Abstract
Previous and ongoing studies have incriminated bats as reservoirs of several emerging and re-emerging zoonoses. Most of these studies, however, have focused on viral agents and neglected important bacterial pathogens. To date, there has been no report investigating the prevalence of Bartonella spp. in bats and bat flies from Nigeria, despite the fact that bats are used as food and for cultural ritual purposes by some ethnic groups in Nigeria. To elucidate the role of bats as reservoirs of bartonellae, we screened by molecular methods 148 bats and 34 bat flies, Diptera:Hippoboscoidea:Nycteribiidae (Cyclopodia greeffi) from Nigeria for Bartonella spp. Overall, Bartonella spp. DNA was detected in 76 out of 148 (51.4%) bat blood samples tested and 10 out of 24 (41.7%) bat flies tested by qPCR targeting the 16S–23S internal transcribed spacer (ITS) locus. Bartonella was isolated from 23 of 148 (15.5%) bat blood samples, and the isolates were genetically characterized. Prevalence of Bartonella spp. culture-positive samples ranged from 0% to 45.5% among five bat species. Micropterus spp. bats had a significantly higher relative risk of 3.45 for being culture positive compared to Eidolon helvum, Epomophorus spp., Rhinolophus spp., and Chaerephon nigeriae. Bartonella spp. detected in this study fall into three distinct clusters along with other Bartonella spp. isolated from bats and bat flies from Kenya and Ghana, respectively. The isolation of Bartonella spp. in 10.0–45.5% of four out of five bat species screened in this study indicates a widespread infection in bat population in Nigeria. Further investigation is warranted to determine the role of these bacteria as a cause of human and animal diseases in Nigeria.
Key Words: : Bartonella, Bats, Vector, Reservoir host, Flies
Introduction
The order Chiroptera is the most diverse and geographically distributed mammalian taxon found on all continents except Antarctica (Schipper et al. 2008). They are the second-most speciose group of mammals after Rodentia (Simmons 2005). In Nigeria, bats form an important part of cultural and traditional ceremonies among some ethnic groups and serve as food and a protein source (Billeter et al. 2012). Bats are unique among mammals in their capability to fly and cover long distances during seasonal migrations and in their potential for living in diverse colonial populations. These characteristics together with the long life span of some species and the ability to inhabit a multitude of diverse ecological niches make bats among the most successful species adapting and surviving in different climates on earth. As a result, there is a renewed global interest in bats as potential reservoir hosts and vectors of zoonotic pathogens (Calisher et al. 2006, Wong et al. 2007, Kuzmin et al. 2011, Wang et al. 2011).
Multiple studies have highlighted the potential role of bats as natural reservoirs for numerous pathogens including viruses and bacteria (Hanson 1970, Messenger et al. 2003, Loftis et al. 2005, Matthias et al. 2005, Gibson et al. 2005, Calisher et al. 2006, Reeves et al. 2007, Evans et al. 2009, Bai et al. 2011, Lin et al. 2012). Research studies have confirmed the presence of Bartonella spp. in bats from Peru, Guatemala, Kenya, and the United Kingdom (Concannon et al. 2005, Kosoy et al. 2010, Bai et al. 2011, 2012), and in bat ectoparasites from Egypt, the United States, and Ghana (Loftis et al. 2005, Reeves et al. 2005, 2007, Billeter et al. 2012). However, knowledge on bat bartonellae is preliminary and further research is warranted. Moreover, no study on Bartonella spp. in bats and bat flies from Nigeria has been carried out to date. The objectives of this study were therefore to: (1) Screen bats and bat flies from Nigeria for bartonellae, and determine the prevalence of Bartonella spp. in bat populations from Nigeria and (2) genetically characterize bat-associated Bartonella spp. from Nigeria and evaluate their genetic diversity.
Materials and Methods
Ethics statement
This study and sampling protocol were approved by the Institutional (National Veterinary Research Institute [NVRI], Vom Nigeria), Animal Care and Use Committee (approval number AEC/03/01/13). Oral permission to place hand nets in the study area was granted by village heads and residents. Animals were treated in a humane manner and in accordance with authorizations and guidelines for Ethical Conduct in the Care and Use of Nonhuman Animals in Research of the American Psychological Association (APA) for use by scientists working with nonhuman animals (American Psychological Association Committee on Animal Research and Ethics) in 2010.
Study area
Blood and ectoparasites from bats were collected from Bauchi (10°18′57″N, 09°50′39″E) and Gboko (7°19′30″N, 9°0′18″E) towns, Nigeria (Fig. 1). Sampling sites were chosen on the basis of available information on bat roosts and foraging sites.
FIG. 1.
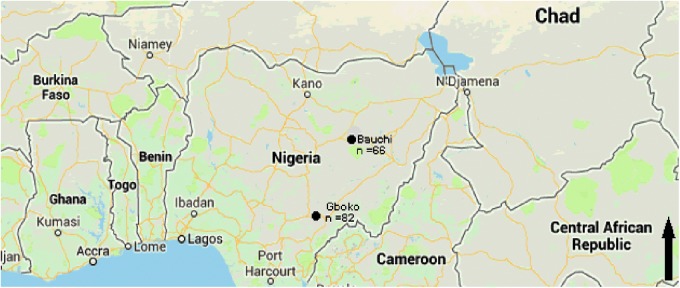
Map of Nigeria, West Africa showing the two sampling sites, Bauchi and Gboko, and the number (n) of bats sampled at each site. Color images available online at www.liebertpub.com/vbz
Samples collection
Bats were collected by hand or mist nets around roosts or in locations of nocturnal foraging. In each sampling site, six mist nets (9×3 meters each) were set up for 18–24 h, and checked every hour. Both adult and subadult bats were randomly selected; however, preference was given to adults. Trapped bats were collected and inspected for the presence of ectoparasites. When present, the ectoparasites from each bat were collected and placed in labeled vials containing 70% ethanol.
The bats were then morphologically identified to genus and when possible to species level (Bergmans 1988) by field experts (Gablong P. Litwat, NVRI Outstation Laboratory, Bauchi, Nigeria, and Nyor Gelahan, College of Agriculture, Yandev, Nigeria) and exsanguinated by cardiac puncture following anesthesia using isoflurane gas. About 0.5–2 mL of blood (depending on the bat size) was collected from each bat into EDTA tubes and labeled. Blood samples were transported from the field in cool boxes to the Parasitology Laboratory, NVRI, Vom, and stored at −20°C until shipped to the Koret School of Veterinary Medicine in Israel for further analysis.
Identification of bat flies
Initial examination of each specimen was done at 10× and 30× with a Meiji stereo microscope with the specimen in an excavated glass block and 70% ethanol and quickly keyed using Theodor (1967) to the genus Cyclopodia. Further detailed examination was performed at 60× of the chaetotaxy (bristles) of the female abdomen. Females are distinctive and are easy to identify. Males, however, are difficult, needing very careful examination of male claspers, which on some specimens, needed to be levered gently out of the male abdomen. All materials were further compared to the Cyclopodia specimens in the entomology collection of the Natural History Museum, London, which confirmed the initial identification of C. greeffi.
Bartonella spp. culture from bat blood
Fifty microliters of thawed EDTA–blood was plated onto chocolate agar and sealed with a shrink seal. The plates were incubated at 35°C and 5% carbon dioxide. Bacterial growth was monitored at the end of the first week and then on alternate days for up to 45 days. Bacterial colonies were presumptively identified as Bartonella spp. based on their morphology. Suspected Bartonella colonies were randomly selected from the original agar plate, streaked onto secondary chocolate agar plates, and incubated at the same conditions until sufficient growth was observed. Pure cultures were harvested and stored in 10% glycerol at −80°C.
DNA extraction from Bartonella cultures
A heavy suspension of the microorganisms was heated for 10 min at 95°C followed by 1 min of centrifugation at 8000 rpm to precipitate the lysed bacteria. The supernatant containing the genomic DNA to be used as DNA template in the PCR amplification assay was then pipetted into a clean 1.5-mL centrifuge tube.
DNA extraction from bat flies
Prior to DNA extraction, the bat flies were removed from the ethanol vials and rinsed in sterile phosphate-buffered saline (PBS). Bat flies were treated individually, except in cases where more than one fly was identified on a host (five bats), and then treated as a pool of two to four flies. The flies were first triturated using a sterile scalpel blade and manually homogenized using a plastic pestle. DNA extraction was performed using a DNeasy blood and tissue kit (QIAGEN, GmbH Germany) according to the manufacturer's instructions.
DNA extraction from blood of bats
DNA from bats' blood was extracted using the DNeasy blood and tissue kit (QIAGEN, GmbH Germany) according to the manufacturer's instructions. DNA quality and concentrations were assessed with a NanoVue spectrometer (GE Healthcare, UK Limited).
Real-time PCR amplification of the rpoB gene fragment and 16S–23S internal transcribed space locus
Initial detection of Bartonella spp. was performed using real-time PCR analysis targeting the rpoB and 16S–23S internal transcribed spacer (ITS) region in genomic DNA extracted directly from blood of bats and bat flies using oligonucleotide primers listed in Table 1. Real-time PCR was carried out using the StepOne (Applied Biosystems) real-time system under the following conditions, one cycle of 95°C for 15 min followed by 50 cycles of 15 s at 95°C, 30 s at 57°C, and 10 s at 76°C with data collection in the FAM channel. The PCR was performed in 20- μL reaction volumes containing 3 μL of DNA, 1 μL of 10 mM of each primer, 10 μL ABsolute Blue dye (Thermo Scientific, Surrey, UK), and 5 μL ultrapure PCR water (Thermo Scientific, Surrey, UK). Bartonella henselae DNA extracted from blood of a naturally infected cat was used as a positive control, and DNA from a cat negative by PCR for Bartonella spp. was used as a negative control. One sample containing all the ingredients of the reaction except DNA was used as a nontemplate control (NTC). All of the controls were included in each PCR reaction to evaluate the presence of appropriate melting curves and possible contamination.
Table 1.
Oligonucleotide Primer Pairs Used in Polymerase Chain Reaction Amplifications for the Detection of Bartonella Species in Bats and Bat Flies
| Primer pairs | 5′-primer sequence-3′ | Target locus | Amplicon (bp) | Reference |
|---|---|---|---|---|
| BhCS781.p BhCS1137.n CSH1fa BhCS1137.na |
GGGGACCAGCTCATGGTGG AATGCAAAAAGAACAGTAAACA GCGAATGAAGCGTGCCTAAA AATGCAAAAAGAACAGTAAACA |
Citrate synthase gene (gltA) |
379 350 |
Norman et al. 1995 Birtles and Raoult 1996 |
| 600f 800r |
CGTGCAACAGAAGATTTAGATC CGATTCGCATCATCATTTTC |
RNA polymerase beta subunit gene (rpoB) | 191 | Morick et al. 2009 |
| 1400f 2300r |
CGCATTGGCTTACTTCGTATG GTAGACTGATTAGAACGCTG |
RNA polymerase beta subunit gene (rpoB) | 830 | Renesto et al. 2001 |
| 321s 493as |
AGATGATGATCCCAAGCCTTCTGG TGAACCTCCGACCTCACGCTTATC |
16S-23S (ITS) | 208 | Maggi et al. 2005 |
Primer pair used for amplification of gltA sequences from DNA extracted from bat blood and flies.
Conventional PCR amplification of the gltA and rpoB fragments
PCR amplifications were performed using oligonucleotide primers (Table 1) to amplify a 379-bp fragment of the gltA and a 830-bp fragment of the rpoB genes of Bartonella spp. under the cycling conditions, as previously described (Norman et al. 1995, Birtles and Raoult 1996, Renesto et al. 2001). PCR was performed using Syntezza PCR-Ready High Specificity (Syntezza Bioscience, Israel) PCR kit using a programmable conventional thermocycler (Biometra, Goettingen, Germany) as described previously (Kamani et al. 2013).
PCR products were electrophoresed on 1.5% agarose gel stained with ethidium bromide and checked under ultraviolet (UV) light for the size of amplified fragments by comparison to a 100-bp DNA molecular weight marker.
Sequencing
PCR products of appropriate size were purified using ExoSap (New England Biolabs Inc., USA) according to the manufacturer's instructions. Sequencing was performed in both directions (forward and reverse) at the Center for Genomic Technologies, Hebrew University of Jerusalem, Israel. DNA sequences obtained were evaluated with MEGA 5.2 software and compared for similarity to sequences deposited in GenBank, using the BLAST program hosted by the National Center for Biotechnology Information (NCBI), National Institutes of Health, Bethesda, MD (www.ncbi.nlm.nih.gov/BLAST). Selected sequences were deposited in GenBank and were assigned the following accession numbers KF418806–KF418812 and KF533081–KF533103.
Results
Bats and bat flies
From November, 2012, to March, 2013, 148 bats belonging to five species were collected for this study. Eidolon helvum (the straw-colored fruit bat) constituted 53.4% (79/148) of bats captured, followed by Epomophorus spp. (the epauletted fruit bat) (20.3%), Chaerephon nigeriae (the free-tailed bat) (10.8%), Rhinolophus spp. (the horseshoe bat) (8.1%), and Micropterus spp. (the dwarf epauletted fruit bat) (7.4%). All of the bat species captured in this study were found living in mixed colonies except C. nigeriae, which was found as a single colony in the ceiling of a school building. A total of 34 (19 male and 15 female) bat flies belonging to the family Nycteribiidae were collected from 24 (16%) of the bats sampled and were all identified as C. greeffi, Karsch, 1884. Bat flies were removed from 11 E. helvum, three Micropterus spp., and five each of Rhinolophus spp. and Epomophorus spp.
Bartonella species DNA in bats' blood and bat flies as detected by PCR
When tested by ITS locus and rpoB gene-based qPCR, 76/148 (51.4%) and 28/148 (18.9%) bat blood samples were positive, respectively. However, only 14/148 (9.5%) of the same samples were found positive for the Bartonella gltA gene by conventional PCR. None of the blood samples was positive for the rpoB gene by conventional PCR (Table 2). Bartonella spp. DNA was detected in 10 out of 24 (41.7%) and 7 out of 24 (29.2%) bat flies by real-time PCR targeting the 16S–23S ITS locus and rpoB gene, respectively. Similarly, both gltA and rpoB genes were detected in seven out of 24 (29.2%) of the bat fly samples screened in the study (Table 2).
Table 2.
Bartonella DNA in Nigerian Bats and Bat Flies According to Their Species
| Bat species/bat fly species | Samples tested | Number positive by real- time PCR | Number positive by conventional PCR | ||
|---|---|---|---|---|---|
| Bat species | ITS (205bp) | rpoB (190 bp) | gltA (350 bp) | rpoB (830 bp) | |
| Eidolon helvum | 79 | 44 | 14 | 9 | 0 |
| Epomorphorus spp. | 30 | 16 | 9 | 3 | 0 |
| Micropterus spp. | 11 | 6 | 2 | 1 | 0 |
| Chaerephon nigeriae | 16 | 2 | 0 | 0 | 0 |
| Rhinolophus spp. | 12 | 8 | 3 | 1 | 0 |
| Total | 148 | 76 | 28 | 14 | 0 |
| Bat fly species | |||||
| Cyclopodia greeffi | 24 | 10 | 7 | 7 | 7 |
ITS, internal transcribed spacer.
Several of the bats infested with bat flies at the time of sampling were positive for Bartonella spp. DNA targeting 16S—23S ITS locus, E. helvum (7/11, 63.6%), Micropterus spp. (2/3, 66.7%), Rhinolophus spp. (3/5, 60%), and Epomophorus spp. (1/5, 20%). Furthermore, of the infested bats that were positive for Bartonella spp. DNA 16S–23S ITS locus, bat flies removed from 71.4% of E. helvum and 100% of both Micropterus spp. and Rhinolophus spp. were also positive. However, the bat flies removed from the only infested and positive Epomophorus spp. was negative for Bartonella spp. DNA targeting 16S–23S ITS locus.
Bartonella isolation from bat blood
Bartonella colonies were cultured from 23 of 148 (15.5%) bat blood samples. Prevalence of Bartonella spp. culture-positive samples ranged from 0% to 45.5% among the five bat species. Micropterus spp. bats had the highest prevalence (45.5%) of Bartonella spp. culture positive, followed by Rhinolophus spp. (25%), E. helvum (15.2%), and Epomophorus spp. (10%). None of the 16 C. nigeriae bats was Bartonella spp. culture positive. Micropterus spp. bats had a significantly higher relative risk of 3.45 for being Bartonella spp. culture positive compared to other bat species sampled (Table 3). No significant difference was noticed in the prevalence of Bartonella spp. culture-positive bats between the two different sampling sites (14.8% vs. 15.9%; χ2=0.03, p=0.87, 95% confidence interval [CI]=0.36–2.33).
Table 3.
Prevalence of Culture-Positive Bartonella spp. among Bat Species from Nigeria
| Bat species | Number of samples cultured | Number of positive samples | Percent | Relative riska(CI 95%) | p value |
|---|---|---|---|---|---|
| Eidolon helvum | 79 | 12 | 15.2 | 0.95 (0.45–2.02) | 0.90 |
| Epomophorus spp. | 30 | 3 | 10.0 | 0.59 (0.19–1.85) | 0.37 |
| Micropterus spp. | 11 | 5 | 45.5 | 3.45 (1.59–7.53) | 0.0018b |
| Chaerephon nigeriae | 16 | 0 | 0 | Not definedc | 1.27 |
| Rhinolophus spp. | 12 | 3 | 25.0 | 1.709 (0.58–4.90) | 0.33 |
Relative risk was calculated by comparing proportions of positive for each bat species with the number of positive for all other bat species.
Statistically significant relative risk.
Relative risk is less than 1 but is not defined because of 0 in one of the cells.
CI, confidence interval.
Distribution and diversity of Bartonella spp. in bats and bat flies based on citrate synthase (gltA) sequences
Two gltA genotypes of Bartonella spp. with 98%–100% identity to Bartonella sp. E3-106 (Eidolon helvum, Kenya) and Bartonella sp. EW-111 (Eidolon helvum, Kenya) were detected and deposited in the GenBank from the three bat genera, E. helvum, Epomophorus s., and Micropterus s. sampled in Bauchi state. In samples collected from four bat genera in Gboko, Benue state, three Bartonella spp. gltA genotypes with 98%–100% identity to Bartonella sp. E3-106, Bartonella sp. E-124 (Eidolon helvum, Kenya), and Bartonella sp. EW-111 were detected and deposited in the GenBank (Table 4).
Table 4.
Distribution and Diversity of Bartonella Strains from Bats and Bartonella Spp. from Bat Flies, Nigeria Based on Citrate Synthase (Glta) Gene Fragments
| Species | Place captured | GenBank first match (no.) | % identity |
|---|---|---|---|
| Bat species | |||
| Eidolon helvum | Bauchi | Bartonella sp. E3-106 (4) | 99 |
| Eidolon helvum | Bauchi | Bartonella sp. EW-111 (1) | 99 |
| Epomophorus spp. | Bauchi | Bartonella sp. EW-111 (1) | 99 |
| Micropterus spp. | Bauchi | Bartonella sp. EW-111 (1) | 100 |
| Micropterus spp. | Bauchi | Bartonella sp. EW-111 (1) | 99 |
| Micropterus spp. | Bauchi | Bartonella sp. EW-111 (1) | 98 |
| Eidolon helvum | Gboko | Bartonella sp. EW-111 (3) | 99 |
| Eidolon helvum | Gboko | Bartonella sp. EW-111 (2) | 98 |
| Eidolon helvum | Gboko | Bartonella sp. E3-106 (1) | 99 |
| Epomophorus spp. | Gboko | Bartonella sp. E3-106 (1) | 99 |
| Epomophorus spp. | Gboko | Bartonella sp. E3-106 (1) | 98 |
| Micropterus spp. | Gboko | Bartonella sp. E3-106 (1) | 99 |
| Micropterus spp. | Gboko | Bartonella sp. E-124 (2) | 98 |
| Rhinolophus spp. | Gboko | Bartonella sp. E3-106 (2) | 99 |
| Rhinolophus spp. | Gboko | Bartonella sp. EW-111 (1) | 99 |
| Bat flies | |||
| Cyclopodia greeffi | Bauchi | Bartonella sp. E3-106 (1) | 97 |
| Cyclopodia greeffi | Gboko | Bartonella sp. E3-106 (3) | 99 |
| Cyclopodia greeffi | Gboko | Bartonella sp. E-124 (2) | 97 |
| Cyclopodia greeffi | Gboko | Bartonella sp. E-124 (1) | 99 |
In both study areas, Bartonella genotypes with 98%–100% identity with Bartonella sp. E3-106 and Bartonella sp. EW-111 in the GenBank were detected in the Bartonella spp. culture-positive bat blood examined in this study. In addition, a Bartonella genotype with 97%–99% identity to Bartonella sp. E-124 was also detected in Micropterus spp. bats and C. greeffi flies captured in Gboko, Benue state, Nigeria (Table 4).
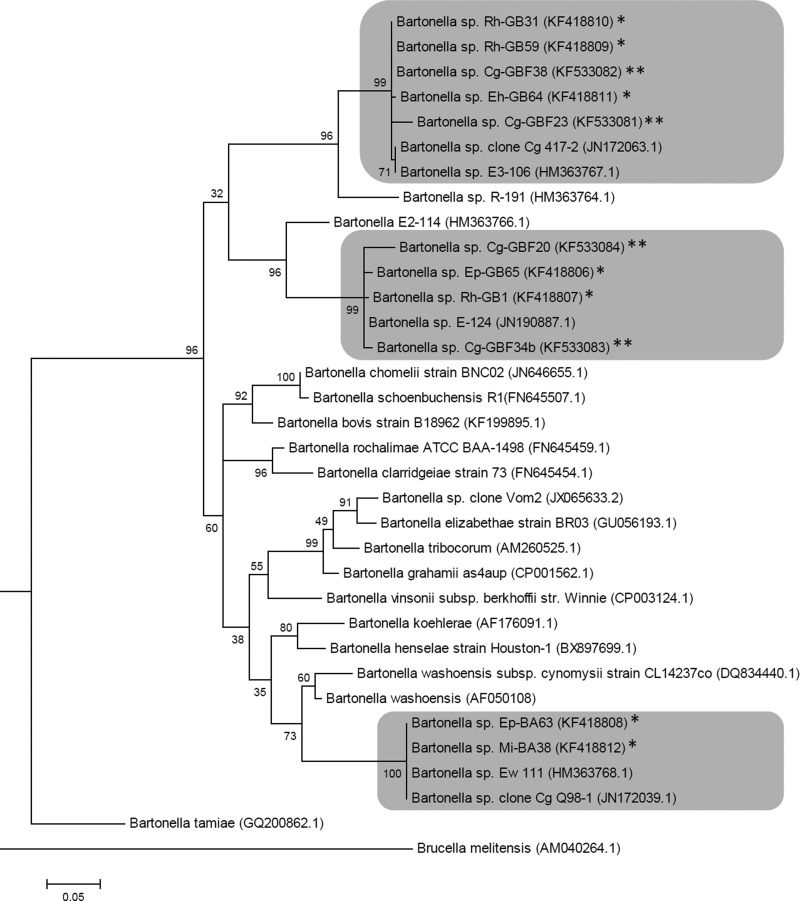
Phylogenetic analysis of Bartonella spp. detected in this study shows that they fall into three distinct clusters along with other Bartonella genotypes isolated from bats and bat flies from Kenya and Ghana, respectively. The first cluster consists of Bartonella spp. genotypes cultured from blood of E. helvum, Rhinolophus spp. bats, and C. greeffi bat flies along with Bartonella sp. E3-106 isolated from bats from Kenya and Bartonella sp. clone Cg 417-2 from bat flies, Ghana. The second cluster is made up of Bartonella genotypes isolated from the blood of Epomophorus spp. and Rhinolophus spp. bats and C. greeffi bat flies along with Bartonella sp. E-124 from bats in Kenya. The third cluster consisted of Bartonella spp. cultured from blood of Epomophorus spp. and Micropterus spp. along with Bartonella EW-111 isolated from bats in Kenya and Bartonella sp. clone Cg Q98-1 isolated from bat flies, Ghana (Fig. 2).
FIG. 2.
Maximun likelihood phylogenetic tree of partial (322 bp) gltA-Bartonella sequences obtained from bats (*) and their nycteribiids (**) from Nigeria, common Bartonella species, and Bartonella genotypes detected from bats and bat flies from Africa (sequences from GenBank database), with accession numbers in brackets. Brucella melitensis gltA sequence was used as an outgroup. Gray boxes indicate clusters that are phylogenetically closely related.
Discussion
Bats represent about 20% of all mammalian species (Calisher et al. 2006) and are acknowledged to harbor several pathogenic organisms (Gardner et al 1987, Bennett, 2006, van der Poel et al. 2006, Schneider et al. 2009, Dzikwi et al 2010). The present study investigated the presence, prevalence, and genetic diversity of Bartonella spp. in five bat species and one bat fly species from Nigeria, West Africa. A total of 132 fruit-eating bats belonging to four genera constituted 89.2% of the study population, while the remaining 16 bats belonged to an insectivorous bat species (C. nigeriae). Previous reports documented the presence of bartonellae in bats from Kenya (Kosoy et al. 2010) and bat flies from Ghana, Western Africa (Billeter et al. 2012). To the best of our knowledge, this is the first report of Bartonella spp. detection and isolation in bats and bat flies from Nigeria.
The isolation of Bartonella spp. in 10%–45.5% of four out of five bat species in this study is an indication of a high prevalence of this bacterium in bat communities in Nigeria. The mean prevalence of 15.5% Bartonella spp. culture-positive bats reported in this study is lower than the 33% reported in Guatemala (Bai et al. 2011), 30.2% reported from Kenya, eastern Africa (Kosoy et al. 2010), and 24.1% in Peru (Bai et al. 2012), but closer to 18% from Europe (Gardner et al. 1987). Gardner et al. (1987) initially reported the organisms detected in blood smears in European samples as Grahamella, but they are now regarded as Bartonella spp., and are higher than the 8.3% reported in United Kingdom bats (Concannon et al. 2005). It is noteworthy that the prevalence found in this study varied greatly (0%–45.5%) between bat species. Greater differences (0%–88%) were previously reported in Kenyan bat species (Kosoy et al. 2010), Guatemalan bat species (05–90%) (Bai et al. 2011), and Peruvian bat species (0%–100%) (Bai et al. 2012). In our study, Micropterus spp. had the highest prevalence (45.5%) with a statistically significant increased risk of Bartonella spp. infection compared to the other bat species. The prevalence of 15.2% of Bartonella spp. in E. helvum bats in this study is lower than that of 26% reported in Kenya; however, both studies showed that this species is at lower risk of Bartonella spp. infection compared to other bat species. We report a prevalence of 25% and 10% in Rhinolophus spp. and Epomophorus spp., respectively, as opposed to 0% reported for both species in bats from Kenya (Kosoy et al. 2010).
The high prevalence of Bartonella infection among bats has been attributed to their lifestyle, the colonial structure of their populations, the close physical contact between individuals, the aggressive interactions, and the typically heavy ectoparasite infestations that probably contribute to the frequent transmission of Bartonella spp. among individual animals (Kosoy et al. 2010). The insectivorous bat C. nigeriae is a locally abundant species, normally found in ceiling roofs of residential and public buildings such as schools, hospitals, and churches; these bats were all culture negative for Bartonella spp., and only two out of 16 bats belonging to this species were found positive by real-time PCR. This finding highlights the need to screen samples by more than one diagnostic method. The low positive prevalence results in C. nigeriae can be attributed to the existence of this species in a single colony without cohabitation and intermingling with other bat species as in the case of fruit-eating bats, as was shown in this study.
Bartonella spp. DNA was detected in 29.2% (7/24) C. greeffi flies removed from bats in this study. This rate of infection is much lower than that of 66.4% reported for the same bat fly species removed from E. helvum bats from Ghana (Billeter et al. 2012), a neighboring country to Nigeria. The lower prevalence found in our study may be due to the low number of flies tested in this study that could be a nonrepresentative sample size or due to differences between the screening tests. Bartonella spp. isolated in this study formed three distinct clusters along with Bartonella spp. isolated from bats from Africa and deposited in the GenBank (Fig. 2), suggesting apparent congruence between the phylogeny of Bartonella and their hosts in Africa (Bai et al. 2012).
Conclusions
In conclusion, a high percentage of bats and bat flies were found positive for Bartonella spp. Because the bats sampled in this study were captured in close proximity to human habitations, and due to the fact that bats serve as a protein source or are used for cultural and ritual purposes in some Nigerian communities, there is a need to pay close attention to the role of bats in the epidemiology of Bartonella infections in humans and animals in Nigeria. Although, to date, there is no clinical evidence available to suggest whether strains of Bartonella spp. found in bats globally are associated with human illness (Kosoy et al. 2010), there is a need for a large-scale surveillance of Bartonella spp. in bats across Africa to elucidate the potential role of African bats in public and veterinary health.
Acknowledgments
The authors gratefully acknowledge Oche D. Awulu, Nyor Gelahan, and Iber Christopher (College of Agriculture, Yandev) and Atuman Y. Joel and Gablong Pekap Litwat (NVRI Out Station Laboratory, Bauchi) for assistance during field sampling and identification of the bats. The technical assistance of Yaarit Nachum-Biala of the Koret School of Veterinary Medicine Rehovot, Israel, during the analysis of samples is equally acknowledged. We also thank Dr Duncan Sivell, Curator, Diptera at the Natural History Museum, South Kensington, London for access to the Museum's collections and for help and support in this endeavor.
Author Disclosure Statement
No competing financial interests exist.
References
- Bai Y, Kosoy M, Recuenco S, Alvarez D, et al. Bartonella spp. in bats, Guatemala. Emerg Infect Dis 2011; 17:1269–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Recuenco S, Gilbert AT, Osikowicz LM, et al. Prevalence and diversity of Bartonella spp. in bats in Peru. Am J Trop Med Hyg 2012; 87:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. Bats and human emerging diseases. Epidemiol Infect 2006; 134:905–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans W. Taxonomy and biogeography of African fruit bats (Mammalia, Megachiroptera). 1. General introduction; material and methods; results: the genus Epomophorus Bennett, 1836. Beaufortia 1988; 38:75–146 [Google Scholar]
- Billeter SA, Hayman DTS, Peel AJ, Baker K, et al. Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology 2012; 139:324–329 [DOI] [PubMed] [Google Scholar]
- Birtles RJ, Raoult D Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int Journ Syst Bacteriol 1996. ; 46:891–897 [DOI] [PubMed] [Google Scholar]
- Calisher CH, Childs JE, Field HE, Holmes KV, et al. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev 2006; 19:531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ. Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology 2005; 131:489–496 [DOI] [PubMed] [Google Scholar]
- Dzikwi AA, Kuzmin II, Umoh JU, Kwaga JKP, et al. Evidence of Lagos bat virus circulation among Nigerian Fruit Bats. J Wildlife Dis 2010; 46:267–271 [DOI] [PubMed] [Google Scholar]
- Evans NJ, Bown K, Timofte D, Simpson VR, et al. Fatal borreliosis in bat caused by relapsing fever spirochete, United Kingdom. Emerg Infect Dis 2009; 15:1331–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RA, Molyneux DH, Stebbings RE. Studies on the prevalence of haematozoa of British bats. Mammal Rev 1987; 17:75–80 [Google Scholar]
- Gibson KE, Rikihisa Y, Zhang C, Martin C. Neorickettsia risticii is vertically transmitted in the trematode Acanthatrium oregonense and horizontally transmitted to bats. Environ Microbiol 2005; 7:203–212 [DOI] [PubMed] [Google Scholar]
- Hanson AW. Isolation of spirochaetes from primates and other mammalian species. Br J Vener Dis 1970; 46:303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamani J, Morick D, Mumcuoglu KY, Harrus S. Prevalence and diversity of Bartonella species in commensal rodents and ectoparasites from Nigeria, West Africa. PLoS Negl Trop Dis 2013; 7:e2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy M, Bai Y, Lynch T, Kuzmin IV, et al. Bartonella spp. in bats, Kenya. Emerg Infect Dis 2010; 16:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin IV, Bozick B, Guagliardo SA, Kunkel R, et al. Bats, emerging infectious diseases, and the rabies paradigm revisited. Emerg Health Threats J 2011; 4:7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Hsu YM, Chomel BB, Lin LK, et al. Identification of novel Bartonella spp. in bats and evidence of Asian gray shrew as a new potential reservoir of Bartonella. Vet Microbiol 2012; 156:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftis AD, Gill JS, Schriefer ME, Levin ML, et al. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Argasidae). J Med Entomol 2005; 42:473–480 [DOI] [PubMed] [Google Scholar]
- Maggi RG, Breitschwerdt EB. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J Clin Microbiol 2005; 43:1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias MA, Díaz MM, Campos KJ, Calderon M, et al. Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by babesian phylogenetic analysis of 16S ribosomal DNA sequences. Am J Trop Med Hyg 2005; 73:964–974 [PMC free article] [PubMed] [Google Scholar]
- Messenger SL, Smith JS, Orciari LA, Yager PA, et al. Emerging pattern of rabies deaths and increased viral infectivity. Emerg Infect Dis 2003; 9:151–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morick D, Baneth G, Avidor B, Kosoy MY, et al. Detection of Bartonella spp. in wild rodents in Israel using HRM real-time PCR. Vet Microbiol 2009; 139:293–297 [DOI] [PubMed] [Google Scholar]
- Norman AF, Regnery R, Jameson P, Greene C, et al. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 1995; 33:1797–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WK, Loftis AD, Gore JA, Dasch GA. Molecular evidence for novel Bartonella species in Trichobius major (Diptera: Streblidae) and Cimex adjunctus (Hemiptera: Cimicidae) from two southeastern bat caves, U.S.A. J Vector Ecol 2005; 30:339–341 [PubMed] [Google Scholar]
- Reeves WK, Rogers TE, Durden LA, Dasch GA. Association of Bartonella with the fleas (Siphonaptera) of rodents and bats using molecular techniques. J Vector Ecol 2007; 32:118–122 [DOI] [PubMed] [Google Scholar]
- Renesto P, Gouvernet J, Drancourt M, Roux V, et al. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol 2001; 39:430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper J, Chanson JS, Chiozza F, Cox NA, et al. The status of the world's land and marine mammals: Diversity, threat and knowledge. Science 2008; 322:225–230 [DOI] [PubMed] [Google Scholar]
- Schneider MC, Romijn PC, Uieda W, Tamayo H, et al. Rabies transmitted by vampire bats to humans: An emerging zoonotic disease in Latin America? Rev Panam Salud Publica 2009; 25:260–269 [DOI] [PubMed] [Google Scholar]
- Simmons NB. Order Chiroptera. In: Wilson DE, Reeder DM, eds. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed. Baltimore, MD: Johns Hopkins University Press, 2005; 312–529 [Google Scholar]
- Theodor O. An illustrated catalogue of the Rothschild Collection of Nycteribiidae (Diptera) in the British Museum (Natural History) with keys and short descriptions for the identification of subfamilies, genera, species, and subspecies. London: British Museum (Natural History), 1967. ; 655 VIII 1 506, p. 898, p. 5, Pl, 6 maps. [Google Scholar]
- van der Poel WH, Lina PH, Kramps JA. Public health awareness of emerging zoonotic viruses of bats: A European perspective. Vector Borne Zoonotic Dis 2006; 6:315–324 [DOI] [PubMed] [Google Scholar]
- Wang LF, Walker PJ, Poon LLM. Mass extinctions, biodiversity and mitochondrial function: Are bats ‘special’ as reservoirs for emerging viruses? Curr Opin Virol 2011; 1:649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Lau S, Woo P, Yuen KY. Bats as a continuing source of emerging infections in humans. Rev Med Virol 2007; 17:67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]