Abstract
Tissue sampling for gene expression analysis is usually performed under general anesthesia. Anesthetics are known to modulate hemodynamics, receptor-mediated signaling cascades, and outcome parameters. The present study determined the influence of anesthetic paradigms typically used for euthanization and tissue sampling on cerebral mRNA expression in mice. Naïve mice and animals with acute traumatic brain injury induced by controlled cortical impact (CCI) were randomized to the following euthanasia protocols (n=10–11/group): no anesthesia (NA), 1 min of 4 vol% isoflurane in room air (ISO), 3 min of a combination of 5 mg/kg midazolam, 0.05 mg/kg fentanyl, and 0.5 mg/kg medetomidine intraperitoneally (COMB), or 3 min of 360 mg/kg chloral hydrate intraperitoneally (CH). mRNA expression of actin-1-related gene (Act1), FBJ murine osteosarcoma viral oncogene homolog B (FosB), tumor necrosis factor alpha (TNFα), heat shock protein beta-1 (HspB1), interleukin (IL)-6, tight junction protein 1 (ZO-1), IL-1ß, cyclophilin A, micro RNA 497 (miR497), and small cajal body-specific RNA 17 were determined by real-time polymerase chain reaction (PCR) in hippocampus samples. In naïve animals, Act1 expression was downregulated in the CH group compared with NA. FosB expression was downregulated in COMB and CH groups compared with NA. CCI reduced Act1 and FosB expression, whereas HspB1 and TNFα expression increased. After CCI, HspB1 expression was significantly higher in ISO, COMB, and CH groups, and TNFα expression was elevated in ISO and COMB groups. MiR497, IL-6, and IL-1ß were upregulated after CCI but not affected by anesthetics. Effects were independent of absolute mRNA copy numbers. The data demonstrate that a few minutes of anesthesia before tissue sampling are sufficient to induce immediate mRNA changes, which seem to predominate in the early-regulated gene cluster. Anesthesia-related effects on gene expression might explain limited reproduciblity of real-time PCR data between studies or research groups and should therefore be considered for quantitative PCR data.
Key words: : anesthesia, gene expression, early regulated genes, organ sampling, real-time PCR, traumatic brain injury
Introduction
For determination of mRNA regulation, quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) is the method of choice. In contrast to semi-quantitive RT-PCR, real-time RT-PCR is more accurate and is able to detect even small expression changes. Therefore, a correction of the results for inaccuracies because of the experimental conditions is essential. The optimal preservation of the biological sample is of key importance for all downstream applications in molecular biology. The necessary steps from mRNA isolation and quality control to cDNA synthesis and quantification are well standardized with highly optimized protocols. The impact of the appropriate selection of a housekeeping gene to correct for variations by normalization has been demonstrated.1 PCR data, however, may still differ even within the same research group despite optimized and similar procedures.2,3
Tissue sampling is usually performed under general anesthesia to reduce the stress level of animals. Anesthetics, however, affect systemic and cerebral hemodynamics,4 cerebral metabolism,5 and receptor-mediated signaling cascades. A recent study demonstrated that even 15 min of anesthesia during surgical preparation for experimental brain trauma altered outcome parameters and heightened the awareness for the importance of standardized experimental conditions.6 So far, no focus has been put on changes induced during the tissue removal process. Therefore, the present study investigated the impact of extremely brief episodes of background anesthesia during tissue sampling on cerebral mRNA expression. Three common anesthesia protocols for euthanasia and tissue sampling were applied in two sets of experiments with naïve animals and mice subjected to experimental brain injury to determine the impact on mRNA expression levels of eight representative genes in hippocampus tissue samples.
Methods
All experiments were approved by the local governmental authorities and performed in accordance with the German animal protection law. A total of 84 male C57Bl6 mice (Charles River Laboratories, Sulzfeld, Germany) were included in the study.
Experimental groups
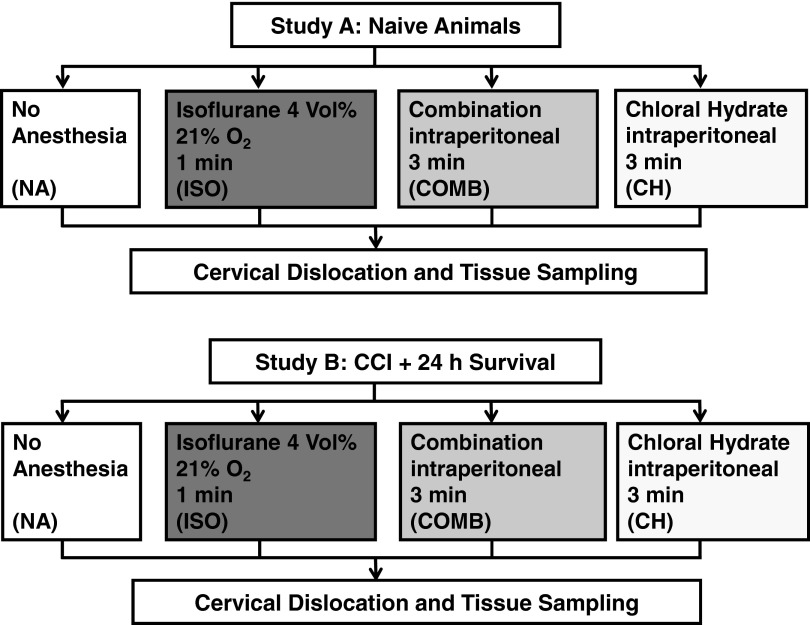
Study A: Naïve animals were randomized to the following groups (n=10/group, Fig. 1): euthanasia without anesthesia (NA), after 1 min exposure to 4 vol% isoflurane (Abbott, Wiesbaden, Germany) in room air (ISO), or 3 min after intraperitoneal (i.p.) injection of a combination (COMB) of 5 mg/kg midazolam (Ratiopharm, Ulm, Germany), 0.05 mg/kg fentanyl (CuraMed, Karlsruhe, Germany), and 0.5 mg/kg medetomidine (Pfizer, Karlsruhe, Germany), or 3 min after i.p. injection of 360 mg/kg chloral hydrate (CH, 3.6 g/100 mL, Pharmacy of the Medical Center of the Johannes Gutenberg-University, Mainz, Germany). Study B: Mice that had received brain trauma 24 h before were randomized to the same euthanasia groups (n=11/group, Fig. 1).
FIG. 1.
Treatment groups. Four common euthanasia protocols were used for RNA analysis in the healthy brain (Study A, naïve animals) and in the traumatized brain (Study B, animals with controlled cortical impact [CCI]).
The effect of anesthetics on hippocampal mRNA expression of actin-1-related gene (Act1), FBJ murine osteosarcoma viral oncogene homolog B (FosB), tumor necrosis factor alpha (TNFα), heat shock protein beta-1 (HspB1), interleukin 6 (IL-6), tight junction protein 1 isoform 1 (TJP1=ZO-1), interleukin 1 beta (IL-1ß), cyclophilin A (PPIA, control gene), micro RNA 497 (miR497), and small cajal body-specific RNA 17 (scaRNA17, miRNA control gene) were determined by real-time RT-PCR (Lightcycler).
Traumatic brain injury
Anesthesia was induced in a bell jar filled with 4% sevoflurane (Abbott, Wiesbaden, Germany) and maintained by inhalation via face mask (2% sevoflurane in 40% O2 and 60% N2). A thermostatically regulated, feedback-controlled heating pad was used to maintain body temperature at 37°C (Hugo Sachs, March-Hugstetten, Germany). The brain trauma model was performed as described previously.7 The skull was fixed in a stereotactic frame (Kopf Instruments, Tujunga, CA), and a craniotomy was performed above the right parietal cortex between the sagittal, lambdoid, and coronal sutures and the insertion of the temporal muscle with a saline cooled high-speed drill.
The lesion was induced perpendicular to the surface of the brain with a custom fabricated pneumatic controlled cortical impactor device (L. Kopacz, Mainz, Germany; diameter of the impactor tip: 3 mm; impact velocity: 8 m/sec; impact duration: 150 msec; displacement: 1 mm). The craniotomy was closed with the initially removed bone flap using conventional tissue glue (Histoacryl,® Braun-Melsungen, Germany). The skin was carefully closed, sevoflurane discontinued, and the animals transferred into their cages. For a maximum of 6 h, animals were placed in a neonatal incubator (IC8000, Draeger, Luebeck, Germany) with controlled air temperature (33°C) and ambient humidity (35%) to maintain constant body temperature.
Euthanasia and tissue preparation
The animals were killed by cervical dislocation without anesthesia (NA), ISO via face mask, COMB, or CH. After immediate decapitation, the brain was cut in coronal orientation and the right hippocampus was collected, frozen in liquid nitrogen, and stored at −80°C. At the same time, a second investigator performed arterial blood gas analysis.
RNA isolation, quality control, and cDNA synthesis
Brain tissue samples of hippocampus were homogenized and lysed with Qiazol-reagent (Qiagen, Hilden, Germany) and with a MM300 mill mixer (Retsch, Haan, Germany) operated at 20 Hz for 2 min. Total RNA was isolated using RNeasy Lipid Tissue Mini Kit (Qiagen) according to the manufacturer's instructions and eluted with 30–50 μL of RNase-free water. RNA concentration was calculated using the Quanti-iT™ RNA Assay Kit with the Qubit™ fluorimeter (Molecular Probes,™ Invitrogen, Karlsruhe, Germany) and with the NanoVue system (GE Healthcare Europe, Munich, Germany). 0.5 μg extracted RNA was reverse-transcribed into cDNA by Verso™ cDNA Kit (ABgene, Hamburg, Germany) and miScript RT Kit (Qiagen) according to the manufacturer's instructions.
PCR standard generation for absolute qPCR quantification
PCR fragments of all applied genes were generated by PCR on an Eppendorf Thermocycler gradient. PCR cycling parameters were as follows: thermal activation for 10 min at 95°C and 50 cycles of PCR (melting for 45 sec at 94°C, annealing for 45 sec at 55–65°C, and extension for 60 sec at 72°C). Applied primers are listed in Table 1. To verify the specificity of the PCR reaction, PCR products underwent electrophoresis alongside the 50 bp DNA Molecular Weight Marker XIII (Roche) through a 2% (w/v) agarose gel (Invitrogen). The gels were stained with SYBR green (Roche), and images were captured using a Kodak EDAS 120 Image System (Eastman Kodak Sàrl, Genève, Switzerland). The PCR products were purified with QIA quick PCR Purification Kit (Qiagen) according the manufacturer's instructions, and the DNA concentration was determined using NanoVue. The copy number was calculated and serial 10-fold dilutions were made in the range of 1×107 to 1×101 copies. A standard curve for absolute quantification was generated for each PCR product, showing good efficiency and linearity. For miRNA assays, a standard curve was generated for miR497 and scaRNA17 expression by serial 10-fold dilutions of a highly concentrated cDNA.
Table 1.
Specific Primer and Probes and Optimized Temperature Conditions for Real-Time Polymerase Chain Reaction
| PCR assay (amplicon size, annealing temp) | Oligonucleotide sequence (5′-3′) | GenBank No. |
|---|---|---|
| Cyclophilin A (PPIA) | Forw: 5′-GCGTCTSCTTCGAGCTGTT-3′ | NM_008907 |
| (146 bp, 55°C) | Rev: 5′-RAAGTCACCACCCTGGCA-3′ | |
| R670: Red-TTGGCTATAAGGGTTCCTCCTTTCACAG-Phos | ||
| FL: 5′-GCTCTGAGCACTGGRGAGAAAGGA-FL | ||
| IL-1ß | Forw: 5′-GTGCTGTCGGACCCATATGAG-3′ | NM_008361 |
| (348 bp, 55°C) | Rev: 5′-CAGGAAGACAGGCTTGTGCTC-3′ | |
| R640: Red-CAGCTGGAGAGTGTGGATCCCAAGC-Phos | ||
| FL: 5′-TAATGAAAGACGGCACACCCACCC-FL | ||
| IL-6 | Forw: 5′-TCGTGGAAATGAGAAAAGAGTTG-3′ | NM_031168.1 |
| (471 bp, 55°C) | Rev: 5′-TATGCTTAGGCATAACGCACTAG-3′ | |
| R640: Red-TGCTCTCCTAACAGATAAGCTGGAGTCAC-Phos | ||
| FL: 5′-CATAAAATAGTCCTTCCTACCCCAATTTCC-FL | ||
| TJP1 (=ZO-1) | Forw: 5′-TGTCCCTGTGAGTCCTTCAG-3′ | NM_009386 |
| (331 bp, 55°C) | Rev: 5′-CCAGGTTTTAGGGTCACAGT-3′ | |
| R640: Red-ATGCCACGAGCTGTAGCCACTAC-Phos | ||
| FL: 5′-CTCAACACACCACCATTgCTgTT-FL | ||
| TNFα | Forw: 5′-TCTCATCAGTTCTATGGCCC-3′ | NM_013693 |
| (212 bp, 58°C) | Rev: 5′-GGG AGTAGACAAGGTACAAC-3′ | |
| FosB | Forw: 5′- CCGAGAAGAGACACTTACCC-3′ | NM_008036 |
| (142 bp, 58°C) | Rev: 5′- CTCTTCAAGCTGATCAGTTTCC-3′ | |
| HspB1 | Forw: 5′- GACGAACATGGCTACATCTC -′ | NM_013560 |
| (244 bp, 58°C) | Rev: 5′- CTGATGGCTTCTACTTGGCT-3′ | |
| Act1 | Forw: 5′- TGATCAAGATGACAGCATGG-3′ | NM_009652 |
| (224 bp, 58°C) | Rev: 5′- GCCCACAGTAGAAACATCCT-3′ | |
| Mir497 | Mir497 miScript Primer Assay (Qiagen) | Mm_miR-497_1 |
| SCARNA17 | SCARNA17 miScript Primer Assay (Qiagen) | Hs_SCARNA17_1 |
PCR, polymerase chain reaction; Forw, sense primer; Rev, anti-sense primer; R640/670, Lightcycler® Red640/670; Phos, phosphate; FL, fluorescein.
Quantitative PCR
Equal amounts of cDNA (1 μL) were analyzed of each sample and in duplicates by real-time Lightcycler 480 PCR System (Roche). For SYBR green runs Absolute Blue qPCR Sybrgreen Mix (Thermo Scientific), HybProbe runs Light Cycler 480 Probes Master (Roche), and for miRNA quantification runs miScript PCR Kit (Qiagen) was used for real-time PCR according to the manufacturer's instructions. Applied primers and probes are listed in Table 1. The specificity for SYBR green assays was confirmed by gel electrophoresis and melting curve analysis. The used HybProbe probes were designed by TIB-Molbiol (Berlin, Germany). The absolute copy numbers of the target genes were normalized against the absolute copy numbers of cyclophilin A (PPIA) as control gene. The relative miR497 expression was normalized against scaRNA17 expression as control gene.
Statistical analysis
Statistical analysis was performed with Sigma Stat 12.5 statistical software (Systat Software, Chicago, IL). mRNA and miRNA expression profiles are presented as median and quartiles and were compared with Wilcoxon Mann-Whitney rank sum tests. The p values were adjusted for multiple comparisons (Bonferroni Holmes adjustment). Correlations between RNA concentrations were tested with Spearman product moment correlation. Physiological parameters are presented as mean±standard deviation and were analyzed with Wilcoxon Mann-Whitney rank sum tests. The p values were adjusted for multiple comparisons (Bonferroni Holmes adjustment). Differences were considered significant at the p<0.05 level.
Results
Physiological data
Physiological variables were measured immediately after decapitation in arterial blood samples (Table 2). In animals without controlled cortical impact (CCI), blood glucose was higher in the COMB group and CH group compared with the NA group. In animals with experimental brain trauma, PaO2 was higher in anesthetized animals and PaCO2 was higher in the COMB group compared with the NA group. Also, hemoglobin concentration was slightly higher in anesthetized animals compared with the NA group. Blood glucose was higher in the COMB group and sampling time was longer in the CH group compared with the NA group. Rectal temperature was higher in animals after CCI compared with naïve mice.
Table 2.
Physiological Parameters at Decapitation
| NA | ISO | COMB | CH | |
|---|---|---|---|---|
| Study A: Naïve mice without brain injury | ||||
| pH | 7.21±0.10 | 7.22±0.07 | 7.22±0.11 | 7.14±0.11 |
| paO2 (mm Hg) | 58±24 | 69±11 | 57±10 | 77±16 |
| paCO2 (mm Hg) | 46.7±5.6 | 45.7±5.3 | 51.4±7.7 | 46.7±5.7 |
| Hemoglobin (mg/dL) | 13.4±0.5 | 13.4±0.5 | 12.9±1.6 | 12.8±1.1 |
| Glucose (mg/dL) | 217±33 | 214±24 | 262±4a | 271±39a |
| Sampling duration (min) | 7.3±0.9 | 7.2±0.8 | 7.1±0.8 | 6.9±0.6 |
| Body weight (g) | 20.5±0.9 | 20.4±1.4 | 10.3±1.5 | 19.8±1.1 |
| Rectal temperature (°C) | 36.0±0.6 | 36.0±0.8 | 36.0±0.6 | 36.1±0.5 |
| Study B: Mice 24 h after CCI | ||||
| pH | 7.21±0.31 | 7.17±0.37 | 7.13±0.34 | 7.18±0.16 |
| paO2 (mm Hg) | 38±9 | 78±29a | 63±13a | 70±11a |
| paCO2 (mm Hg) | 42.8±3.9 | 39.6±5.2 | 53.9±6.9a | 45.6±7.7 |
| Hemoglobin (mg/dL) | 15.3±0.5 | 14.5±0.4a | 13.8±0.8a | 14.0±1.1a |
| Glucose (mg/dL) | 234±29 | 254±49 | 290±54a | 265±36 |
| Sampling duration (min) | 6.2±0.5 | 6.8±0.5 | 6.2±0.6 | 7.2±0.8a |
| Body weight (g) | 20.9±0.8 | 20.7±2.1 | 20.5±0.5 | 20.7±0.8 |
| Rectal temperature (°C) | 38.1±0.9 | 38.0±0.9 | 38.4±0.6 | 38.0±0.7 |
| CCI anesthesia (min) | 19±2 | 19±1 | 20±3 | 19±4 |
NA, no anesthesia; ISO, isoflurane in room air; COMB, intraperitoneal (i.p.) injection of midazolam, fentanyl, and medetomidine; CH, i.p. injection of chloral hydrate. Physiological parameters were determined at the time of decapitation. PaO2, arterial oxygen tension; PaCO2, arterial carbon dioxide tension; glucose, plasma glucose concentration; CCI, controlled cortical impact; CCI anesthesia, time of anesthesia exposure during CCI. Data are presented as mean±standard deviation. a, p<0.05 vs. NA.
RNA quantification and quality control
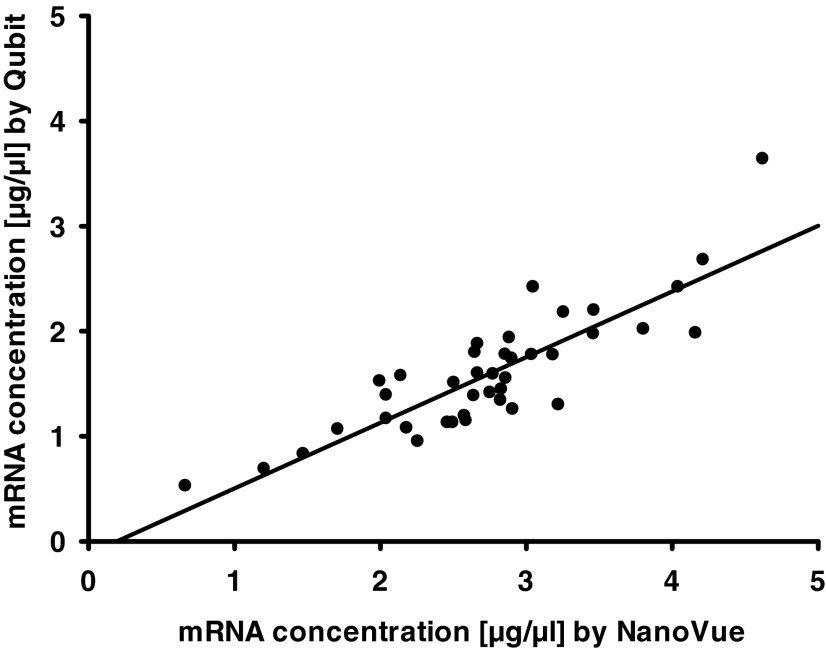
RNA concentration was determined by two techniques: the Qubit fluorimeter and ultraviolet absorbance with the NanoVue system. Determined concentrations correlated well between both techniques (r=0.827; p<0.01; Fig. 2). Qubit results were lower in comparison with the NanoVue system. NanoVue system concentration data were applied for the RNA RT into cDNA. RNA quality was investigated with the Bio-Rad Experion™ automated electrophoresis system and rated according to the mean RNA integrity number (RIN) system.8 The mean RIN for all samples was categorized 8.6±0.6 indicating a good RNA quality.
FIG. 2.
RNA concentrations, as determined by Qubit, correlated with values determined by NanoVue (r=0.827, p<0.01).
Differential regulation of cerebral mRNA expression
Study A: Naïve mice
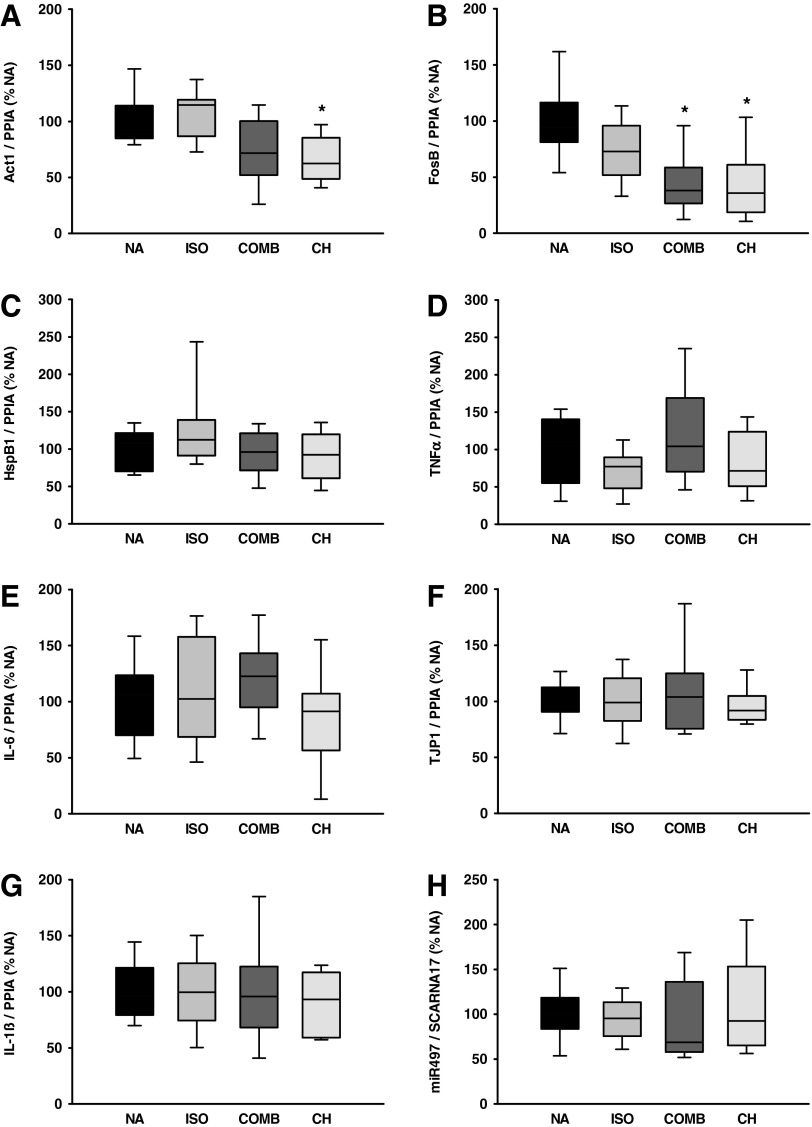
Median normalized expression of Act1, FosB, HspB1, TNFα, IL-6, TJP1, IL-1ß, and miR497 in naïve animals is shown in Figure 3 A–H. Act1 was significantly downregulated in the CH group (1.6-fold, p<0.01) whereas expression data in the ISO group and COMB group remained insignificant compared with NA. FosB expression decreased with COMB (2.6-fold, p<0.01) and CH (3.4-fold, p<0.01) whereas expression data in the ISO group remained insignificant compared with NA. HspB1, TNFα, TJP1, IL-1ß, IL-6, and miR497 mRNA expression were not significantly different compared with NA. The observed effects were independent of absolute mRNA copy numbers (Table 3).
FIG. 3.
mRNA expression of actin-1-related gene (Act1, A), FBJ murine osteosarcoma viral oncogene homolog B (FosB, B), heat shock protein beta-1 (HspB1, C), tumor necrosis factor alpha (TNFα, D), interleukin 6 (IL-6, E), tight junction protein 1 (TJP1=ZO-1, F), interleukin 1 beta (IL-1ß, G), micro RNA 497 (miR497, H) in naïve animals. RNA-data were normalized to cyclophillin A (PPIA), and microRNA data were normalized to small cajal body-specific RNA 17 (scaRNA17). Data are presented as median and quartiles. NA, no anesthesia; ISO, isoflurane anesthesia; COMB, intraperitoneal (i.p.) injection of fentanyl, midazolam, and medetomidine; CH, i.p. injection of chloral hydrate. *p<0.05 vs. NA.
Table 3.
Absolute Copy Number of Investigated Genes
| Naive (copy number/PPIA) | CCI (copy number/PPIA) | |
|---|---|---|
| Act1 | 0.045±0.014 | 0.032±0.020 |
| FosB | 0.0015±0.0008 | 0.0010±0.00004 |
| HspB1 | 0.0067±0.0024 | 0.054±0.044 |
| TNFα | 0.00003±0.00002 | 0.0008±0.0003 |
| IL-6 | 0.00031±0.00016 | 0.0053±0.0096 |
| TJP1/ZO-1 | 0.030±0.006 | 0.043±0.008 |
| IL-1ß | 0.003±0.001 | 0.020±0.008 |
PPIA, cyclophilin A; CCI, controlled cortical impact.
Absolute normalized mRNA copy numbers for actin-1-related gene (Act1), FBJ murine osteosarcoma viral oncogene homolog B (FosB), heat shock protein beta-1 (HspB1), tumor necrosis factor alpha (TNFα), interleukin 6 (IL-6), tight junction protein 1 (TJP1), and interleukin 1 beta (IL-1ß). Values are presented as mean±standard deviation.
Study B: Mice with acute brain injury
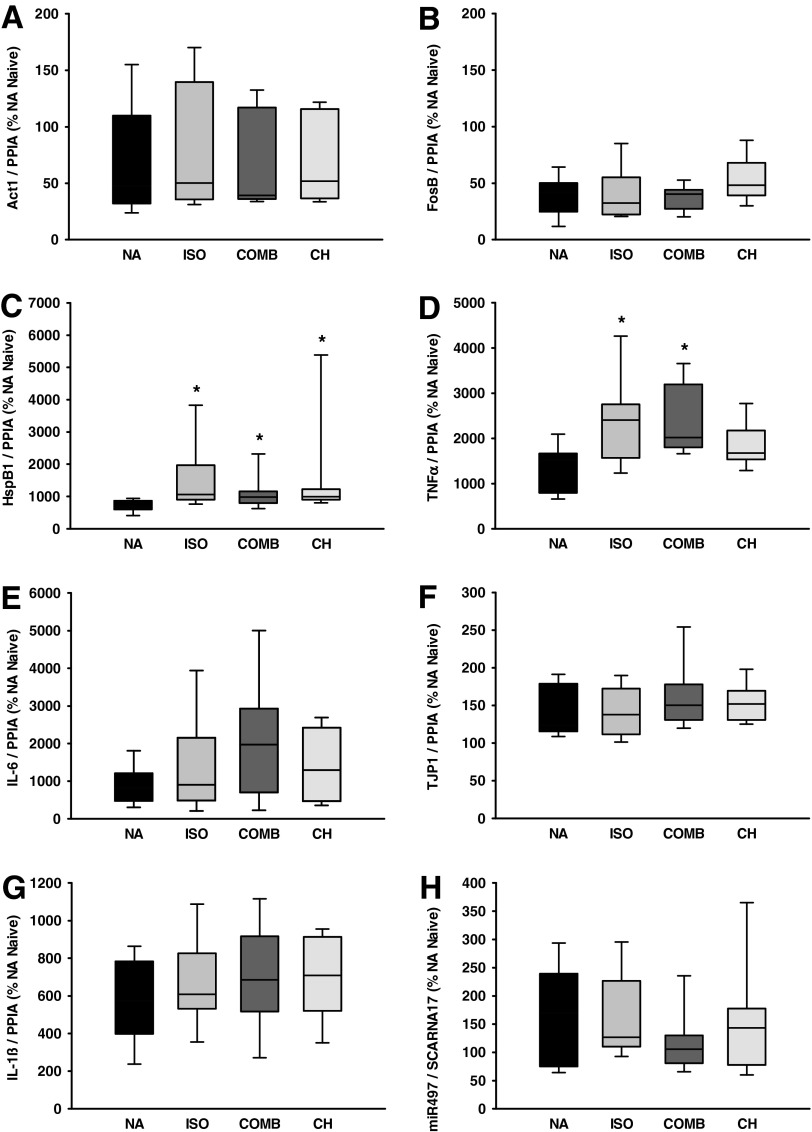
Median normalized expression of Act1, FosB, HspB1, TNFα, IL-6, TJP1, IL-1ß, and mi497 in animals with traumatic brain injury is shown in Figure 4 A–H. In animals after CCI and without anesthesia, Act1 expression decreased to 47% and FosB expression decreased to 42% of naïve animals without CCI. In contrast, HspB1 and TNFα expression increased to 664% and 1525% of naïve animals without CCI, respectively. Compared with NA, expression of Act1 and FosB remained stable with minor, nonsignificant changes in CCI animals with anesthesia. In contrast, HspB1 expression increased with anesthetic treatment (ISO 1.6-fold, p<0.01, COMB 1.5-fold, p=0.01, CH 1.5-fold, p<0.01). Likewise, TNFα expression levels increased with anesthetic treatment (ISO 1.6-fold, p=0.013, COMB 1.3-fold, p<0.01). IL-6, IL-1ß, and miR497 mRNA expression increased after CCI compared with naïve animals (IL-6 823%, IL-1ß 573%, miR497 169%); however, anesthetic treatment did not have significant impact on these mRNA expression levels compared with CCI without anesthetic treatment. TJP1 was neither affected by CCI nor by anesthetic treatment. All observed effects were independent of absolute mRNA copy numbers.
FIG. 4.
mRNA expression of actin-1-related gene (Act1, A), FBJ murine osteosarcoma viral oncogene homolog B (FosB, B), heat shock protein beta-1 (HspB1, C), tumor necrosis factor alpha (TNFα, D), interleukin 6 (IL-6, E), tight junction protein 1 (TJP1=ZO-1, F), interleukin 1 beta (IL-1ß, G), micro RNA 497 (miR497, H) in animals 24 hours after controlled cortical impact. RNA-data were normalized to cyclophillin A (PPIA) and microRNA data was normalized to small cajal body-specific RNA 17 (scaRNA17). Data are presented as median and quartiles. NA, no anesthesia; ISO, isoflurane anesthesia; COMB, intraperitoneal (i.p.) injection of fentanyl, midazolam, and medetomidine; CH, i.p. injection of chloral hydrate. *p<0.05 vs. NA.
Discussion
The key finding of the present study is that even a few minutes of anesthesia before euthanasia and tissue sampling are sufficient to induce immediate cerebral mRNA expression changes. This effect seems to predominate in the early-regulated gene cluster. Genes induced by acute cerebral injury seem to be especially susceptible to anesthesia-related changes in mRNA expression levels and independent of absolute mRNA copy numbers.
To date, there is very little information on the effects of anesthesia on immediate tissue gene expression. Data in salmon demonstrate that sedation of 30 min to 2 h before euthanasia and tissue sampling resulted in differential regulation of fast-responding genes.9,10 Data in rodents are mostly limited to comparison of anesthesia effects during experimental induction of cerebral pathologies.6,11 There is evidence, however, that the use of isoflurane for euthanasia induces immediate changes in electrophysiology and neuroplasticity compared with decapitation without anesthesia or decapitation with anesthesia12; and that isoflurane anesthesia before CO2 euthanasia increased c-fos expression in mice brains.13 This is in accordance with results of the present study where Act1 and FosB, as examples of early regulated genes,14 were downregulated by anesthesia whereas delayed-regulated genes remained unaffected. The stability of delayed-regulated genes in the present study, such as the inflammatory marker genes IL-1ß and IL-6, is in accordance with data showing that 15 min after anesthesia, expression of IL-6, cyclooxygenase-2, and inducible nitric oxide synthase remained unchanged by sevoflurane, isoflurane, and an intraperitoneal combination of fentanyl, midazolam, and medetomidine.6 Anesthesia-related effects, however, also seem to be dependent on duration of the administration period, as recently demonstrated in naïve mice for isoflurane, which increased IL-1ß and IL-6 expression after 2-h isoflurane exposure.15
The presence or absence of an acute brain damage is a major influencing factor of cerebral mRNA expression.1,16 Mechanical injury by CCI increased HspB1 and TNFα expression, two genes taking part in the quick response to ischemic stimuli in brain tissue17–19 and belonging to the early regulated gene cluster.20,21 Anesthetics induced a further increase in HspB1 and TNFα expression. Administration of isoflurane induces an elevated TNFα expression in naïve mice after 2 h of administration.22 In the present study, isoflurane did not influence TNFα levels in naïve animals. This may be attributed to the administration time of 1 min, which was probably too short to induce these changes. In contrast, when basal TNFα levels were elevated 24 h after brain trauma, 1 min of isoflurane administration was sufficient to induce a further increase in TNFα expression levels. In contrast, the early regulated genes Act-1 and FosB were downregulated after cerebral injury and not further influenced by the anesthesia techniques. Again, the delayed-regulated inflammatory genes IL-1ß and IL-6 were not modulated by anesthetics, although volatile anesthetics are known to suppress inflammation; however, for this effect, longer application duration seems to be essential.23
Protein levels are not only influenced by mRNA levels, but also by miRNA. In the present study, micro RNA 497 was therefore quantified as a representative miRNA, which is known to be upregulated on ischemic insults. Accordingly, miR497 levels are increased in brain samples after CCI injury, but expression levels were not influenced by anesthesia during organ sampling. In the present study, only one representative miRNA was quantified; therefore, the study can only provide insufficient information on the stability of expression of miRNA after anesthesia exposure.
The differential effects of anesthetics on gene expression in the brain may be explained by their effects on cerebral metabolism reflected in the change of vigilance and brain function. As a result of direct receptor interaction, anesthetics could directly influence gene expression. On the other hand, anesthetics are known to reduce cerebral metabolism reflected in a reduction of cerebral metabolic rate of oxygen5; reduction of cell metabolisms could result in changes of mRNA turnover by modulation of mRNA synthesis or mRNA degradation. In the present study, direct changes of mRNA turnover were not quantified—therefore, underlying mechanisms remain to be speculative.
The most important limitation of the present study is that it is not possible to design the perfect control group, which allows the exact unchanged quantification of RNA levels in living animals. Euthanasia without the use of anesthetics cannot be considered as a perfect control group because this euthanasia approach may induce stress in the animals. Stress and anxiety can also cause gene expression changes because it has been demonstrated for the immediate early gene c-fos after CO2 euthanasia without anesthesia.24 Therefore, the question of adequate euthanasia techniques cannot be fully answered, because it is impossible to create the perfect control group. The present study also did not assess a sham-operated control group or a control group exposed to sevoflurane 24 h before euthanasia. Therefore, no statement can be made regarding the effects of the sham operation, the exposure to anesthesia on two occasions, as well as the interaction of two different anesthetics.
The euthanasia protocol may influence physiological variables, which in turn may affect mRNA expression data. In the present study, higher plasma glucose concentrations and lower hemoglobin concentrations were observed in the majority of intraperitoneally anesthetized mice, most likely from elevated stress levels and hemodilution after resorption of intraperitoneally administered liquids. Euthanasia without anesthesia resulted in a lower arterial oxygen tension in animals previously subjected to brain trauma, presumably caused by animal handling, which may compromise thorax excursions. Because an influence on cerebral gene expression has been demonstrated,25–27 it cannot be excluded that outcome parameters were affected by inhomogeneous physiological data.
A further study limitation may be the focus of analysis on the hippocampus, which was chosen as the target region because of its high sensitivity to ischemic insults.28 Further, no other organs besides the brain were analyzed despite evidence that immediate early gene expression is also affected by anesthetics in other organs such as the heart, liver, or kidney.29 The focus of the present study, however, was the mRNA expression changes in the healthy and the damaged brain. The present data suggest that mRNA expression changes occur predominantly in the early-regulated gene cluster. Because no microarrays with specific pathway analyses have been performed, however, the presented genes have to be considered as exemplary.
Conclusion
The present study demonstrates that even a few minutes of anesthesia results in significant alterations of cerebral mRNA expression levels compared with NA-treated animals. These anesthesia-related changes seem to be detectible primarily in the early-regulated gene cluster. Especially early-regulated genes, which show increased expression levels after CCI, seem to be strongly affected by anesthesia during organ sampling. The data indicate that the choice of anesthetics during organ sampling influences cerebral mRNA expression and therefore needs to be considered for a proper interpretation of qPCR results.
Acknowledgments
This work was supported by the German Research Foundation (DFG grant TH1430/3-1 to Dr. Thal) and by the Federal Ministry of Education and Research (BMBF 01EO1003). The data presented in this manuscript are part of the doctoral theses presented by Oliver Kriege to the Medical Faculty of the Johannes Gutenberg-University, Mainz, Germany.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thal S.C., Wyschkon S., Pieter D., Engelhard K., and Werner C. (2008). Selection of endogenous control genes for normalization of gene expression analysis after experimental brain trauma in mice. J. Neurotrauma 25, 785–794 [DOI] [PubMed] [Google Scholar]
- 2.Thal S.C., Heinemann M., Luh C., Pieter D., Werner C., and Engelhard K. (2011). Pioglitazone reduces secondary brain damage after experimental brain trauma by PPAR-gamma-independent mechanisms. J. Neurotrauma 28, 983–993 [DOI] [PubMed] [Google Scholar]
- 3.Timaru-Kast R., Luh C., Gotthardt P., Huang C., Schäfer M.K., Engelhard K., and Thal S.C. (2012). Influence of age on brain edema formation, secondary brain damage and inflammatory response after brain trauma in mice. PloS One 7, e43829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen B.J., De Celle T., Debets J.J., Brouns A.E., Callahan M.F., and Smith T.L. (2004). Effects of anesthetics on systemic hemodynamics in mice. Am. J. Physiol. Heart Circ. Physiol. 287, H1618–1624 [DOI] [PubMed] [Google Scholar]
- 5.Kaisti K.K., Langsjo J.W., Aalto S., Oikonen V., Sipila H., Teras M., Hinkka S., Metsahonkala L., and Scheinin H. (2003). Effects of sevoflurane, propofol, and adjunct nitrous oxide on regional cerebral blood flow, oxygen consumption, and blood volume in humans. Anesthesiology 99, 603–613 [DOI] [PubMed] [Google Scholar]
- 6.Luh C., Gierth K., Timaru-Kast R., Engelhard K., Werner C., and Thal S.C. (2011). Influence of a brief episode of anesthesia during the induction of experimental brain trauma on secondary brain damage and inflammation. PloS One 6, e19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zweckberger K., Stoffel M., Baethmann A., and Plesnila N. (2003). Effect of decompression craniotomy on increase of contusion volume and functional outcome after controlled cortical impact in mice. J. Neurotrauma 20, 1307–1314 [DOI] [PubMed] [Google Scholar]
- 8.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., and Ragg T. (2006). The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsvik P.A., Lie K.K., and Hevroy E.M. (2007). Do anesthetics and sampling strategies affect transcription analysis of fish tissues? BMC Mol. Biol. 8, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trösse C., Waagbo R., Breck O., and Olsvik P.A. (2010). Optimisation of gene expression analysis in Atlantic salmon lenses by refining sampling strategy and tissue storage. Fish Physiol. Biochem 36, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 11.Tecoult E., Mesenge H., Stutzmann A.M., Plotkine M., and Wahl F. (2000). Influence of anesthesia protocol in experimental traumatic brain injury. J. Neurosurg Anesthesiol 12, 255–261 [DOI] [PubMed] [Google Scholar]
- 12.Kulisch C., Eckers N., and Albrecht D. (2011). Method of euthanasia affects amygdala plasticity in horizontal brain slices from mice. J. Neurosci. Meth. 201, 340–345 [DOI] [PubMed] [Google Scholar]
- 13.Valentine H., Williams W.O., and Maurer K.J. (2012). Sedation or inhalant anesthesia before euthanasia with CO2 does not reduce behavioral or physiologic signs of pain and stress in mice. J. Am. Assoc. Lab. Anim. Sci. 51, 50–57 [PMC free article] [PubMed] [Google Scholar]
- 14.Ebihara K., Ishida Y., Takeda R., Abe H., Matsuo H., Kawai K., Magata Y., and Nishimori T. (2011). Differential expression of Fosb, c-Fos, and Zif268 in forebrain regions after acute or chronic L-DOPA treatment in a rat model of Parkinson's disease. Neurosci. Lett. 496, 90–94 [DOI] [PubMed] [Google Scholar]
- 15.Cao L., Li L., Lin D., and Zuo Z. (2012). Isoflurane induces learning impairment that is mediated by interleukin 1beta in rodents. PloS One 7, e51431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michael D.B., Byers D.M., and Irwin L.N. (2005). Gene expression following traumatic brain injury in humans: Analysis by microarray. J. Clin. Neurosci. 12, 284–290 [DOI] [PubMed] [Google Scholar]
- 17.Franklin T.B., Krueger-Naug A.M., Clarke D.B., Arrigo A.P., and Currie R.W. (2005). The role of heat shock proteins Hsp70 and Hsp27 in cellular protection of the central nervous system. Int. J. Hyperthermia 21, 379–392 [DOI] [PubMed] [Google Scholar]
- 18.Lasarzik I., Cordier J., Peetz D., von Landenberg P., Werner C., Engelhard K., and Thal S.C. (2011). Effect of autologous blood transfusion on cerebral cytokine expression. J. Neurosurg. Anesthesiol. 23, 215–221 [DOI] [PubMed] [Google Scholar]
- 19.Stetler R.A., Gao Y., Zhang L., Wenig Z., Zhang F., Hu X., Wang S., Vosler P., Cao G., Sun D., Graham S.H., and Chen J. (2012). Phosphorylation of HSP27 by protein kinase D is essential for mediating neuroprotection against ischemic neuronal injury. J. Neurosci. 32, 2667–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falvo J.V., Tsytsykova A.V., and Goldfeld A.E. (2010). Transcriptional control of the TNF gene. Curr. Dir. Autoimmun. 11, 27–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp F.R., and Sagar S.M. (1994). Alterations in gene expression as an index of neuronal injury: Heat shock and the immediate early gene response. Neurotoxicology 15, 51–59 [PubMed] [Google Scholar]
- 22.Wu X., Lu Y, Dong Y., Zhang G., Zhang Y., Xu Z., Culley D.J., Crosby G., Marcantonio E.R., Tanzi R.E., and Xie Z. (2012). The inhalation anesthetic isoflurane increases levels of proinflammatory TNF-alpha, IL-6, and IL-1beta. Neurobiol. Aging 33, 1364–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Yin J., Li L., Deng J., Feng C., and Zuo Z. (2013). Isoflurane postconditioning reduces ischemia-induced nuclear factor-kappaB activation and interleukin 1beta production to provide neuroprotection in rats and mice. Neurobiol. Dis. 54, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson P.L., Fitz S.D., Hollis J.H., Moratalla R., Lightman S.L., Shekhar A., and Lowry C.A.Induction of c-Fos in ‘panic/defence’-related brain circuits following brief hypercarbic gas exposure. J. Psychopharmacol. 2011;25:26–36 [DOI] [PubMed] [Google Scholar]
- 25.Kim G.S., Jung J.E., Narasimhan P., Sakata H., and Chan P.H. (2012). Induction of thioredoxin-interacting protein is mediated by oxidative stress, calcium, and glucose after brain injury in mice. Neurobiol. Dis. 46, 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C., Peng Z., Zhang N., Yu L., Han S., Li D. and Li J. (2012). Identification of differentially expressed microRNAs and their PKC-isoform specific gene network prediction during hypoxic pre-conditioning and focal cerebral ischemia of mice. J. Neurochem. 120, 830–841 [DOI] [PubMed] [Google Scholar]
- 27.Briet F., Mazer C.D., Tsui A.K., Zhang H., Khang J., Pang V., Baker A.J., and Hare G.M. (2009). Cerebral cortical gene expression in acutely anemic rats: A microarray analysis. Can. J. Anaesth. 56, 921–934 [DOI] [PubMed] [Google Scholar]
- 28.Nellgard B., Mackensen G.B., Massey G., Pearlstein R.D., and Warner D.S. (2000). The effects of anesthetics on stress responses to forebrain ischemia and reperfusion in the rat. Anesth. Analg. 91, 145–151 [DOI] [PubMed] [Google Scholar]
- 29.Hamaya Y., Takeda T., Dohi S., Nakashima S., and Nozawa Y. (2000). The effects of pentobarbital, isoflurane, and propofol on immediate-early gene expression in the vital organs of the rat. Anesth. Analg. 90, 1177–1183 [DOI] [PubMed] [Google Scholar]