analysis using a DNA autosequencer (Applied Biosystems 373A sequencer, Applied Biosystems, Foster City, CA, USA). We used five microsatellite markers (BAT 25, BAT 26, D5S346, D17S250, D2S123), as recommended by the National Cancer Institute (NCI; Bethesda, MD, USA) [12].
hMLH1 promoter methylation using methylation-specific PCR
By comparing the signals from tumor-derived tissues with signals from normal tissues, we determined hMLH1 promoter methylation. Methylationspecific PCR (MS-PCR) was used to distinguish unmethylated from methylated DNA based on sequence alterations produced by bisulfate treatment of DNA, which converted unmethylated cytosine to uracil (EZ DNA methylation kit, Zymo Research, Burlington, ON, Canada). These changes were identified by subsequent PCR using primers specific to the methylated (unchanged) or unmethylated (changed) DNA. The forward primer for the methylated hMLH1 promoter was 5'-GATAGCGATTTTTAACGC. The reverse primer was 5'-TCTATAAATTACTAAATCTCTTCG. In unmethylated hMLH1 promoter, the forward primer was 5'-AGAGTGGATAGTGATTTTTAATGT, and the reverse primer was 5'-ACTCTATAA ATTACTAAATCTCTTCA.
PCR reactions were performed using the primer pairs described below in the following reaction mix: 10× PCR buffer; deoxynucleotide triphosphates (each at 2.5 mM/L); primers (10 µM/L each per reaction); 0.5 unit of AmpliTaq Gold DNA polymerase (Applied Systems); and 50 ng of bisulfate-modified DNA (from paraffin-embedded tissue) in a final volume of 10 µL. PCR cycling conditions were as follows: 30 seconds at 94℃; 30 seconds at 94℃; 30 seconds at 54℃; 30 seconds at 72℃; and 10 minutes at 72℃ for 40 cycles. The presence of a band in the unmethylated tumor and matched normal tissue with the absence of a methylation band in the tumor was defined as unmethylated. However, when a methylated band was present for tumor and absent for the normal tissue, we defined the sample as methylated.
We used control methylation DNA (Millpore CpGenome universal methylated DNA, Millipore Co., Billerica, MA, USA) and control unmethylated DNA (Millipore CpGenome universal unmethylated DNA set, Millipore Co.). All resections were in duplicate.
BRAF mutation analysis
The fragment encompassing exon 15 was amplified by PCR in 33 paraffin-embedded carcinoma samples. Primer sequences and PCR conditions were based on those reported previously [11]. The BRAF p.Val600Glu primer of exon 15 was 10 and 1 pmole (forward and reverse, respectively). PCR was performed with 10 pmole of Mut R. Genomic DNA (100 ng) was amplified by PCR using the following cycling conditions: 30 seconds at 94℃; 30 seconds at 54℃; and 30 seconds at 72℃ for 40 cycles. Genomic DNA was placed in a 100v PAGE-gel running for 40-60 minutes. We used DLD-1, which is a colon cancer cell line, as a negative control. Colorectal cancer cell line (RKO) was a positive control in BRAF-controlled DNA. All the resections were in duplicate.
Immunohistochemistry
DNA MMR protein expression (MLH1, MSH2, and MSH6) was evaluated by IHC. IHC was performed on paraffin sections of normal and tumor tissues (4 µm thick) using mouse monoclonal antibodies specific for each MMR protein as follows: MLH1 (clone G168-15, 1:200; BD Pharmingen, San Diego, CA, USA), MSH2 (clone FE11, 1:400; Calbiochem, La Jolla, CA, USA), and MSH6 (clone 44, 1:400; BD Transduction Laboratories, San Diego, CA, USA). MMR protein expression was described as negative for absent or <10% nuclear staining, and positive for ≥10% nuclear staining. Normal colonic epithelium adjacent to the tumor and lymphocytes served as positive controls.
Statistical analysis
We used Fisher exact test to compare MSI status and BGs with clinicopathologic factors. All P-values were two-tailed and P-values <0.05 were considered statistically significant. All statistical analyses were carried out using SPSS ver. 17.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Among 1,867 patients, MSI-H existed in 141 (7.5%). Nine hundred and nine patients (48.7%) were BGs (+). Of the 909 patients, 94 (10.3%) had MSI-H tumors. Nine hundred and fifty-eight patients (51.3%) were BGs (-), 47 (4.9%) of whom had MSI-H.
hMLH1 promoter methylation and protein expression of MLH1, MSH2, and MSH6
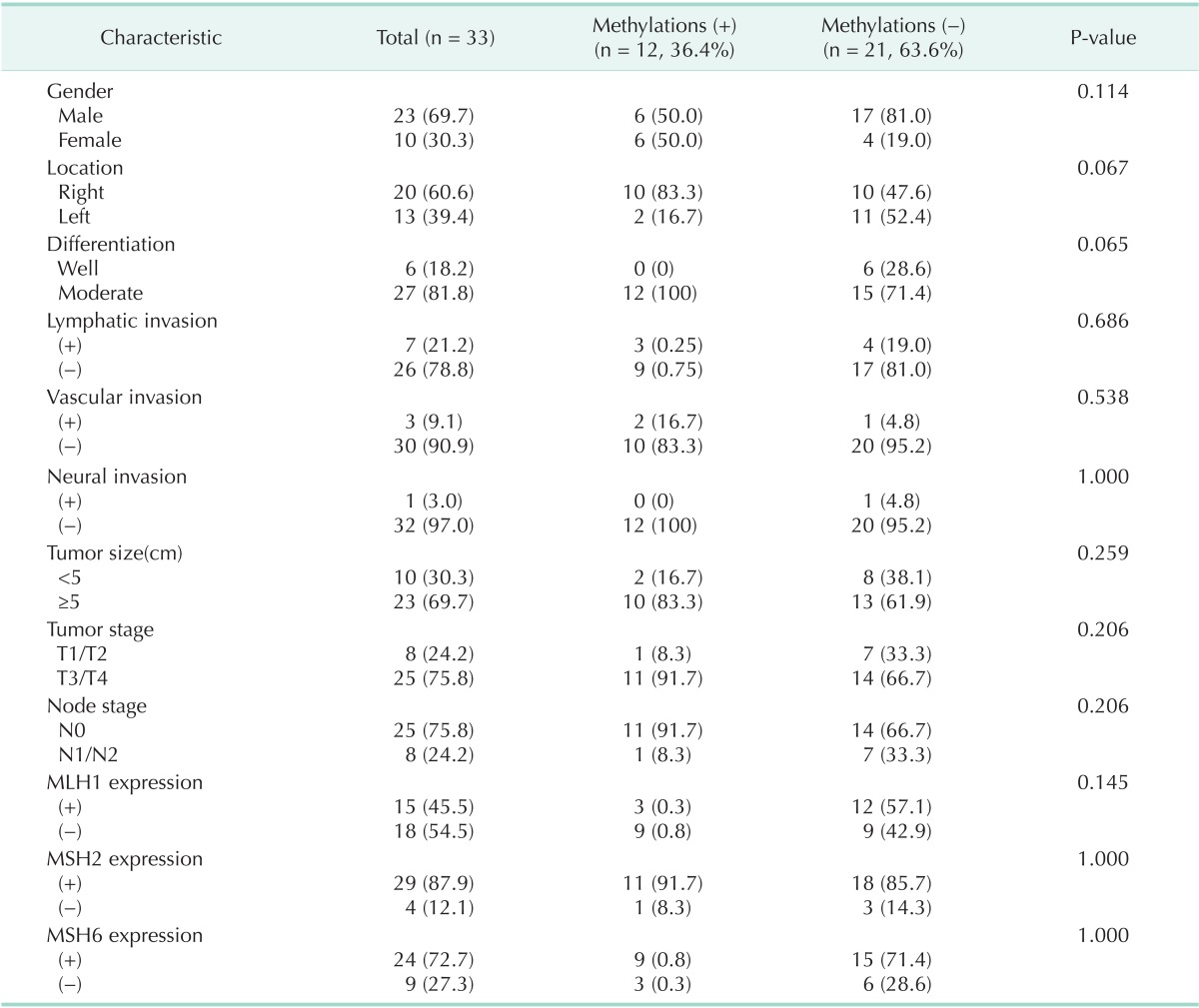
Among 33 patients, hMLH1 promoter methylation existed in 36.4% (n = 12). Of the 12 patients, 3 had MLH1 expression and 9 did not express MLH1. Eleven patients expressed MSH2 and 9 patients expressed MSH6. Among 21 patients (63.7%) without hMLH1 promoter methylation, 12 (36.4%) had expression of MLH1 and the remaining patients did not express MLH1. Eighteen patients expressed MSH2 and 15 patients expressed MSH6 (Table 1).
Table 1.
Clinicopathologic characteristics according to hMLH1 methylations
Values are presented as number (%).
There was no statistical significance between hMLH1 promoter methylation and clinicopathologic variables (Table 1). Fifteen patients (45.5%) had MLH1 expression, but did not demonstrate a relationship between clinicopathologic features and MLH1 protein expression. Even if there was no significant difference, hMLH1 promoter methylation was more frequent with respect to loss of MLH1 (Table 1).
BRAF mutations and hMLH1 promoter methylation
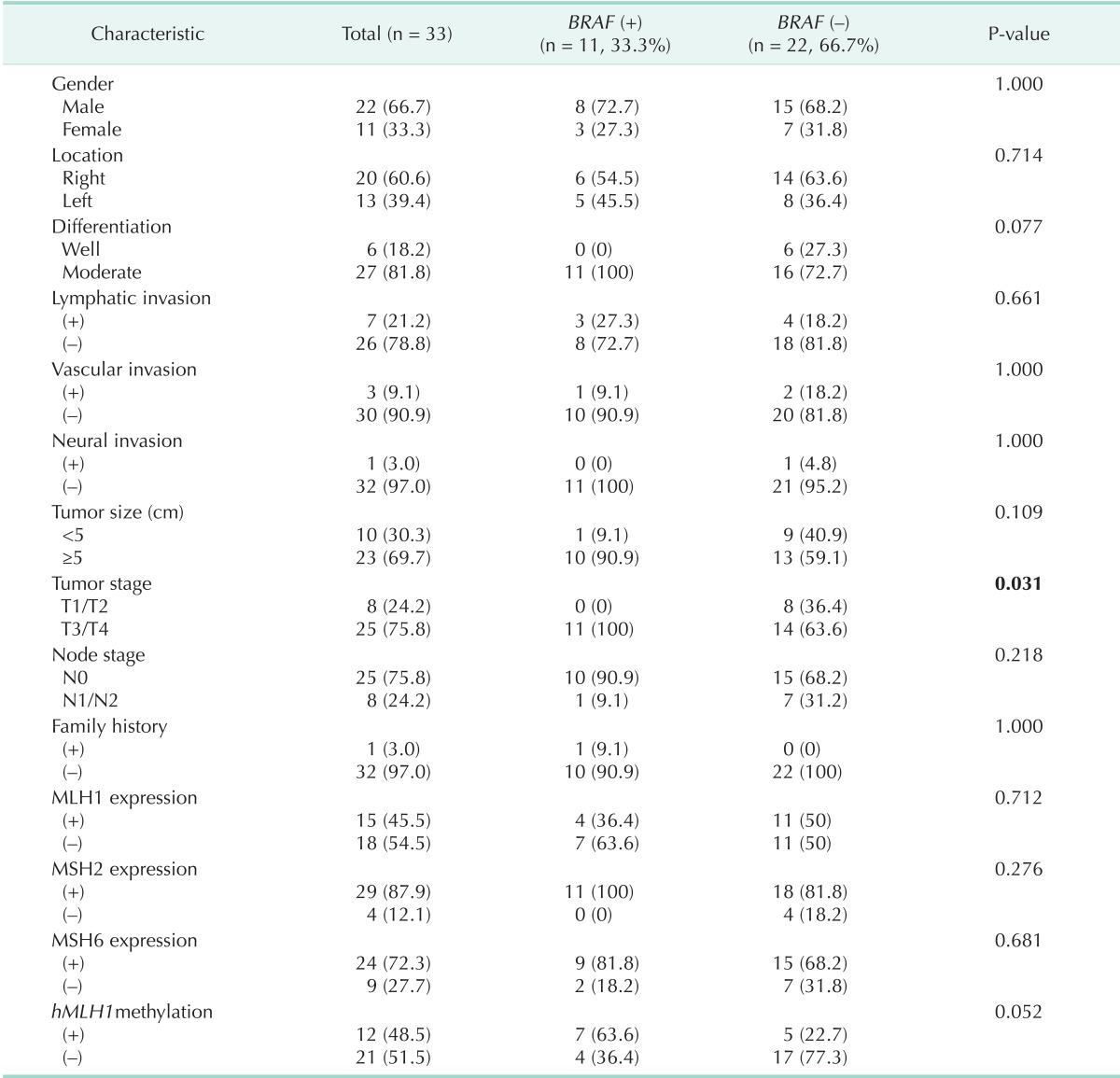
We identified 11 patients who had BRAF mutations (33.3%) and 22 patients (66.7%) who had no BRAF mutations. BRAF mutations were more frequent in T3/T4 cancers than T1/T2 (P = 0.031) (Table 2).
Table 2.
Clinicopathologic characteristics according to BRAF mutations
Values are presented as number (%).
Of the 11 patients with BRAF mutations, 7 (21.2%) had hMLH1 promoter methylation. In the 22 patients without BRAF mutation, 5 (15.2%) had hMLH1 promoter methylation and 17 (51.5%) had no hMLH1 promoter methylation. The incidence of BRAF mutations was higher in patients with methylated hMLH1 promoters, but there was not a significance difference between BRAF mutations and hMLH1 promoter methylation (P = 0.052) (Table 2).
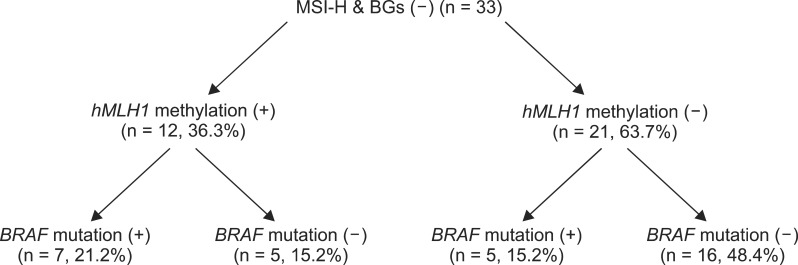
Seven patients (21.2%) had hMLH1 promoter methylation (+)/BRAF mutation (+), which was indicative of sporadic colorectal cancer. Five patients (15.2%) had MSI colon cancers with hMLH1 promoter methylation (+)/BRAF mutation (-) or hMLH1 promoter methylation (-)/BRAF mutation (+). The remaining 16 patients (48.4%) had hMLH1 promoter methylation (-)/BRAF mutation (-), which was suggestive of Lynch syndrome (Fig. 1).
Fig. 1.
Patient distribution according to hMLH1 promoter methylation and BRAF mutations. MSI-H, high frequency microsatellite instability; BGs, Bethesda guidelines.
Table 3 shows the profile of BRAF mutations, hMLH1 promoter methylation, and MMR expression.
Table 3.
BRAF mutation, hMLH1 promoter methylation, and MMR expression profile
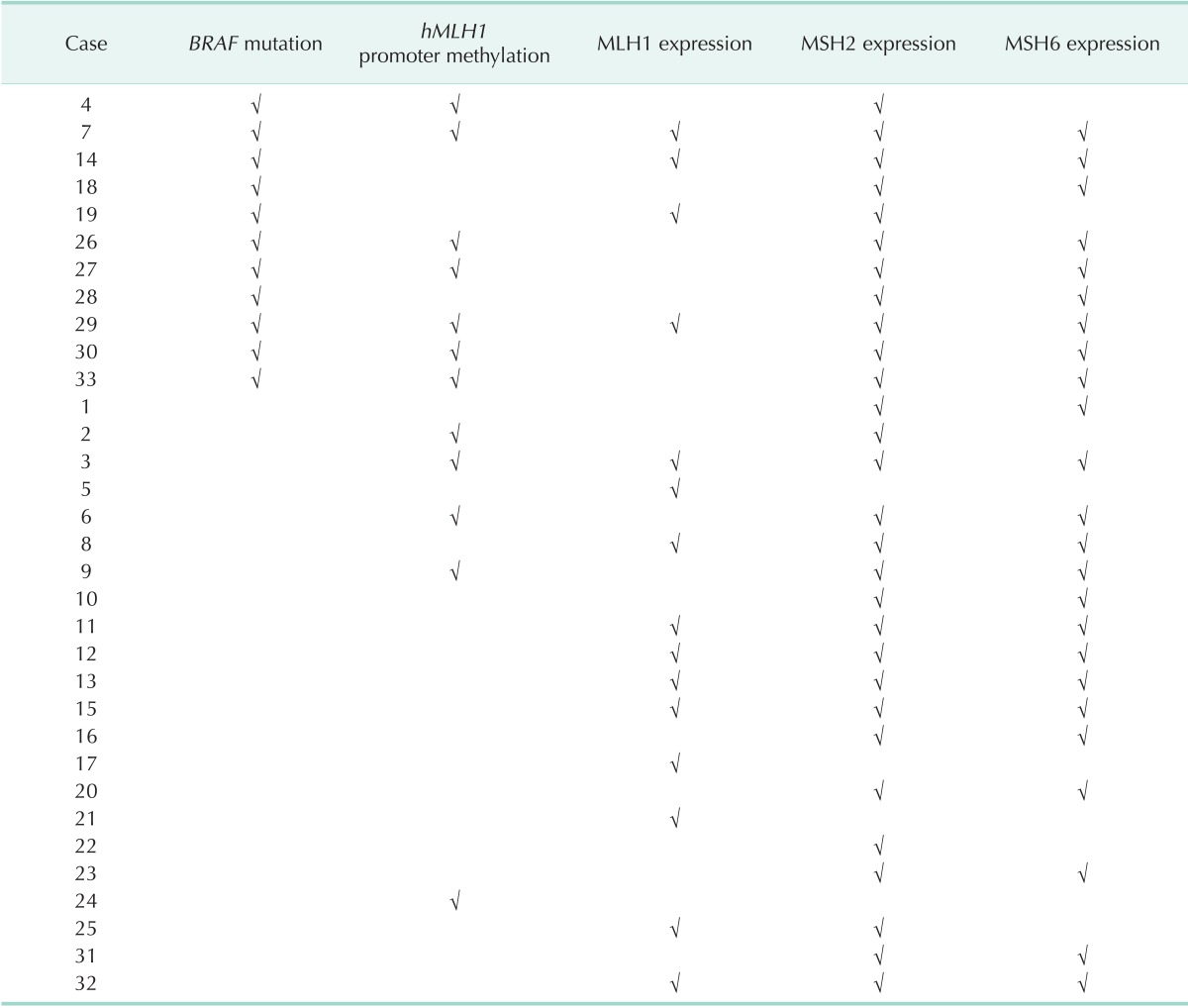
Check mark (√) indicate presence of BRAF mutation, hMLH1 promoter methylation, and MMR expression.
DISCUSSION
Detection of patients or families with hereditary colorectal cancer is requisite of treatment, surveillance provision, and adequate counseling of family members. The Amsterdam criteria are widely used to identify putative patients with Lynch syndrome, but these criteria are strictly defined and do not take into account most suspected hereditary colorectal cancers without a strong family history [15]. The 2002 revision of the BGs was developed to select patients for molecular analysis of MSI [15]. If the results of MSI suggest Lynch syndrome, it is recommended to carry out germ line testing of MMR genes [7]. The majority of the patients who do not fulfill the BGs cannot be considered to have Lynch syndrome, but rather sporadic colon cancer. However, there are several reports showing that the Lynch syndrome is related to colorectal cancer and can be diagnosed at 60 years of age, suggesting the BGs are not entirety adequate [16,17].
In the present study, about one-half of patients did not have any components of the BGs; approximately 5% of the patients had MSI colorectal cancers. One-third of MSI colorectal cancer patients did not have any components of the BGs. With respect to Korean patients with colon cancer, if only patients who satisfied the BGs were genetically tested, one-third of MSI colon cancers would be missed, which had the possibility of hereditary colon cancers. MSI colorectal cancer has been reported in approximately 95% of patients with Lynch syndrome-related colon cancers and 10%-20% of patients with sporadic colon cancer [17,18]. The proportion of MSI tumors was less compared to other series [19,20,21]. This discrepancy may be attributed, in part, to ethnic differences [22].
The detection of germ line mutations in MMR is an important supplement to clinical criteria and crucial for the definitive diagnosis of Lynch syndrome, especially for patients with an uncertain family history and small family numbers [17,23]. However, germ line tests have several obstacles to overcome, including high cost and time-consumption in performing the tests [10].
Many researchers [10,11,24] have reported a high frequency of BRAF mutations in patients with sporadic colorectal cancer with MSI and a methylated hMLH1 promoter, and the lack of BRAF mutations in patients with Lynch syndrome is useful in detecting families with Lynch syndrome. BRAF mutations have rarely been found in colorectal cancers and cell lines of Lynch syndrome related with mutations of hMLH1 [7,10]. However, BRAF mutations are closely correlated to hMLH1 promoter methylation in patients with sporadic colon cancer [10,25,26]. MSI colon cancers with hMLH1 promoter methylation, as well as BRAF mutations, can be regarded as sporadic colon cancers and MSI tumors without hMLH1 promoter methylation or BRAF mutations can be regarded as hereditary colon cancer. MSI colon cancers with hMLH1 promoter methylation (+)/BRAF mutation (-) or hMLH1 promoter methylation (-)/BRAF mutation (+) are difficult to distinguish from hereditary colon cancer and sporadic MSI tumors. However, because hMLH1 promoter methylation is frequently reported in Lynch syndrome-related colorectal cancer, the subset of tumors with hMLH1 promoter methylation (+)/BRAF mutation (-) might be Lynch syndrome-related colorectal cancer. All these things require the analysis of germ line mutations to confirm Lynch syndrome. In the present study, BRAF mutations were found in 33.3% of MSI colon cancers that did not meet the BGs. Among 33 patients, 48.5% (16/33; hMLH1 promoter methylation (-)/BRAF mutation (-)) to 63.6% (21/33; hMLH1 promoter methylation (-)/BRAF mutation (-) and hMLH1 promoter methylation (+)/BRAF mutation (-)) might be a Lynch syndrome-related colon cancer among MSI tumors that do not satisfy the BGs criteria.
The combination of three molecular tests (MSI, BRAF mutations, and hMLH1 promoter methylation) must be validated against analysis of MMR germ line mutations. It was a limitation of the current study that mutational analysis was not available.
In conclusion, even though the patients do not fulfill the BGs criteria, there may be a high likelihood that colon cancer results from germ line mutations of MMR. Thus, MSI testing has been suggested as necessary for late-onset colon cancer patients or patients not satisfying BGs. Adding analysis of BRAF mutations and hMLH1 promoter methylation to MSI could be an easy and efficient way to have the information of whether or not the patients have Lynch syndrome without full sequencing of MMR genes.
ACKNOWLEDGEMENTS
This study was supported by Samsung Biomedical Research Institute (CB11093).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 2.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 3.Vasen HF, Wijnen JT, Menko FH, Kleibeuker JH, Taal BG, Griffioen G, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 5.Peltomaki P, Vasen HF The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 6.Lipton LR, Johnson V, Cummings C, Fisher S, Risby P, Eftekhar Sadat AT, et al. Refining the Amsterdam Criteria and Bethesda Guidelines: testing algorithms for the prediction of mismatch repair mutation status in the familial cancer clinic. J Clin Oncol. 2004;22:4934–4943. doi: 10.1200/JCO.2004.11.084. [DOI] [PubMed] [Google Scholar]
- 7.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, et al. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 8.Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 10.Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, et al. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10(1 Pt 1):191–195. doi: 10.1158/1078-0432.ccr-1118-3. [DOI] [PubMed] [Google Scholar]
- 11.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 12.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 13.Odenthal M, Barta N, Lohfink D, Drebber U, Schulze F, Dienes HP, et al. Analysis of microsatellite instability in colorectal carcinoma by microfluidic-based chip electrophoresis. J Clin Pathol. 2009;62:850–852. doi: 10.1136/jcp.2008.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wijnen JT, Vasen HF, Khan PM, Zwinderman AH, van der Klift H, Mulder A, et al. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med. 1998;339:511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 15.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urso E, Pucciarelli S, Agostini M, Maretto I, Mescoli C, Bertorelle R, et al. Proximal colon cancer in patients aged 51-60 years of age should be tested for microsatellites instability. A comment on the Revised Bethesda Guidelines. Int J Colorectal Dis. 2008;23:801–806. doi: 10.1007/s00384-008-0484-2. [DOI] [PubMed] [Google Scholar]
- 17.Berginc G, Bracko M, Ravnik-Glavac M, Glavac D. Screening for germline mutations of MLH1, MSH2, MSH6 and PMS2 genes in Slovenian colorectal cancer patients: implications for a population specific detection strategy of Lynch syndrome. Fam Cancer. 2009;8:421–429. doi: 10.1007/s10689-009-9258-4. [DOI] [PubMed] [Google Scholar]
- 18.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 19.Lawes DA, Pearson T, Sengupta S, Boulos PB. The role of MLH1, MSH2 and MSH6 in the development of multiple colorectal cancers. Br J Cancer. 2005;93:472–477. doi: 10.1038/sj.bjc.6602708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatch SB, Lightfoot HM, Jr, Garwacki CP, Moore DT, Calvo BF, Woosley JT, et al. Microsatellite instability testing in colorectal carcinoma: choice of markers affects sensitivity of detection of mismatch repair-deficient tumors. Clin Cancer Res. 2005;11:2180–2187. doi: 10.1158/1078-0432.CCR-04-0234. [DOI] [PubMed] [Google Scholar]
- 21.Rigau V, Sebbagh N, Olschwang S, Paraf F, Mourra N, Parc Y, et al. Microsatellite instability in colorectal carcinoma. The comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 [correction of hMLH6] immunostaining. Arch Pathol Lab Med. 2003;127:694–700. doi: 10.5858/2003-127-694-MIICC. [DOI] [PubMed] [Google Scholar]
- 22.Chan AO, Soliman AS, Zhang Q, Rashid A, Bedeir A, Houlihan PS, et al. Differing DNA methylation patterns and gene mutation frequencies in colorectal carcinomas from Middle Eastern countries. Clin Cancer Res. 2005;11:8281–8287. doi: 10.1158/1078-0432.CCR-05-1000. [DOI] [PubMed] [Google Scholar]
- 23.Mead LJ, Jenkins MA, Young J, Royce SG, Smith L, St John DJ, et al. Microsatellite instability markers for identifying early-onset colorectal cancers caused by germ-line mutations in DNA mismatch repair genes. Clin Cancer Res. 2007;13:2865–2869. doi: 10.1158/1078-0432.CCR-06-2174. [DOI] [PubMed] [Google Scholar]
- 24.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 25.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, et al. BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGivern A, Wynter CV, Whitehall VL, Kambara T, Spring KJ, Walsh MD, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Fam Cancer. 2004;3:101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]