Abstract
Identifying the peptidases that inactivate bioactive peptides (e.g. peptide hormones and neuropeptides) in mammals is an important unmet challenge. This protocol describes a recent approach that combines liquid chromatography-mass spectrometry peptidomics to identify endogenous cleavage sites of a bioactive peptide, the subsequent biochemical purification of a candidate peptidase based on these cleavage sites, and validation of the candidate peptidase’s role in the physiological regulation of the bioactive peptide by examining a peptidase knockout mouse. We highlight successful application of this protocol to discover that insulin-degrading enzyme (IDE) regulates physiological calcitonin gene-related peptide (CGRP) levels and detail the key stages and steps in this approach. This protocol requires 7 days of work; however, the total time for this protocol is highly variable because of its dependence on the availability of biological reagents, namely purified enzymes and knockout mice. The protocol is valuable because it expedites the characterization of mammalian peptidases, such as IDE, which in certain instances can be used to develop novel therapeutics.
Introduction
Human bioactive peptides, such as insulin1 and glucagon2,3, have a rich scientific history and represent a class of important medicines4. Prior to the advent of molecular biology techniques5,6, such molecules were isolated and identified by biochemical purification guided by activity assays7–9. Testing for compounds that reduced glucose levels in dogs, for example, led to the isolation of insulin1. While biochemical purification is a powerful strategy for identifying bioactive peptides, this approach reveals nothing about the regulation, biosynthesis, catabolism and/or post-translational regulation of these molecules10–12. While insulin can be used acutely to treat diabetes, through injections when blood glucose levels are elevated, the levels of other bioactive peptides must be perturbed for much longer periods to achieve therapeutic value13,14. Unfortunately, the instability of peptides in vivo, where they are susceptible to inactivation by proteolytic cleavage15, makes this a challenging task.
Targeting the biochemical pathways that regulate the production and degradation of bioactive peptides can also control endogenous peptide levels. Most bioactive peptides are synthesized as longer preproproteins and then undergo proteolysis by a class of proteases called prohormone convertases (PCs) to produce the mature bioactive peptides 16,17. Disruption of the proteases in these pathways leads to deficiencies in peptide production, but these pathways cannot be used to regulate the levels of individual peptides because PCs are responsible for the production of multiple bioactive peptides. The deletion of prohormone convertase 2 (PC2) in mice, for example, results in higher proinsulin levels and lower insulin levels but these animals do not show a diabetic phenotype because PC2 deletion also results in the loss of glucagon11,18,19.
Peptidases have also been found to regulate the activity of some bioactive peptides after their release from cells, and targeting these enzymes has enabled control over the levels of specific bioactive peptides. A classic example is the proteolysis of angiotensin I to produce angiotensin II, a potent vasoconstrictor14, by the enzyme angiotensin-converting enzyme (ACE). Small-molecule ACE inhibitors have been used in the clinic for many years and lower blood pressure by reducing angiotensin II levels. More recently, small-molecule inhibitors of renin, the protease responsible for the production of angiotensin I, have been developed and these compounds have successfully been used to reduce blood pressure because they also lead to lower angiotensin II levels20.
This strategy has also found success in the treatment of metabolic disease. Glucagon-like peptide 1 (GLP-1), an intestinal peptide produced from the glucagon gene, promotes glucose stimulated-insulin secretion (GSIS) from pancreatic islets21. Administration of GLP-1 in mammals results in elevated insulin levels and reduced blood glucose levels22, suggesting that this peptide may be a valuable therapeutic. Unfortunately, GLP-1 has a short half-life due to proteolysis12,23, which prevents it from being an effective drug. Consequently, the focus shifted towards the identification of the enzyme(s) responsible for GLP-1 proteolysis, which revealed that dipeptidyl peptidase 4 (DPP4) is responsible for GLP-1 inactivation24. This insight resulted in the development of small-molecule DPP4 inhibitors as anti-diabetic drugs. The ACE and DPP4 examples highlight the value of identifying peptidases that regulate bioactive peptides in characterizing the role of these enzymes in vivo14 and providing new opportunities in therapeutic development25,26.
Moreover, these examples suggest that the partial proteolysis of bioactive peptides is a common form of biochemical regulation in mammals. Though there are many known bioactive peptides, we know relatively little about the physiological proteolysis of these molecules. This dearth of knowledge is primarily due to the lack of an effective protocol for elucidating the enzymes that regulate bioactive peptides. The typical approach tries to connect peptidases to particular bioactive peptides based on their substrate specificity. For example, the neuropeptide vasopressin is a terrific substrate for the enzyme prolyl endopeptidase in vitro27, suggesting that vasopressin may be regulated by prolyl endopeptidase. However, inhibition of prolyl peptidase does not change vasopressin levels in the nervous system. Because this approach does not rely on any in vivo information about the regulation of the bioactive peptide it is prone to making mistakes. Since the effort it takes to validate the regulation of a bioactive peptide through selective inhibitors and/or knockout mice is so great, a high failure rate is unacceptable. Thus, this vasopressin example and others highlights the need for a more effective protocol for the discovery of the peptidases that regulate bioactive peptides.
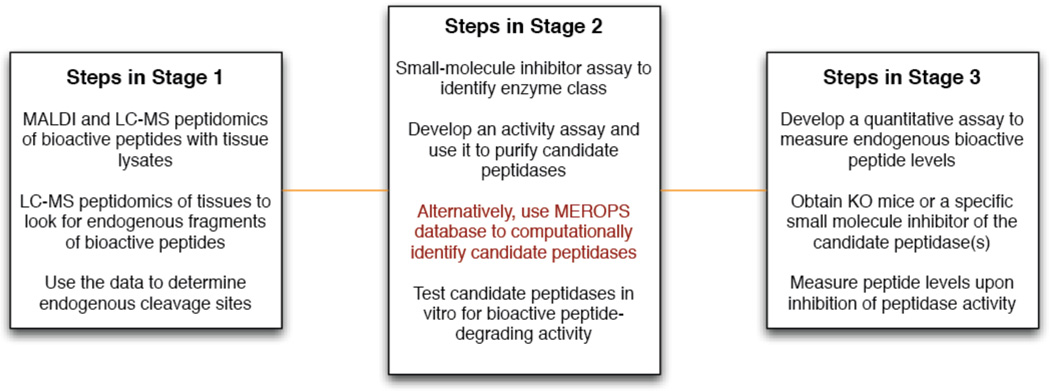
There are three key stages in our protocol for identifying peptidases that cleave and inactivate bioactive peptides (Fig. 1). The first is the use of mass spectrometry peptidomics to identify endogenous fragments of bioactive peptides to reveal the natural cleavage sites (i.e. the proteolytic pathway). The second stage is the development of a peptidase-isolation assay using tissue lysates to isolate the candidate peptidase(s) that are able to produce these fragments. The final step is the physiological validation of proteolysis to ensure that the pathway and enzyme actually regulate the endogenous peptide. This is accomplished by inhibition of the enzyme and quantitative measurements of the endogenous peptide levels. Importantly, this approach overcomes the challenges with current methods because it utilizes knowledge of endogenous peptide fragments to drive discovery, which removes any doubt about the relevance of particular cleavage sites.
Figure 1.
The protocol is divided into three key stages with a number of individual steps making up each stage. In stage 1, LC-MS peptidomics of tissues is used to identify fragments of a bioactive peptide, which reveal the endogenous cleavage sites of the bioactive peptide. This information is used in stage 2 to identify candidate peptidases responsible for the cleavage of the bioactive peptide by experimental and computational methods. Finally, in stage 3 the peptidase or protease is validated in vivo through quantitative measurements of the peptide as a function of the peptidase activity using knockout mice or by treating the knockout mice with small-molecule inhibitors.
Experimental Design
In developing this protocol we wanted to answer the following two questions concerning bioactive peptide proteolysis: 1) Can we identify the endogenous cleavage sites for a given bioactive peptide? 2) Can we use this information to quickly identify and validate the peptidase(s) responsible for endogenous bioactive peptide proteolysis? We highlight our work with two bioactive peptides, peptide histidine isoleucine (PHI)28–30 and calcitonin gene-related peptide (CGRP)31–38. These peptides were selected due to their interesting biological activities and lack of knowledge about their proteolytic regulation. In theory, the peptidase that cleaves any bioactive peptide can be identified using this protocol.
In stage 1 of this protocol (Fig. 1), we use LC-MS peptidomics to identify all the fragments of a given bioactive peptide, such as PHI or CGRP, in tissues where they are found. In the case of CGRP, for example, we isolated mouse spinal cord peptides smaller than 10 kilodaltons using molecular weight cutoff (MWCO) filters and analyzed these peptides by mass spectrometry using a standard setup for shotgun liquid chromatography-mass spectrometry (LC-MS) proteomics39. Peptidomics experiments are distinguished from proteomics experiments in that they include this MWCO step and in this case also lack a trypsin digest step, since we are trying to identify cleavage sites. Since the detection of low abundant peptides can be stochastic, different peptide fragments can occasionally be identified in different runs. To reduce the possibility that a peptide is missed, we recommend performing at least three replicates (biological or technical replicates) and using the data from all three experiments.
After LC-MS, the data is analyzed with SEQUEST40, a tandem mass spectrometry peptide identification program. With SEQUEST, peptides that are detected in a sample are grouped by the gene or protein from which they derive, so any CGRP fragments could easily be identified41. While we have not used other peptide identification programs such as MASCOT42 or MaxQuant43, these programs provide similar information to SEQUEST, and can be substituted. We recommend using stringent scoring criteria for the spectra and visually inspecting spectra to improve confidence in the fragment identification. Since all subsequent steps depend on this dataset, this additional validation step is worth the modest time it adds to the analysis.
With CGRP, there have been several attempts to identify its cleavage sites using immunoassays41. In these experiments, tissue samples were fractionated using gel chromatography (Sephadex G50) and individual fractions were tested for CGRP immunoreactivity using an antibody specific to the C-terminal epitope of CGRP. This analysis identified two predominant fragments, CGRP18–37 and CGRP19–37, and the cleavage site was predicted to be between amino acids 18–19 because CGRP19–37 is more abundant. By contrast, peptidomics revealed ten CGRP fragments including novel fragments as well the previously identified fragments, which provided further confidence in the dataset. Most importantly, the unbiased nature of the method allowed identification of two N-terminal peptides, CGRP1–17 and CGRP1–26, which revealed that CGRP actually contains two cleavage sites: between amino acids 17–18 and 26–27. These peptides could not be detected using the published immunoassay because of the c-terminal specificity of the antibody, which demonstrates the advantages of peptidomics over immunoassays for the identification of endogenous cleavage sites.
CGRP is a widely used marker for staining different parts of the spinal cord and is known to be quite abundant44. Consequently, its fragments are likely also abundant, making them easier to detect. However, if the peptide under investigation is of low abundance, then fragments of this peptide may not be detected by shotgun LC-MS, which is generally biased towards more abundant peptides45–48. To overcome this issue, different types of LC-MS experiments that target peptide fragments in vivo can be performed to improve the sensitivity of the detection. However, to carry out these experiments, the structures of candidate peptide fragments must be known ahead of time.
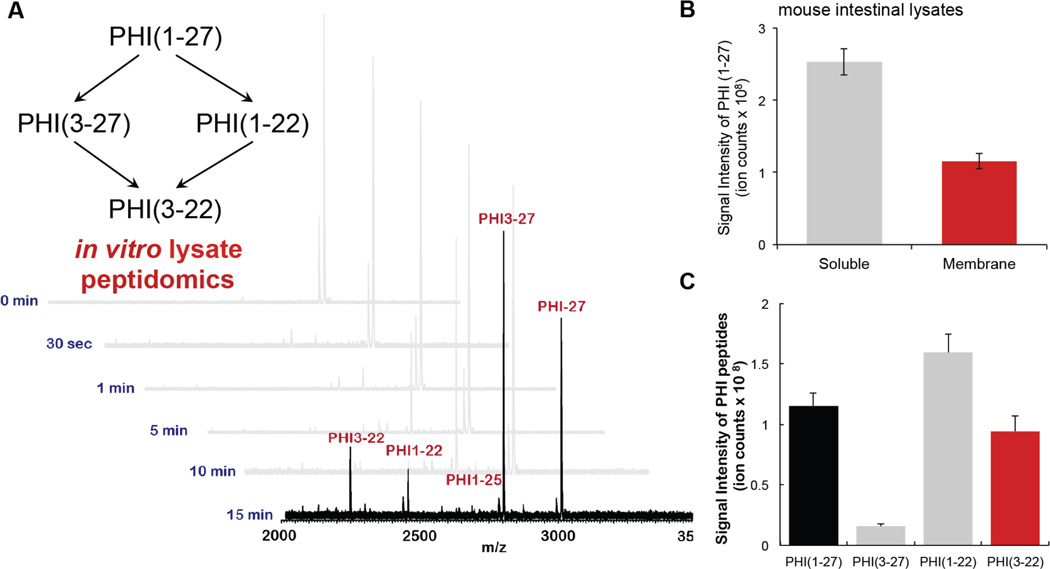
Therefore, we also recommend that in vitro assays on lysates to identify candidate peptides, which can be targeted by LC-MS for more sensitive detection, are performed. These in vitro assays are also useful in that they provide more confidence in the endogenous results (Fig. 1). For example, in our investigation of PHI proteolysis, we incubated synthetic PHI with tissue lysate and identified the fragments generated by matrix-assisted laser desorption/ionization (MALDI) (Fig. 2)49. Since the concentration of the synthetic peptide added in this experiment is high (10–100 µM) relative to endogenous peptides (lysate is added at 1mg/mL), only the synthetic peptide will be detected by MALDI, allowing the use of unlabeled peptide for this experiment (Fig. 2A). After incubating the peptide for 15–60 min with the lysate, a few microliters of the mixture is desalted using a ZipTip and then analyzed by MALDI to identify candidate peptide fragments to look for in tissue lysate experiments. In the case of PHI, incubation of the full-length peptide with intestinal lysate resulted in the production of PHI3–27 and PHI1–21 fragments (Fig. 2). In addition to analyzing this sample by MALDI, we also performed an LC-MS shotgun proteomics experiment to obtain information about the retention time, mass-to-charge ratios and tandem mass spectra of the relevant PHI fragments (Fig. 2).
Figure 2.
Lysate peptidomics. A. MALDI profiles of in vitro assay using gut membrane lysates with gut peptide PHI1–27 for optimization of reaction time. B. PHI1–27 (100 µM) was incubated with the soluble (1 mg/mL) or membrane (1 mg/mL) fraction of the intestinal proteome (lysate) for 15 min. The sample was then quenched and analyzed by LC–MS to quantify PHI1–27 levels. A majority of the PHI-degrading activity was found in the membrane fraction, as evidenced by greatly diminished PHI1–27 levels after 15 min in the membrane fraction, and largely unchanged levels after exposure to the soluble fraction. C. PHI1–27 proteolytic fragments after incubation with intestinal membrane lysates.
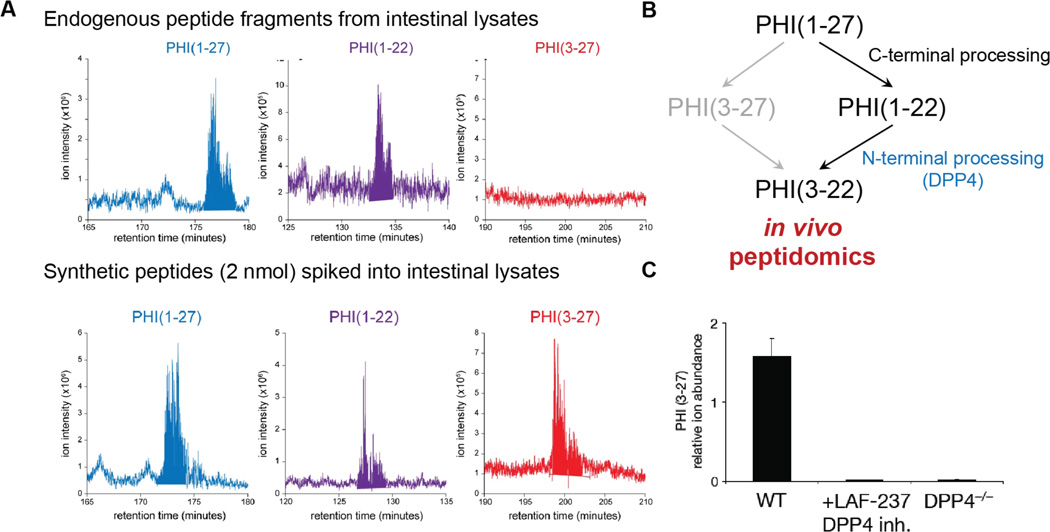
This information about the properties of the different candidate peptide fragments can be used to design a number of different experiments to improve sensitivity of detection of endogenous peptides. In one approach, the known retention times and mass-to-charge ratios of peptide fragments are used to improve the searching of shotgun proteomics data to identify the presence of peptides that were present but were unidentified by SEQUEST, for example. In addition, since tandem MS uses data-dependent acquisition it is possible that certain peptides are missed because they are not selected for subsequent tandem MS. Knowing the mass-to-charge and retention time of a given bioactive peptide fragment can be used to target peptides for tandem MS, providing a modified experimental workflow to identify the peptide. Using this targeted approach, for example, we were able to detect PHI1–21, but not PHI3–27 in vivo, indicating that PHI3–27 is not an endogenous PHI fragment in the gut (Fig. 3)49. Lastly, the same information can also be used to design a targeted multiple-reaction monitoring (MRM) LC-MS experiment for each candidate fragment50. MRM experiments require a triple quadrupole (QQQ) mass spectrometer, and provide superior sensitivity for molecules in complex mixtures50. While no approach can guarantee that a fragment will not be missed, performing lysate experiments and targeting those peptides when performing the analysis of samples from in vivo experiment using targeted mass spectrometry experiments provides an effective way of maximizing sensitivity and reducing the possibility that a bioactive peptide fragment is missed.
Figure 3.
In vivo processing of a bioactive peptide. A. in vivo peptidomics of the intestine revealed the presence of PHI1–27 and PHI1–22, but not PHI3–27. Synthetic peptides spiked into the sample prior to LC-MS/MS analysis. B. A model for the in vivo proteolysis of PHI1–27 based on physiological fragments of PHI1–27 detected by peptidomics. C. The addition of the DPP4 inhibitor LAF-237 to intestinal lysates or the use of lysates from DPP4−/− mice prevent the production of PHI3–27 to implicate DPP4 in the N-terminal processing of PHI1–27 in these tissue lysates.
Once the endogenous fragments of a peptide have been identified in Stage 1, this information is then used to develop a biochemical model to determine the endogenous cleavage sites (Fig. 1). For example, the identification of CGRP1–17, CGRP18–37, CGRP1–26 and CGRP27–37 indicated that CGRP is cleaved between amino acids 17–18 and 26–27. Thus, in Stage 2, we sought to identify candidate enzyme(s) that were able to cleave CGRP at these endogenous cleavage sites (Fig. 1). For CGRP, we decided to purify candidate enzymes directly from spinal cords to ensure that we identify the candidate peptidase directly from the tissue source.
To be able to isolate the CGRP-degrading enzyme, we needed to be able to follow its activity through various purification steps. We noted that addition of CGRP to spinal cord lysates produced CGRP1–17 and CGRP18–37, which was easily detectable by MALDI (Fig. 4). The CGRP1–26 and CGRP27–37 fragments were also produced in this assay but were less prominent in MALDI spectra, though readily detected by LC-MS. With this MALDI assay, we could simply incubate proteome fractions from the fractionation procedure with synthetic CGRP and follow, by MALDI, which fractions retained CGRP-degrading activity. If desired, different peptide substrate assays could be developed at this stage (e.g. a FRET-based peptide analog for measuring cleavage) depending on the type of equipment readily available.
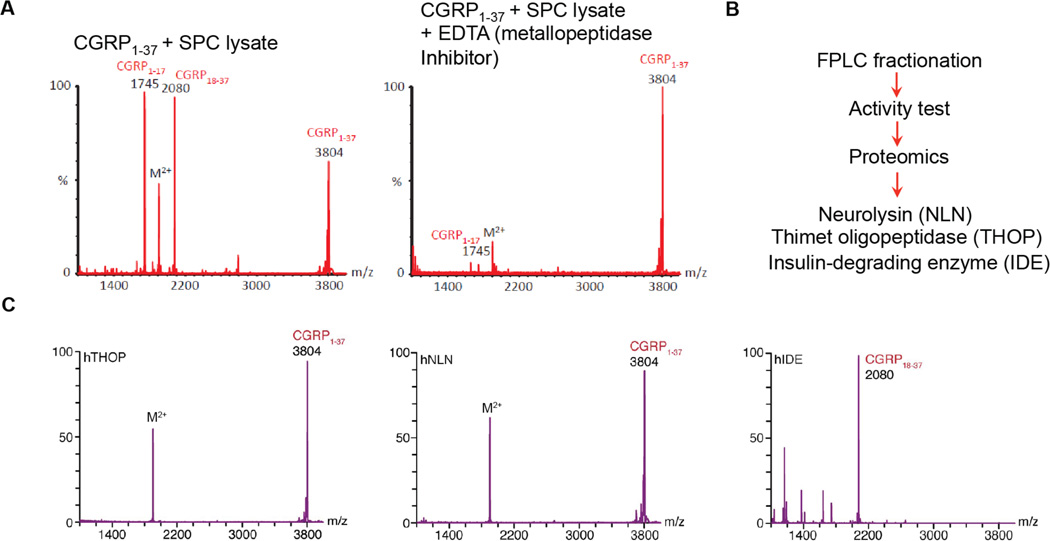
Figure 4. Identification of IDE as a candidate CGRP-degrading enzyme.
A. MALDI assay for classification of bioactive peptide regulating peptidase using different peptidase inhibitors (WT vs. EDTA metallopeptidase inhibitor) B. Proteomic analysis of the FPLC fractions showing the highest level of CGRP-degrading activity to identify candidate peptidases. Proteomics identified three metallopeptidases in these fractions; THOP, NLN, and IDE. C. Verification of CGRP-degrading enzyme from the candidate memtallopeptidase using purified human THOP, NLN and IDE (0.05 mg/mL) at 37 °C for 15 min.
To isolate candidate CGRP-degrading enzymes, we began by fractionating the proteome into soluble and membrane fractions using a simple centrifugation protocol. The membrane fraction was then solubilized. Using the same concentration of total protein from either the membrane or soluble fraction, we then assessed their relative activities by incubating CGRP with each lysate and measuring the production of the key CGRP fragments identified in Stage 1, CGRP1–17 and CGRP18–37, by MALDI and LC-MS. The relevant CGRP fragments were only produced in the soluble fraction, which allowed us to focus on the soluble proteome when searching for candidate CGRP-degrading enzymes.
Next, we used a series of peptidase class-selective small molecule inhibitors to determine what class of peptidase (metallo, serine, cysteine or aspartyl) is responsible for the CGRP-cleaving activity41,49 (Fig. 4). Each inhibitor was preincubated with the soluble fraction of spinal cord lysates followed by addition of CGRP. The degradation of CGRP was then monitored by the amount of CGRP18–37 generated, as measured by LC-MS. In this case, we found that the metallopeptidase inhibitor (1,10-phenanthroline and EDTA) strongly inhibited CGRP cleavage, indicating that the CGRP-degrading enzyme is a metalloprotease (Fig. 4). This information on peptidase class was highly useful because it allowed us to focus on candidate metallopeptidases.
To further narrow down candidate CGRP-degrading enzymes, we then proceeded to fractionate the soluble spinal cord proteome, which contained all of the CGRP-degrading activity, by fast protein liquid chromatography (FPLC) (Fig. 4). Specifically, we utilized an anion exchange column to separate the proteome into multiple fractions, and then tested each of these fractions for their CGRP-degrading activity. We proceeded to identify the candidate enzymes by analyzing active fractions by proteomics. This proteomics analysis identified three metallopeptidases present in the active fraction, namely insulin-degrading enzyme (IDE)51, thimet oligopeptidase (THOP)52 and neurolysin (NLN)52, which were thereafter considered candidate CGRP-degrading enzymes (Fig. 4).
Depending on the peptidase and its location (i.e. soluble versus membrane), the fractionation step will have to be optimized on a case-by-case basis. For example, a soluble peptidase, like the CGRP-degrading enzyme, is easier to work with than a membrane protein, which requires a solubilization step prior to purification53. For membrane peptidases, we recommend solubilizing the membrane fraction using triton or another similar surfactant53. After solubilizing the membrane, the activity of the enzyme must be checked with the activity assay, such as the CGRP proteolysis assay. If the activity of the enzyme is retained after solubilization of the membrane fraction, then the purification of a membrane protein can proceed in a similar manner to that of a soluble protein, but every individual case will vary.
In the case of the candidate CGRP-degrading enzymes, a single FPLC purification yielded active fractions with only three candidate enzymes, which could easily be tested individually. In some cases, however, many more peptidases could be identified in the active fractions, which would require further optimization and possible further fractionations. In particular, if a large number (> 8–10) of candidate peptidases are associated with the active fractions, it may be beneficial to perform a second, orthogonal, fractionation step. This additional fractionation step could be size exclusion chromatography (SEC)54, strong cation exchange chromatography (SCX)55,56 or hydrophobic interaction chromatography (HIC)57. All of these would further fractionate the proteome so that the active fractions have a smaller number of candidate peptidases to validate in subsequent steps.
After identifying IDE, THOP and NLN as metallopeptidases present in the active CGRP-degrading fraction, it was necessary to test each of these peptidases for their ability to cleave full-length CGRP to generate CGRP1–17, CGRP18–37, CGRP1–26 and CGRP27–37 (Fig. 1 and Fig. 4). These proteins are currently all commercially available, but at the time of our work, THOP and NLN could not be purchased and so were instead produced by bacterial overexpression based on published procedures for their expression and purification41. To ensure that the expressed enzymes were active, we tested each against a known substrate and assessed cleavage. If commercially available, we highly recommend purchasing candidate proteins, though they are expensive, because it is fast and ultimately more cost effective when researcher time and reagent cost is taken into account. Each candidate peptidase is then incubated with the full-length peptide, in this case CGRP, and the proteolysis of this peptide is monitored by LC-MS. THOP and NLN did not cleave CGRP, while IDE readily cleaved CGRP to afford the endogenous fragments CGRP1–17, CGRP18–37, CGRP1–26 and CGRP27–37 (Fig. 4). Thus, IDE was the only one of our candidate CGRP-degrading enzyme capable of cleaving this peptide at the physiologically relevant sites and we therefore proceeded to take it to the next stage of the protocol, namely Stage 3, validation in vivo (Fig. 1).
As a potential alternative to extensive fractionation, databases can also be used to produce a list of candidate enzymes capable of cleaving at a given site in a peptide of interest. The MEROPS database58,59 contains information on all proteases and peptidases from a variety of different organisms and offers the opportunity to search this database in a number of different ways, including a search based on enzymes that are predicted to be able to cleave a particular bond. For instance, using the cleavage sites Ser17-Arg18 and Asn26-Phe27 in CGRP, we obtained lists of candidate peptidases capable of cleaving at each bond. The results from the Ser17-Arg18 search included five metallopeptidases: THOP, IDE, matrix metalloprotease (MMP) 19, meprin and nardilysin. The peptidases from the Asn26-Phe27 search included THOP, NLN, MMP1, MMP2, MMP3, MMP8, MMP9, MMP13, MMP20, ADAMTS4 and ADAMTS1. Of these candidate enzymes only THOP, IDE, NLN and nardilysin are soluble and therefore likely to be responsible for the CGRP-degrading activity. All of these enzymes are commercially available, which would make testing them for CGRP-cleaving activity straightforward. We recommend using the MEROPS database to identify candidate peptidases due to the speed and relative inexpensiveness of this approach relative to enzyme purification from lysates. A caveat to relying solely on databases, however, is that that they may not be comprehensive and may miss some enzymes. For example, IDE is not listed as one of the enzymes capable of cleaving CGRP between amino acids 26–27, but it does so readily. Given that many peptidases and proteases are in fact well characterized, however, this database can be a very valuable tool for predicting candidate peptidases.
Having identified IDE as a candidate CGRP-degrading enzyme capable of cleaving this peptide at the physiologically relevant sites in vitro in Stage 2 of this protocol, we then set out to validate IDE as an endogenous CGRP-degrading enzyme in Stage 3 (Fig. 1). To do this, we needed to perturb the activity of this enzyme in vivo and quantitatively measure changes in CGRP levels. IDE heterozygote mice (IDE−/+) were commercially available and we bred these animals using standard animal husbandry techniques to isolate the IDE knockout (IDE−/−) mice we utilized in our experiments60. If an in vivo model is unavailable, the decision must be made to either create a knockout mouse or identify alternative methods for testing the connection between an enzyme and a substrate. We believed IDE was cleaving CGRP extracellularly and this required us to use an in vivo model; however, if the enzyme-substrate interaction is believed to take place in the cytosol, or if the peptidase and peptide are both secreted, a cell culture model should suffice. It is also possible to use other more genetically tractable organisms, such as Caenorhabditis elegans or Drosophila melanogaster, if available and applicable.
The fastest option, if available, is to use a selective small molecule inhibitor of the peptidase25. Such inhibitors would enable much faster testing of the hypothesis because wildtype mice can be purchased and tested in a very short time compared to the breeding necessary to get sufficient numbers of knockout mice, especially when homozygote knockouts are not commercially available. In addition, knockout animals sometimes suffer from the emergence of compensatory mechanisms that could mask the role of an enzyme in the regulation of a bioactive peptide. For example, the loss of a peptidase may result in the upregulation of another peptidase to compensate for the knocked out enzyme61,62. The use of a small molecule inhibitor overcomes these challenges because it can acutely inhibit the enzyme, allowing measurements to be made prior to the onset of any compensation63. Therefore, we highly recommend the use of small molecule peptidase inhibitors when available. These compounds are currently not available for IDE, which prompted our use of knockout mice.
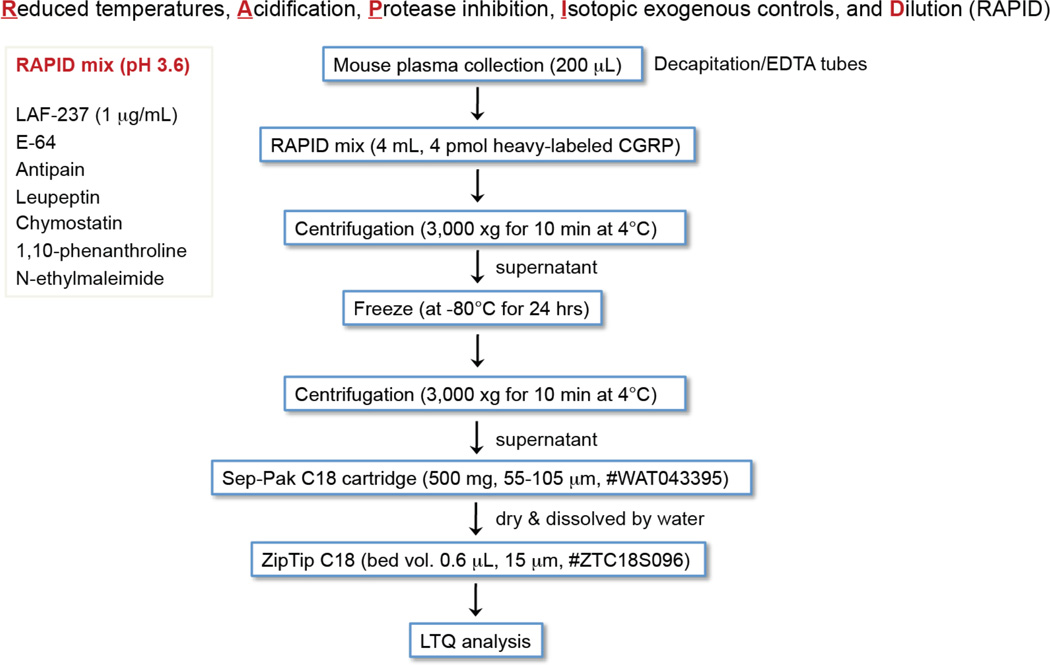
During initial experiments to measure changes in CGRP levels in IDE knockout mice, we noticed a great deal of variability in our data from the ELISA assay we were using. We suspected that this may be due to variability in the peptide isolation and this led us to use a reported protocol termed RAPID64 that limit proteolysis of peptide hormones for immunoassays (Fig. 5). The data was better but not good enough, so we surmised that the issue was the immunoassay. While the data always trended towards higher levels of CGRP in IDE knockout mice, the precision of the measurement was not good enough. Therefore, we decided to develop an isotope dilution mass spectrometry (IDMS) assay41 to reliably and accurately quantitate endogenous CGRP levels.
Figure 5.
Overall steps of RAPID method64 for IDMS studies.
In cases where a good immunoassay is commercially available, we do recommend using these assays to save time.
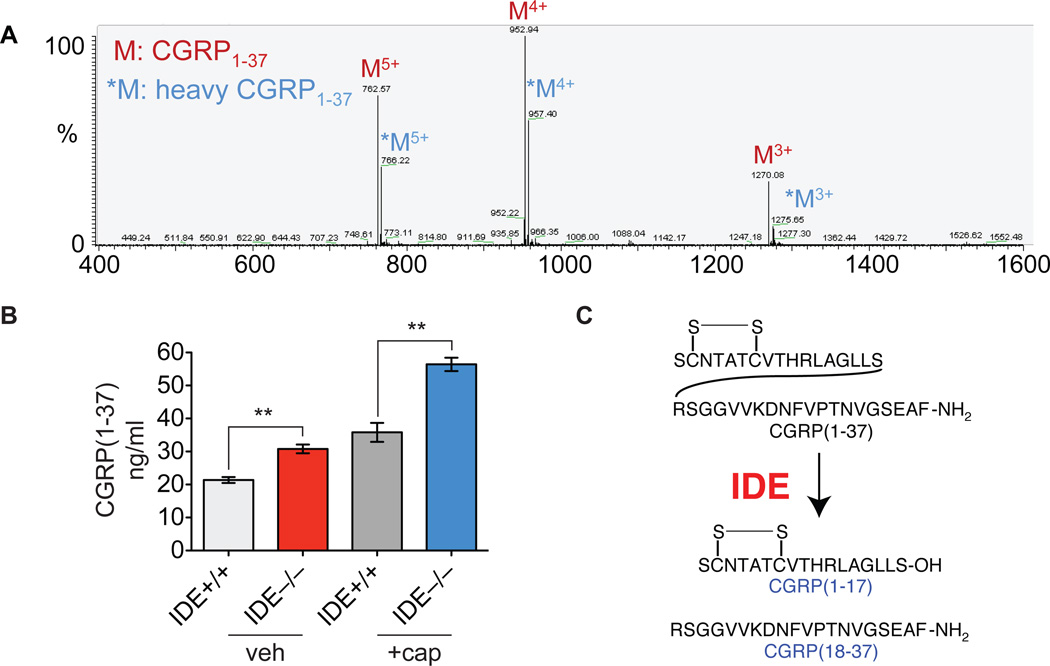
In an IDMS assay, an isotopically labeled standard, in this case deuterated CGRP (d18-CGRP), is added at a known concentration to the mouse tissue or plasma prior to peptide extraction. After LC-MS, the endogenous molecule and the labeled standard can be distinguished in the mass spectrometer because of the difference in their molecular weights. The ratio of the peak intensities for the two molecules (endogenous peptide and standard) enables the concentration of the endogenous peptide to be calculated (Fig. 6). This assay provided a much more reproducible measurement of endogenous CGRP levels and demonstrated that IDE is in fact regulating endogenous CGRP levels (Fig. 6). This protocol successfully identified the peptidase that regulates CGRP in mammals (Fig. 6), and should be applicable to other important peptides moving forward.
Figure 6.
Bioactive peptide degradation in mouse spinal cord and plasma. A. LC-MS profile of multiple-charged endogenous CGRP1–37 and heavy-labeled CGRP1–37 in the RAPID method64 B. IDE regulates CGRP levels in mice as demonstrated by elevated levels of CGRP in animals lacking IDE. Capsaicin causes CGRP secretion leading to higher absolute levels of CGRP but the difference in CGRP levels between genotypes is consistent indicating that IDE can regulate CGRP under different conditions. C. CGRP proteolysis pathway revealed through the application of this protocol.
MATERIALS
REAGENTS
<CAUTION>
Use gloves and wear goggles when handling corrosive substances
Use a fume hood when doing experiments with toxic chemicals
Never taste any chemicals and never directly smell the source of any vapor or gas
Do not wear contact lenses even when worn under safety goggles
Ice-cold acetic acid, 0.25% (v/v) in water ! CAUTION Combustible, Corrosive
Formic acid, 0.1% (v/v) in water or in acetonitrile ! CAUTION Combustible liquid/Target organ (kidney, liver, central nerve system, blood) effect/Harmful by ingestion/Corrosive
Acetonitrile for LC-MS (Honeywell, cat. no. LC015-1) ! CAUTION Flammable liquid/Harmful by ingestion/Harmful by skin absorption/Irritant
Water for LC-MS (Honeywell, cat. no. 365-4)
Trifluoroacetic acid (TFA; Sigma-Aldrich, cat. no. T6508) ! CAUTION TFA is extremely hazardous in case of skin contact, of eye contact, of ingestion, of inhalation.
Tris-HCl (Sigma-Aldrich, cat. no. T5941)
Sodium chloride (Sigma-Aldrich, cat. no. S3014)
Potassium phosphate monobasic (Sigma-Aldrich, cat. no. P0662)
Potassium chloride (Sigma-Aldrich, cat. no. P9333)
Sterile 1× phosphate-buffered saline (PBS), pH 7.4 (Thermo Scientific, cat. no. SH30256.01)
Guanidinium-HCl, 8 M (Sigma-Aldrich, cat. no. G9284) ! CAUTION Irritant/Toxic by ingestion
α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich, cat. no. C2020)
Deuterated amino acids (Cambridge isotope laboratories)
E-64 (Sigma-Aldrich, cat. no. E3132)
Iodoacetamide (Sigma-Aldrich, cat. no. I1149) ! CAUTION Toxic by ingestion/Respiratory sensitizer/Skin sensitizer
N-ethylmaleimide (Sigma-Aldrich, cat. no. E3876) ! CAUTION Highly toxic by ingestion/Toxic by skin absorption/Skin sensitizer/Corrosive
EDTA disodium salt (Sigma-Aldrich, cat. no. ED2SS)
1,10-phenanthroline (Sigma-Aldrich, cat. no. P9375) ! CAUTION Toxic by ingestion
Phosphoamidon (Sigma-Aldrich, cat. no. R7385) ! CRITICAL Phosphoamidon solution (10 mg per mL in water) can be stored in aliquots at −20°C with an expected shelf life of at least one month
Pepstatin A (Sigma-Aldrich, cat. no. P5318) ! CRITICAL If stock solution become more yellow the reagent is hydrolyzing
Phenylmethanesulfonyl fluoride (Sigma-Aldrich, cat. no. 78830) ! CAUTION Toxic by ingestion/Corrosive
Ammonium acetate (Sigma-Aldrich, cat. no. A1542)
Antipain (Sigma-Aldrich, cat. no. A6191) ! CRITICAL Dilute solutions should be stored on ice and kept for only a day because of the terminal aldehyde, which is subject to oxidation and racemization
Leupeptin (Sigma-Aldrich, cat. no. L2884) ! CRITICAL At working concentrations (10 to 100 µM) a solution is stable only a few hours
Chymostatin (Sigma-Aldrich, cat. no. C7268) ! CRITICAL Stock solutions (10 mM) are stable for months at −20°C
LAF-237 (Axon Medchem BV, cat. no. Axon 1631)
Bestatin (Sigma-Aldrich, cat. no. B8385) ! CRITICAL Stock dilutions at 1 mM are expected to be stable at least 1 month if stored at −20°C
Amastatin (Sigma-Aldrich, cat. no. A1276) ! CRITICAL A stock solution of amastatin in methanol is stable for at least one month at −20°C
Synthetic peptides should be purchased when available and isotopically labeled peptides can either be purchased from a custom synthesis company or prepared by solid phase-peptide synthesis (SPPS) using standard approaches65.
Recombinant peptidases should be purchased (http://www.origene.com) for these assays or expressed and purified using standard techniques in molecular biology and biochemistry39.
EQUIPMENT
MALDI-TOF MS (for in vitro peptidomics; Waters)
Waters nanoAcquity HPLC configured with in-house packed 75-µm reverse phase (RP) capillary trapping and analytical column (New Objective) coupled with LTQ-Orbitrap mass spectrometer (for in vitro and in vivo peptidomics; ThermoFisher Scientific)
ÄKTA FPLC using a HiTrap Capto Q column (for proteome fractionation; GE Healthcare)
Dounce tissue grinder set (2, 7 and 15 mL complete; Sigma-Aldrich)
10-kDa molecular weight cutoff (MWCO) filter (Microcon YM-10)
Bio-Rad (Bradford) protein assay kit
Speed vacuum concentrator (Savant SC110A)
Ultracentrifuge (Beckman Coulter Inc., Optima™ TLX)
C18 Sep-Pak cartridge (Waters, Oasis HLB 1 cc, cat. no. WAT094225)
ZipTip with 0.6 µL C18 resin (Millipore, cat. no. ZTC18S960)
MALDI target plate (Waters, cat. no. M880675CD1-S)
EDTA-coated tube (BD Biosciences, cat. no. 367835)
Sep-Pak C18 cartridges (6 mL; Waters, cat. no. WAT043395)
Extraction manifold (20 position, 16 × 100 mm tubes; Waters, cat. no. WAT200609)
Protein LoBind Tubes (Eppendorf, cat. no. 0030 108.132)
Protein sequencer (Applied Biosystems, cat. no. 477A)
REAGENT SETUP
Peptidase inhibitors. Make 10 µM E-64, 1 mM iodoacetamide, 10 µM N-ethylmaleimide, 1 mM EDTA, 1 mM 1,10-phenanthroline, 10 µM phosphoamidon, 10 µM pepstatin A, 1 mM phenylfluorophosphate and vehicle [1× PBS with 25% (vol/vol) ethanol for 1% final concentration in reaction], respectively.
-
Aminopeptidase inhibitor cocktail. Mix 100 µM 2',3-dinitroflavone-8-acetic acid, 10 µM bestatin, and 10 µM amastatin
IDMS mix contains 1 nM heavy-labeled target peptide (d18-peptide), 0.1 M ammonium acetate, 0.5 M NaCl, and enzyme inhibitors (1 µg/mL of LAF-237, E-64, antipain, leupeptin, chymostatin, 1,10-phenanthroline, N-ethylmaleimide)
Wildtype (WT) mice (C57BL/6) used in this study are either purchased (Jackson Laboratories, Bar Harbor, ME) or taken from a breeding colony. Animals are kept on a 12 h light-12 h dark schedule and fed ad libitum. For tissue collection, animals are euthanized with CO2, and their tissues are dissected, flash-frozen with liquid nitrogen, and stored at −80 °C. All animal care and use procedures are in strict accordance with the standing committee on the use of animals in research and teaching. ! CAUTION Research protocols involving the use of animals should be reviewed by the investigator's institutional ethical review board to avoid any unnecessary discomfort or pain to the animals and to determine whether alternatives exist to animal research. All animal experiments should be performed in accordance with relevant guidelines and regulations of protocols approved by the investigator's institutional animal research review committee. Personnel should be trained in animal handling.
EQUIPMENT SETUP
Waters MALDI micro MX (Waters MS Technologies, Milford, MA) operate in reflectron positive mode for MALDI spectra of the peptides. detailed operating conditions is as follows: source voltage, 12 kV; matrix suppression delay, 800 amu; pulse voltage, 1.9 kV; reflectron voltage, 5.2 kV; laser firing rate, 5 Hz. Instrument control, spectral acquisition, and processing were performed with MassLynx software (version 4.1).
ÄKTA FPLC using a HiTrap Capto Q column (GE Healthcare) is applied to fractionate animal tissue lysates. Buffer A is 20 mM Tris HCl pH 8, and buffer B is 20 mM Tris HCl and 1 M NaCl pH 8. The gradient is 0–50% B over 30 min at a 1 mL/min flow rate.
Self-pack picofrit analytical column (75 µm i.d.; New Objective, cat. no. PF360-75-30-N-5) is packed 15 cm with 3 µm of C18 (Michrom Bioresources Inc., Magic C18 AQ 200A 3U;). The trap column is obtained prepacked from New Objective Inc. (Integrafrit sample trap, C18 5 µm, 100 µm column i.d.).
Nano flow LC (Nano LC-2D; Eksigent Technologies) system coupled to a linear ion trap mass spectrometer (LTQ; ThermoFinnigan) is set from m/z 400 to m/z 1600 for data acquisition. The peptides are detected in the positive mode, and the LC/MS/MS experiment is performed in a data-dependent acquisition (DDA) mode. The MS/MS data are collected for the most intense peak from the MS chromatogram. Dynamic exclusion is set for 30 s, the exclusion size list is set to 200, and the normalized collision energy for CID was 35%. The capillary spray voltage is set to 2.5 kV.
SEQUEST is a tandem mass spectrometry data analysis program used for peptide identification40. Peptides are accepted within 1Da of the expected mass, meeting a series of custom filters on ScoreFinal (Sf),-10 Log P, and charge state that attained an average peptide FDR of <2% across data sets.
Strong cation exchange (SCX) chromatography is performed using a PolySULFOETHYL A column (200 × 2.1 mm, 5 µm, 300 Å; PolyLC Inc.) connected to an Agilent Technologies 1200 series LC. All runs are operated at 0.3 mL/min. The SCX buffers consist of (A) 7 mM KH2PO4, pH 2.6, 25% ACN (v/v); (B) 40 mM KCl, 7 mM KH2PO4, pH2.6, 25% ACN (v/v); (C) 100 mM KCl, 7 mM KH2PO4, pH 2.6, 25% ACN (v/v); (D) 600 mM KCl, 7 mM KH2PO4, pH2.6, 25% ACN (v/v). A step gradient is applied that included 60 min with buffer A, 40 min with buffer B, 40 min with buffer C, and 40 min with buffer D, with 20 min transitions between the different buffer conditions.
PROCEDURE
Peptide extraction from Tissues • TIMING 1d
-
1.
Weigh the frozen tissue samples and then place them in 500 µL of ice-cold water prior to boiling for 15 min. The proteome is denatured in this step and doing so inactivates any proteolytic activity prior to tissue homogenization.
▲ CRITICAL The frozen tissues should be weighed prior to addition to the ice-cold water to enable absolute quantification.
-
2.
After boiling, separate the aqueous fraction and save on ice or in a cold room.
-
3.
Dounce-homogenize the heat-denatured tissues in ice-cold aqueous acetic acid (0.25% v/v). We perform at least 10–15 strokes with the each of the A (loose) and B (tight) douncers to ensure that the sample is completely homogenized.
▲ CRITICAL STEP Use the same number of strokes with the A (loose) and B (tight) douncer for more consistent results.
-
4.
Combine the aqueous fraction from Step 2 and the homogenate from Step 3 and centrifuge at 20,000 × g for 20 min at 4 °C.
-
5.
Separate the supernatant from the insoluble pellet and pass through a 10-kDa molecular weight cutoff (MWCO) filter.
-
6.
Apply the filtrate (i.e. < 10-kDa fraction) into a C18 Sep-Pak cartridge and washing with water (10 mL).
-
7.
Elute the peptides from the C18 Sep-Pak with 1 mL of 70:30 H2O/ACN (vol/vol). This sample is then concentrated under vacuum using a Speedvac.
-
8.
Dissolve sample with water and then fractionate it with strong cation exchange chromatography (SCX) (see EQUIPMENT SETUP). We use a step gradient of four different salt concentrations (A, B, C and D). After SCX, each of these fractions is desalted using a C18 Sep-Pak cartridge. Each sample (N = 4) is washed with water (25 mL) to remove salts and then the peptides are eluted from the C18 Sep-Pak with 1 mL of 70:30 H2O/ACN and then concentrated under vacuum using a Speedvac.
-
9.
Dissolve each of these samples in 0.1% aqueous formic acid prior to LC-MS/MS analysis. The amount of formic acid solution used to dissolve the sample is calculated using the weight of the tissue used to generate the sample (40 µL of formic acid solution per 75 mg of tissue).
□ PAUSE POINT These samples can be used right away or stored at −80 °C. We typically use samples within one to two weeks but they should be stable for much longer periods.
LC–MS peptidomics of tissue samples • TIMING 1d
-
10.
(See EQUIPMENT SETUP for details of the LC–MS setup) Inject the samples (2 µL) using the autosampler. The analytes are trapped onto a self-pack picofrit C18 column with an isocratic flow rate of 2 µL/min for 10 min. The use of a trapping column enables a larger amount of sample to be loaded more quickly, affording greater coverage of the peptidome. During the data collection by the mass spectrometer the mobile phase passes through the trapping column to the analytical column, eluting and separating the peptides in process. The flow rate is 300 nL/min and binary gradient of 5–40% B in 227 min (mobile phase A, 0.1% formic acid in water; mobile phase B, 0.1% formic acid in acetonitrile).
-
11.
Collect data in full MS mode (N = 4, i.e. no tandem MS spectra) for peptide quantitation by using the area under the peak. By not switching between full MS and tandem MS the LC-MS chromatograms are much smoother, which affords better quantitation. To identify the peptides, we then perform a Top 6 MS2 experiment, where the largest 6 ions during an full MS scan are then subjected to tandem MS, which generate tandem MS spectra that are used to identify the peptide sequences. Importantly, in cases where peptides are not being quantified, we simply rely on a Top 6 experiment to identify these fragments.
-
12.
Set the dynamic exclusion for these experiments to 30 s for the Top 6 analysis and set the exclusion size list to 200. Set the capillary spray voltage at 2.5 kV and the normalized collision energy for CID to 35%.
-
13.
Identify peptide sequences using SEQUEST with differential modification of methionine to its sulfoxide as described in EQUIPMENT SETUP.
Preparation of tissue lysates • TIMING 3h
-
14.
Thaw the frozen tissues on ice (these can be isolated or purchased from various tissues banks).
-
15.
Dounce-homogenize the animal tissue in 1× PBS (100 µL per 100 mg tissue). We perform at least 10–15 strokes with each of the A (loose) and B (tight) douncers to ensure that the sample is completely homogenized.
-
16.
Centrifuge the homogenate at 1000 × g at 4 °C for 5 min to remove large cellular debris.
-
17.
Transfer the supernatant from this spin to a thick walled centrifuge tube and then centrifuge at 100,000 × g at 4 °C for 45 min using a table top ultracentrifuge. This separates the sample into the soluble (supernatant) and membrane (pellet) proteomes.
-
18.
Transfer the resulting supernatant to a new Eppendorf tube and use as the soluble fraction.
▲ CRITICAL Be careful not to contaminate with membrane fraction in transferring the soluble fraction
-
19.
Wash the pellet twice with 1× PBS and then suspend in 100 µL of 1× PBS. This is then used as the membrane fraction.
□ PAUSE POINT Aliquot the samples (10 µL) and store at −80 °C until further use
-
20.
Determine the protein concentrations of these lysates using a Bradford assay.
-
21.
Dilute all lysates using 1× PBS to a final concentration of 1 mg/mL for subsequent activity assays.
Bioactive peptide proteolysis in tissue lysates • TIMING 2h
-
22.
Incubate synthetic peptides (20 µL, 100 µM final concentration) with soluble and membrane lysates (1 mg/mL) at 37 °C for 15 min.
▲ CRITICAL The reaction time varies for each peptide (5 – 60 min) and must be optimized to ensure the optimal amount of degradation, since too little or too much proteolysis is not informative.
-
23.
Quench the reaction. After several attempts at developing a quench, we found that the addition of 8 M GndHCl (20 µL) to the reaction provided the best quench. The addition of acid led to precipitation of certain peptides but the 8 M GndHCl seems to overcome this issue while effectively quenching the reaction. The addition of the 8 M GndHCl requires a desalting step prior to analysis with a ZipTip prior to LC–MS or MALDI analysis.
? TROUBLESHOOTING
MALDI analysis of lysate experiments • TIMING 3 h
-
24.
Mix 1 µL of the desalted sample with 1 µL of the matrix, a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile.
-
25.
Spot 1 µL of the mixture solution on the MALDI target plate and dry at room temperature or under a gentle stream of nitrogen.
-
26.
Acquire MALDI spectra of the proteolytic fragments from m/z 500 to 5,000 with an average of 200 laser shots.
-
27.
Analyze the mass values of the peptide fragments using MassLynx software (version 4.1) and Peptide Mass Calculator v3.2 (http://immweb.vet.uu.nl/P&P_fac/pepcalc.htm).
LC-MS/MS analysis of lysate experiments • TIMING 1d
-
28.
(See EQUIPMENT SETUP for details of the LC–MS setup) Inject samples onto the LC. The samples are trapped onto a self-pack picofrit C18 column with an isocratic flow rate of 2 µL/min for 10 min. The use of a trapping column enables a larger amount of sample to be loaded more quickly, affording greater coverage of the peptidome. During the actual data collection by the mass spectrometer, the mobile phase passes through the trapping column to the analytical column, eluting and separating the peptides in process. The flow rate is 300 nL/min and binary gradient of 5–40% B in 227 min (mobile phase A, 0.1% (vol/vol) formic acid in water; mobile phase B, 0.1% (vol/vol) formic acid in acetonitrile).
-
29.
Identify peptide sequences using SEQUEST with differential modification of methionine to its sulfoxide as described in EQUIPMENT SETUP. If other types of modifications are being studied (i.e. phosphorylation, acetylation, etc.) the search will be adapted at this step to enable the detection of differentially modified peptides.
-
30.
Integrate the area under the curve (AUC) for specific peptides in the LC– MS chromatogram in order to obtain Relative quantitative data for the different peptide fragments generated during the incubation of the full-length peptide with lysates.
▲ CRITICAL STEP To ensure that the quantitation is accurate, make sure to account for all charge states for a given peptide fragment.
Protease inhibitor assays • TIMING 6 h
-
31.
Incubate tissue lysates or plasma (1 mg/mL) at 37 °C for 30 min with each protease inhibitor described in REAGENT SETUP prior to the addition of the synthetic peptide. (Note: if other types of modifications are being studied the choice of inhibitor can vary. For example, if the PTM of interest is a phosphorylation site then different kinase inhibitors can be used at this point.)
-
32.
Add synthetic peptide substrate (500 µM in LC-MS grade water) to the lysates or plasma so that the final concentration of the substrate 100 µM.
-
33.
Allow the reactions to proceed at 37 °C for 15 min to an hour depending on the peptide.
-
34.
Quench the reactions with an equal volume of 8 M GndHCl.
? TROUBLESHOOTING
-
35.
Desalt the peptides using a C18 ZipTip according to the procedure outlined in Box 1 prior to MALDI-TOF MS and LC-MS/MS analysis for peptide-degrading activity.
Box 1 | ZipTip purification for optimal recovery of peptides.
ADDITIONAL REAGENTS
1 µL of TFA in 1 µL water (0.1 % TFA (v/v) in water)
1 µL of TFA in 700 µL ACN + 300 µL water (0.1% TFA (v/v) in 70% ACN)
PROCEDURE
Pre-condition the ZipTip by aspirating 10 µL (maximum volume of ZipTip) of acetonitrile 5 times, followed by aspiration of the ZipTip with 10 µL of 0.1% TFA (v/v) in water 5 times to equilibrate the ZipTip so that you can bind the peptides to the ZipTip.
-
After conditioning, bind the peptides to the ZipTip by placing the ZipTip into the solution containing the sample and gently aspirate the sample 20 times.
▲ CRITICAL Be careful when aspirating the sample and do not introduce air into the ZipTip to maintain optimal desalting conditions.
Desalt the peptide sample. Place the ZipTip into a fresh solution of 0.1% TFA (v/v) and wash the tip by aspirating this solution 5 times using the 10 µL setting on the pipette.
After removal of the 8 M GndHCl and buffer salts, the peptides are ready to be eluted for subsequent LC–MS and MALDI analysis. Elute the peptides using 5 µL of a 70% ACN containing 0.1% TFA (v/v) in an Eppendorf tube. To ensure that the peptides are eluted from the ZipTip, aspirate this sample 20 times.
Assays with recombinant peptidases • TIMING 1 d
-
36.
Add substrate peptides from stocks to the recombinant protease (0.05 – 0.1 mg/mL) in 50 mM Tris HCl pH 7.5 and incubate at 37 °C for 1 h. These conditions can also be adapted to the relevant peptidase as necessary. The final concentration of the peptide in our experiment was 100 µM, but other concentrations can be used as well.
▲ CRITICAL The reaction time should be optimized based on the activity of the recombinant protease
-
37.
Quench the reaction with an equal volume (20 µL) of 8 M GndHCl followed by heating to 95 °C for 5 min.
-
38.
Take 5 µL of the quenched reaction, dilute with an equal volume (5 µL) of proteomics grade water and then desalt the sample using a C18 ZipTip as outlined in Box 1.
-
39.
Perform MALDI-TOF analysis as described in Step 24–27. MALDI-TOF analysis quickly reveals the presence or absence of particular peptide fragments to demonstrate the cleavage site specificity of the peptidase.
-
40.
Analyze the reactions by LC-MS/MS as described in Step 28–30 to obtain quantitative information about the extent of peptide cleavage. Relative quantitation is accomplished by using the AUC within the LC-MS/MS chromatograms. Comparison of peptide degradation from samples with and without inhibitors reveals the peptidase class (i.e. metallo-, serine-, aspartyl, cysteine) responsible for the degradation.
Endogenous peptide quantitation by isotope dilution mass spectrometry (IDMS) • TIMING 1.5 d
-
41.
Kill the mice by CO2 inhalation, decapitate them, and collect their trunk blood into an EDTA-coated tube.
▲ CRITICAL STEP The experimenter should be experienced in animal handling and tissue collection. Ideally, this step should be carried out with two people to increase the speed of sample collection and processing, which will minimize sample degradation. The goal is to have the sample mixed with the RAPID solution in less than 1 minute from the time of decapitation.
-
42.
Quickly invert the tube five times and rapidly transfer aliquots of blood (200 µL) into 4 mL of ice-cold buffer (pH 3.6, 4 mL) containing 1 nM of the stable-isotope labeled heavy peptide (2H18-CGRP, CGRP with 18 deteurium atoms), 0.1 M ammonium acetate, 0.5 M NaCl, and enzyme inhibitors (1 µg/mL of LAF-237, E-64, antipain, leupeptin, chymostatin, 1,10-phenanthroline, N-ethylmaleimide) and then immediately centrifuge at 3,000 × g for 10 min at 4 °C.
-
43.
Transfer the supernatant into a new tube and freeze at −80 °C for 24–72 h. After the centrifugation, the supernatant is clear and very light pink in color.
-
44.
Pre-condition a C18 Sep-Pak cartridge by washing with acetonitrile (3 × 6 mL) followed by 0.1% TFA in water (4 × 6 mL) using extraction manifold.
-
45.
Thaw the frozen samples on ice and apply 2 mL of each sample onto its own pre-conditioned C18 Sep-Pak cartridge.
▲ CRITICAL STEP The flow-through from the column is then added back to the column three times to ensure that all of the material bound to the column.
-
46.
Wash the Sep-Pak with 0.1% TFA in water (24 mL) and elute with a 70:30 mixture of acetonitrile/water containing 0.1% TFA (2 mL total) into Protein LoBind Tubes.
▲ CRITICAL STEP Allow the acetonitrile/water solution to flow through the Sep-Pak by gravity. After a few minutes, apply pressure using a piston to flush out the remaining solution.
-
47.
Dry the sample using a speed vacuum concentrator at 25 °C.
-
48.
Dissolve the sample in water with 0.1% formic acid (100 µL), centrifuge at 13,000 × g (10 min, 4 °C), and desalt 20 µL of this sample using a ZipTip as outlined in Box 1.
? TROUBLESHOOTING
Troubleshooting peptidomics can be found in Table 1
Table 1.
Troubleshooting table.
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 23 | Poor MALDI profiles (weak or no signals) during in vitro assay |
Reaction was not quenched properly |
Pipette the sample 10 to 20 times (Box 1 Step 2) to ensure proper mixing. Alternatively, vortex the samples after addition of 8 M GndHCl quenching solution. |
| 34 | Noisy quantitative data |
Reaction times were not carefully controlled |
Ensure that the reaction start and stop times are synchronized (Step 34) |
TIMING
-
Steps 1–9, Isolation of physiological peptides from tissue for peptidomics analysis: 1 d
Steps 10–13, Tissue peptidomics analysis: 1 d
Steps 14–21, Preparation of animal tissue lysates for in vitro lysate experiments: 3 h
Steps 22–23, In vitro lysate experiments with bioactive peptide: 2 h
Steps 24–27, MALDI analysis of in vitro lysate experiments: 3 h
Steps 28–30, LC-MS/MS analysis of in vitro lysate experiments: 1 d
Steps 31–35, Protease inhibitor assays and tissue proteome fractionation: 6 h
Steps 36–40 Degradation of bioactive peptide by recombinant protease: 1 d
Steps 41–48, Degradation of bioactive peptide in tissue from WT and KO mice (in vitro lysate assay vs. in vivo measurement): 1.5 d
Anticipated results (Conclusions)
This protocol details the approach that we have used to study the proteolysis of bioactive peptides. The identification of these pathways promises to educate the field about the functions of peptidases in mammals and in certain cases provide new opportunities for therapeutic intervention. This protocol can be applied to any peptide and the results demonstrated for PHI and CGRP (Fig. 3–7). Stage 1 and 2 should yield endogenous cleavage sites and candidate peptides, respectively, for any peptide. In particular, we recommend using MEROPS to predict peptidases and purchasing enzymes for testing which will greatly accelerate the identification of candidate enzymes. Stage three requires the development of a quantitative assay to measure changes in peptide abundance to test the relevance of a candidate peptidase. In addition, this stage also requires a model. A great option is to use a selective small molecule inhibitor, if available, which will avoid any issues with compensation63; however, most knockouts should provide the necessary information. Performing the experiments as described in this protocol will reveal the peptidases responsible for the endogenous proteolysis of bioactive peptides, which provides a solution to an important unmet challenge in the biological and biomedical sciences.
Acknowledgments
This work was supported by a Forris Jewitt Moore Fellowship sponsored by Amherst College (A.M.L.), a National Institutes of Health training grant GM007598 (A.M.L), Searle Scholar Award (A.S.), Burroughs Wellcome Fund Career Award in the Biomedical Sciences (A.S.), National Institutes of Health Grant DP2OD002374 (A.S.). A Korea Basic Science Institute (T33617) grant (Y.G.K.) and KIST Institutional Program (2E23720-13-053) grant (Y.G.K.)
Footnotes
Author Contributions. Experimental procedure development and assembly of the manuscript was performed by Y.G.K, A.M.L. and A.S.
Conflicting financial interests statement. The authors declare no competing financial interests.
References
- 1.Bliss M. The discovery of insulin. University of Chicago Press; 2013. [Google Scholar]
- 2.Kimball C, Murlin JR. Aqueous extracts of pancreas III. Some precipitation reactions of insulin. Journal of Biological Chemistry. 1923;58:337–346. [Google Scholar]
- 3.Bromer W, Sinn L, Behrens OK. The amino acid sequence of glucagon. V. Location of amide groups, acid degradation studies and summary of sequential evidence. Journal of the American Chemical Society. 1957;79:2807–2810. [Google Scholar]
- 4.Kastin A. Handbook of biologically active peptides. Academic Press; 2013. [Google Scholar]
- 5.Itakura K, et al. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977;198:1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- 6.Goeddel DV, et al. Expression in Escherichia coli of chemically synthesized genes for human insulin. Proceedings of the National Academy of Sciences. 1979;76:106–110. doi: 10.1073/pnas.76.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y—a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. 1982 doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- 8.Said SI, Mutt V. Polypeptide with broad biological activity: isolation from small intestine. Science. 1970;169:1217–1218. doi: 10.1126/science.169.3951.1217. [DOI] [PubMed] [Google Scholar]
- 9.Tatemoto K, Rökaeus Å, Jörnvall H, McDonald TJ, Mutt V. Galanin—a novel biologically active peptide from porcine intestine. FEBS letters. 1983;164:124–128. doi: 10.1016/0014-5793(83)80033-7. [DOI] [PubMed] [Google Scholar]
- 10.Strand FL. Neuropeptides. Wiley Online Library; 1999. [Google Scholar]
- 11.Steiner DF. The proprotein convertases. Current opinion in chemical biology. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 12.Marguet D, et al. Enhanced insulin secretion and improved glucose tolerance in mice lacking CD26. Proceedings of the National Academy of Sciences. 2000;97:6874–6879. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gradman AH, et al. Aliskiren, a novel orally effective renin inhibitor, provides dose-dependent antihypertensive efficacy and placebo-like tolerability in hypertensive patients. Circulation. 2005;111:1012–1018. doi: 10.1161/01.CIR.0000156466.02908.ED. [DOI] [PubMed] [Google Scholar]
- 14.Patchett AA, et al. A new class of angiotensin-converting enzyme inhibitors. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 15.Deacon CF, et al. Both subcutaneously and intravenously administered glucagon-like peptide I are rapidly degraded from the NH2-terminus in type II diabetic patients and in healthy subjects. Diabetes. 1995;44:1126–1131. doi: 10.2337/diab.44.9.1126. [DOI] [PubMed] [Google Scholar]
- 16.Xu H, Shields D. Prohormone processing in the trans-Golgi network: endoproteolytic cleavage of prosomatostatin and formation of nascent secretory vesicles in permeabilized cells. The Journal of cell biology. 1993;122:1169–1184. doi: 10.1083/jcb.122.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung LJ, Scheller RH. Peptide processing and targeting in the neuronal secretory pathway. Science. 1991;251:1330–1335. doi: 10.1126/science.2003219. [DOI] [PubMed] [Google Scholar]
- 18.Scamuffa N, Calvo F, Chretien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 19.Furuta M, et al. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proceedings of the National Academy of Sciences. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schalekamp MA, Derkx FH, van den Meiracker AH. Renin inhibitors, angiotensin converting enzyme inhibitors and angiotensin II receptor antagonists: relationships between blood pressure responses and effects on the renin-angiotensin system. Journal of hypertension Supplement : official journal of the International Society of Hypertension. 1992;10:S157–S164. [PubMed] [Google Scholar]
- 21.Kjems LL, Holst JJ, Volund A, Madsbad S. The influence of GLP-1 on glucose-stimulated insulin secretion: effects on beta-cell sensitivity in type 2 and nondiabetic subjects. Diabetes. 2003;52:380–386. doi: 10.2337/diabetes.52.2.380. [DOI] [PubMed] [Google Scholar]
- 22.Rachman J, Barrow B, Levy J, Turner R. Near-normalisation of diurnal glucose concentrations by continuous administration of glucagon-like peptide-1 (GLP-1) in subjects with NIDDM. Diabetologia. 1997;40:205–211. doi: 10.1007/s001250050664. [DOI] [PubMed] [Google Scholar]
- 23.Rosenblum JS, Kozarich JW. Prolyl peptidases: a serine protease subfamily with high potential for drug discovery. Curr Opin Chem Biol. 2003;7:496–504. doi: 10.1016/s1367-5931(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 24.Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nature reviews. Molecular cell biology. 2007;8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 25.Thornberry NA, Weber AE. Discovery of JANUVIA (Sitagliptin), a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. Curr Top Med Chem. 2007;7:557–568. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 26.Augeri DJ, et al. Discovery and preclinical profile of Saxagliptin (BMS-477118): a highly potent, long-acting, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes. J Med Chem. 2005;48:5025–5037. doi: 10.1021/jm050261p. [DOI] [PubMed] [Google Scholar]
- 27.Toide K, Okamiya K, Iwamoto Y, Kato T. Effect of a novel prolyl endopeptidase inhibitor, JTP-4819, on prolyl endopeptidase activity and substance P- and arginine-vasopressin-like immunoreactivity in the brains of aged rats. Journal of neurochemistry. 1995;65:234–240. doi: 10.1046/j.1471-4159.1995.65010234.x. [DOI] [PubMed] [Google Scholar]
- 28.Tatemoto K, Mutt V. Isolation and characterization of the intestinal peptide porcine PHI (PHI-27), a new member of the glucagon--secretin family. Proc Natl Acad Sci U S A. 1981;78:6603–6607. doi: 10.1073/pnas.78.11.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szecowka J, Tatemoto K, Mutt V, Efendic S. Interaction of a newly isolated intestinal polypeptide (PHI) with glucose and arginine to effect the secretion of insulin and glucagon. Life Sci. 1980;26:435–438. doi: 10.1016/0024-3205(80)90162-9. [DOI] [PubMed] [Google Scholar]
- 30.Szecowka J, Lins PE, Tatemoto K, Efendic S. Effects of porcine intestinal heptacosapeptide and vasoactive intestinal polypeptide on insulin and glucagon secretion in rats. Endocrinology. 1983;112:1469–1473. doi: 10.1210/endo-112-4-1469. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld MG, Amara SG, Evans RM. Alternative RNA processing: determining neuronal phenotype. Science (New York, NY) 1984;225:1315–1320. doi: 10.1126/science.6089345. [DOI] [PubMed] [Google Scholar]
- 32.Amara S, Arriza J, Leff S, Swanson L. Expression in brain of a messenger RNA encoding a novel neuropeptide homologous to calcitonin gene-related peptide. Science (New York, NY) 1985 doi: 10.1126/science.2994212. [DOI] [PubMed] [Google Scholar]
- 33.Brain S, Williams T, Tippins J, Morris H. Calcitonin gene-related peptide is a potent vasodilator. 1985 doi: 10.1038/313054a0. [DOI] [PubMed] [Google Scholar]
- 34.Ashina M, Bendtsen L, Jensen R, Schifter S, Olesen J. Evidence for increased plasma levels of calcitonin gene-related peptide in migraine outside of attacks. Pain. 2000;86:133–138. doi: 10.1016/s0304-3959(00)00232-3. [DOI] [PubMed] [Google Scholar]
- 35.Ho TW, et al. Randomized controlled trial of an oral CGRP receptor antagonist, MK-0974, in acute treatment of migraine. Neurology. 2008;70:1304–1312. doi: 10.1212/01.WNL.0000286940.29755.61. [DOI] [PubMed] [Google Scholar]
- 36.Lassen LH, et al. CGRP may play a causative role in migraine. Cephalalgia. 2002;22:54–61. doi: 10.1046/j.1468-2982.2002.00310.x. [DOI] [PubMed] [Google Scholar]
- 37.Paone DV, et al. Potent, orally bioavailable calcitonin gene-related peptide receptor antagonists for the treatment of migraine: discovery of N-[(3R,6S)-6-(2,3-difluorophenyl)-2-oxo-1- (2,2,2-trifluoroethyl)azepan-3-yl]-4- (2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin- 1-yl)piperidine-1-carboxamide (MK-0974) Journal of medicinal chemistry. 2007;50:5564–5567. doi: 10.1021/jm070668p. [DOI] [PubMed] [Google Scholar]
- 38.Salvatore CA, et al. Pharmacological Characterization of MK-0974 [N-[(3R,6S)-6-(2,3-Difluorophenyl)-2-oxo-1-(2,2,2-trifluoroethyl)azepan-3-yl]-4-(2-oxo-2,3-dihydro-1H–imidazo[4,5-b]pyridin-1-yl)piperidine-1-carboxamide], a Potent and Orally Active Calcitonin Gene-Related Peptide Receptor Antagonist for the Treatment of Migraine. Journal of Pharmacology and Experimental Therapeutics. 2007;324:416–421. doi: 10.1124/jpet.107.130344. [DOI] [PubMed] [Google Scholar]
- 39.Nolte WM, Tagore DM, Lane WS, Saghatelian A. Peptidomics of prolyl endopeptidase in the central nervous system. Biochemistry. 2009;48:11971–11981. doi: 10.1021/bi901637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eng JK, McCormack AL, Yates Iii JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 41.Kim YG, Lone AM, Nolte WM, Saghatelian A. Peptidomics approach to elucidate the proteolytic regulation of bioactive peptides. Proc Natl Acad Sci U S A. 2012;109:8523–8527. doi: 10.1073/pnas.1203195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Gibson SJ, et al. Calcitonin gene-related peptide immunoreactivity in the spinal cord of man and of eight other species. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1984;4:3101–3111. doi: 10.1523/JNEUROSCI.04-12-03101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Grevès P, Andersson K, Silberring J. Isolation and identification of CGRP C-terminal fragments in the rat spinal cord. Neuropeptides. 1997;31:19–23. doi: 10.1016/s0143-4179(97)90014-7. [DOI] [PubMed] [Google Scholar]
- 46.Katayama M, et al. Catabolism of calcitonin gene-related peptide and substance P by neutral endopeptidase. Peptides. 1991;12:563–567. doi: 10.1016/0196-9781(91)90102-u. [DOI] [PubMed] [Google Scholar]
- 47.Le Greves P, Nyberg F, Hokfelt T, Terenius L. Calcitonin gene-related peptide is metabolized by an endopeptidase hydrolyzing substance P. Regulatory peptides. 1989;25:277–286. doi: 10.1016/0167-0115(89)90176-6. [DOI] [PubMed] [Google Scholar]
- 48.Tam EK, Caughey GH. Degradation of airway neuropeptides by human lung tryptase. American journal of respiratory cell and molecular biology. 1990;3:27–32. doi: 10.1165/ajrcmb/3.1.27. [DOI] [PubMed] [Google Scholar]
- 49.Tinoco AD, et al. A peptidomics strategy to elucidate the proteolytic pathways that inactivate peptide hormones. Biochemistry. 2011;50:2213–2222. doi: 10.1021/bi2000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Duckworth WC, Heinemann MA, Kitabchi AE. Purification of insulin-specific protease by affinity chromatography. Proc Natl Acad Sci U S A. 1972;69:3698–3702. doi: 10.1073/pnas.69.12.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray K, Hines CS, Rodgers DW. Mapping sequence differences between thimet oligopeptidase and neurolysin implicates key residues in substrate recognition. Protein science : a publication of the Protein Society. 2002;11:2237–2246. doi: 10.1110/ps.0216302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, Brophy E. Rat intestinal brush border membrane peptidases. I. Solubilization, purification, and physicochemical properties of two different forms of the enzyme. Journal of Biological Chemistry. 1976;251:3199–3205. [PubMed] [Google Scholar]
- 54.Alvarez-Manilla G, et al. Tools for glycoproteomic analysis: size exclusion chromatography facilitates identification of tryptic glycopeptides with N-linked glycosylation sites. Journal of proteome research. 2006;5:701–708. doi: 10.1021/pr050275j. [DOI] [PubMed] [Google Scholar]
- 55.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature protocols. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slebos RJ, et al. Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. Journal of proteome research. 2008;7:5286–5294. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buckley SJ, Collins PJ, O’Connor BF. The purification and characterisation of novel dipeptidyl peptidase IV-like activity from bovine serum. The international journal of biochemistry & cell biology. 2004;36:1281–1296. doi: 10.1016/j.biocel.2003.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic acids research. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic acids research. 2012;40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farris W, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng TS, et al. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nature medicine. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- 62.Noone D, Howell A, Collery R, Devine KM. YkdA and YvtA, HtrA-Like Serine Proteases inBacillus subtilis, Engage in Negative Autoregulation and Reciprocal Cross-Regulation of ykdA and yvtAGene Expression. Journal of bacteriology. 2001;183:654–663. doi: 10.1128/JB.183.2.654-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chemistry & biology. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Stengel A, et al. The RAPID method for blood processing yields new insight in plasma concentrations and molecular forms of circulating gut peptides. Endocrinology. 2009;150:5113–5118. doi: 10.1210/en.2009-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coin I, Beyermann M, Bienert M. Solid-phase peptide synthesis: from standard procedures to the synthesis of difficult sequences. Nature protocols. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]