Abstract
Background and Aims
Abiotic properties of soil are known to be major drivers of the microbial community within it. Our understanding of how soil microbial properties are related to the functional structure and diversity of plant communities, however, is limited and largely restricted to above-ground plant traits, with the role of below-ground traits being poorly understood. This study investigated the relative contributions of soil abiotic properties and plant traits, both above-ground and below-ground, to variations in microbial processes involved in grassland nitrogen turnover.
Methods
In mountain grasslands distributed across three European sites, a correlative approach was used to examine the role of a large range of plant functional traits and soil abiotic factors on microbial variables, including gene abundance of nitrifiers and denitrifiers and their potential activities.
Key Results
Direct effects of soil abiotic parameters were found to have the most significant influence on the microbial groups investigated. Indirect pathways via plant functional traits contributed substantially to explaining the relative abundance of fungi and bacteria and gene abundances of the investigated microbial communities, while they explained little of the variance in microbial activities. Gene abundances of nitrifiers and denitrifiers were most strongly related to below-ground plant traits, suggesting that they were the most relevant traits for explaining variation in community structure and abundances of soil microbes involved in nitrification and denitrification.
Conclusions
The results suggest that consideration of plant traits, and especially below-ground traits, increases our ability to describe variation in the abundances and the functional characteristics of microbial communities in grassland soils.
Keywords: Ammonia-oxidizing archaea, bacteria, denitrifiers, leaf traits, root traits, nitrite reducers, nitrite oxidizers, nutrient availability, plant functional traits, soil microbial community
INTRODUCTION
In recent years, plant functional trait approaches have emerged as a powerful tool for identifying the mechanisms by which plants are related to ecosystem processes via their interactions with microbial communities (Fig. 1). There is evidence for such plant trait effects at scales ranging from the individual plant to the landscape (Orwin et al., 2010; de Vries et al., 2012; Grigulis et al., 2013), although their quantification remains a challenge. Grigulis et al. (2013) provided a first quantitative estimation of the relative importance of above-ground plant traits and microbial effects for ecosystem nitrogen (N) cycling in grasslands (Fig. 1, grey boxes and arrows). Although some rare studies have analysed plant traits–soil microbe interactions in laboratory conditions in the absence of soil effects (A. Cantarel, Lyon, France, unpubl. res.), similar quantification of other components of plant–soil–microbe interactions in the field is still required, including the respective effects of plant traits and soil abiotic properties on microbial parameters (Fig. 1, dashed black arrows).
Fig. 1.
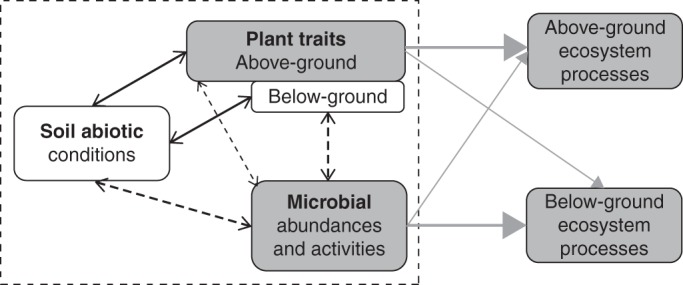
Schematic diagram showing the links between soil, plants and microbial communities, and their relationships to ecosystem processes. The grey boxes and arrows depict the links found by Grigulis et al. (2013). The dashed box and the black dashed arrows represent the links found in the present study.
Numerous inter-relationships of abiotic soil properties with plant and microbial communities have been demonstrated. First, the impact of soil properties on plant traits and some soil microbial functional variables is well established. In particular, it is broadly accepted that soil nutrient availability determines the prevalence of certain plant functional traits, such as specific leaf area (SLA) and leaf N content (LNC) (Grime, 1977; Ordoñez et al., 2009). Nutrient-rich grasslands are dominated by fast-growing exploitative species, characterized by high SLA, LNC (Lavorel and Garnier, 2002) and root N concentration (RNC) (Roumet et al., 2006). Conversely, nutrient-poor grasslands are dominated by slow-growing conservative species, characterized by opposite traits. Soil abiotic properties such as pH, and nutrient and water availability are linked not only to plant traits but also to soil microbial communities such as nitrifiers and denitrifiers, and the N turnover processes that they drive (Bremer et al., 2007; Le Roux et al., 2008; Bannert et al., 2011). Second, there is evidence that plants have a pronounced influence on microbial community structure and function, mainly in the rhizosphere or detritusphere. Interspecific differences in plant litter quality have been shown to affect microbial communities and rates of carbon (C) and N mineralization (Wardle, 1992; De Deyn et al., 2008), as have variations in the quantity (Griffiths et al., 1999; Van der Krift et al., 2001; Kuzyakov, 2006) and quality (De Deyn et al., 2008; Henry et al., 2008) of root exudates of individual species. Moreover, recent studies highlight the role of root exudates in providing easily degradable C sources for soil microbes, which stimulates their growth and the mineralization of soil organic matter (Kuzyakov, 2010). In turn, these impacts of plants on soil microbial communities feedback to plant performance, both through their role in modifying nutrient availability and also through various positive or negative interactions involving symbionts and microbial pathogens (van der Heijden et al., 2008; van der Putten et al., 2013).
Despite these known mechanisms, our understanding of multiple relationships among abiotic soil parameters, the soil microbiome and plants remains fragmented, as individual studies have mostly addressed individual associations, making it difficult to build a generic understanding. In addition, most studies have focused on single plant species or cultivars, and have not considered complex species-rich plant communities as found in the field. A few field-based studies have considered the effects of plant functional traits on soil microbes, focusing either on leaf traits alone (Laughlin 2011; de Vries et al., 2012) or specifically on root traits (Valé et al., 2005; Pohl et al., 2011). But to our knowledge, only a few studies have simultaneously considered the effects of above-ground and below-ground plant traits on soil microbial functional variables, such as root decomposition rate or the fungal to bacterial ratio (Orwin et al., 2010; Birouste et al., 2012), and although no direct relationship with root traits were demonstrated, Legay et al. (2014) suggested that root systems of exploitative species stimulate selected enzyme activities (e.g. aryl-sulphatase) in soil. Despite this, none of these studies has simultaneously considered how plant traits, both above- and below-ground, and abiotic soil properties explain variation in soil microbial properties under field conditions.
Soil N dynamics in mountain grasslands is typically constrained by low rates of N mineralization and low N inputs (Clément et al., 2012), making N one of the most limiting nutrients for primary production (Grigulis et al., 2013). Although studies have explored the role of leaf traits as drivers of microbial communities and ecosystem functioning in these grasslands (Orwin et al., 2010; de Vries et al., 2012; Grigulis et al., 2013), no study to date has tested for the relative effects of leaf and root functional traits, and soil abiotic properties, on soil microbial communities and their activities related to N cycling under field conditions. We aimed to bridge this gap using measures at three European mountain grassland sites, where different land-use intensities within each site provided a broad range of variation in plant functional traits, soil properties and microbial functional variables. We focused in this study on the abundance of nitrifiers and denitrifiers as microbial variables, as those microbes strongly influence N availability and quality in soil and thus drive plant performance to a large extent mainly in unfertilized grassland sites. We applied the conceptual and statistical approach developed by Grigulis et al. (2013) to quantify the contribution of plant above- and below-ground traits to variation in the gene abundance of functional microbial groups and their resulting activity patterns in grassland ecosystems, once direct effects of abiotic soil properties have been taken into account. We did this by expanding the dataset of Grigulis et al. (2013) to include below-ground plant traits and soil abiotic parameters, which allowed us to quantify relative effects of both above- and below-ground traits on microbial functional variables, and assess their importance relative to abiotic effects. We used this approach to test two hypotheses: first, that above- and below-ground plant traits reflecting plant responses to abiotic soil parameters provide unique information about nitrifiers and denitrifiers, which can be considered as important microbial drivers for the inorganic N cycle. This hypothesis was tested by sequentially analysing direct effects of soil abiotic properties on microbial functional variables, and then the additional effects of plant traits. Second, we hypothesized that, whilst above- and below-ground traits may be strongly correlated, root traits are more appropriate for estimating variation in microbial functional variables than the more easily measureable above-ground traits, given the known role of root exudates as drivers of soil microbial communities and their activities. This hypothesis was tested by investigating the relative contributions of above- and below-ground plant traits to the overall effects of plant traits on variations in microbial functional variables.
MATERIALS AND METHODS
Description of field sites
Grasslands were sampled for vegetation and soil properties at three field sites across Europe, at the Lautaret Pass (French Alps), the Stubai Valley (Austrian Alps) and the Yorkshire Dales (UK). These sites were selected to provide a range of grasslands typical of western European upland regions. Moreover, they cover a range of intensities of agricultural activity and their recent changes, including abandonment of management, intensification of grassland production through fertilization and biodiversity restoration for agri-environmental objectives. These sites cover a range of fertilities for semi-natural grasslands, spanning a wide range of N availability and associated vegetation communities (Supplementary Data Table S1). The past and present management of these different grasslands has been fully described in previous studies (see Grigulis et al., 2013). Briefly, at the French site, three grassland types were chosen along a gradient from medium N availability to poor N availability (Robson et al., 2007): two on terraced slopes, of which one is fertilized and mown for hay in early August, and the second is not fertilized and mown but lightly grazed. The third grassland type is unterraced and only very lightly grazed during travel of livestock to summer pastures. The peak standing green biomass (AGB – Garnier et al., 2007) of these three grasslands types was respectively 4·73, 2·99 and 3·76 tons of dry mass per hectare. At the Austrian site, two grassland types were chosen along an intensity gradient; the first is an intensive meadow with fertilizer application and two or three vegetation cuts a year (AGB = 3·44 t ha−1), and the second is an abandoned meadow which was previously mown and grazed and is now colonized by trees and shrubs (AGB = 2·98 t ha−1). The UK site is characterized by a long-term fertilization gradient. The most intensively managed meadow is subject to high rates of inorganic fertilizer application with 1–2 vegetation cuts per year and heavy inter-season grazing (AGB = 2·71 t ha−1). The second is a meadow with inorganic fertilizer application, a single annual hay cut and inter-season grazing (AGB = 2·44 t ha−1). The third type is a traditionally managed species-rich hay meadow with light inter-season grazing, a single annual hay cut and no fertilizer inputs (AGB = 2·24 t ha−1).
Sampling strategy
Within each site, three replicate fields (100 m2) were sampled for each of 2–3 management types. Within each replicate field, soil and vegetation were sampled at the same time in four fixed quadrats (50 × 50 cm) during peak biomass at the end of June, early July and end of July 2010 for the UK, French and Austrian sites, respectively. Vegetation was cut at the centre of each quadrat (30 × 30 cm) and five soil cores (4·5 cm Ø, 10 cm depth) were taken from each quadrat. Four cores from each of the four corners were sampled simultaneously and subsequently pooled for soil analysis, and a single core was taken from the centre for root trait analyses. One extra core (205 cm3) was sampled next to the root core for measurement of bulk density and parameters related to soil water availability (see next section).
Soil analysis
Fresh composite soil samples from each plot were weighed and passed through a 5·6-mm sieve. Fresh sieved soil was stored at –20°C (for further phospholipid analysis, DNA extraction and quantification of gene abundance) or 4°C and immediately (within 48 h) processed for the determination of microbial biomass and potential enzymatic activities, soil moisture, soil organic matter content, pH, soil texture (using soil subsequently passed through a 2-mm sieve) and soil nutrients [ammonium (soil NH4+), nitrate (soil NO3−), total dissolved nitrogen (TDN), and dissolved organic nitrogen (DON)] from 0·5 m K2SO4 soil extracts (Jones and Willett, 2006) using an FS-IV colorimetric chain (OI-Analytical Corp., College Station, TX, USA). In situ available soil inorganic N was measured using ion exchange resin bags. In each plot, resin bags were inserted in the soil (5–15 cm deep at a 45° angle) over the growing season at 6-week intervals from May until October (four times), once over the winter period (November to April), and during the following growing season from May to October 2011 (four times). Resin bags were made using nylon bags (10 × 5 or 5 × 5 cm) containing 5 g of mixed anion cation exchange resin (Amberlite IRN150, VWR International S.A.S., Fontenay-sous-Bois, France). Captured nitrate and ammonium were released from the resins in 1 m KCl and analysed on an FS-IV colorimetric chain (OI-Analytical). Subsamples of fresh soil were dried at 70°C for 1 week to determine soil water content (SWC), followed by 4 h at 550°C to determine soil organic matter content (SOM). Soil subsamples were air dried and ground to measure total soil C and N with a FlashEA 1112 elemental analyser (Fisher Scientific Inc., Waltham, MA, USA) and soil pH was measured in a 1:4 (soil/distilled water) solution. Bulk density and soil porosity were obtained by measuring the dry mass of a fixed-volume (205 cm3) soil core. Prior to drying, 100 mL distilled water was added to saturate each soil core and allowed us to calculate water holding capacity (WHC) and water-filled pore space (WFPS).
Fungal and bacterial biomass were determined using phospholipid fatty acid (PLFA) analysis, using the method of Bligh and Dyer (1959), adapted by White et al. (1979) and described by Bardgett et al. (1996). This allowed us to calculate the fungal to bacterial PLFA ratio (F/B ratio), which is commonly used as a measure of the composition of microbial communities and has been linked to soil biogeochemical processes (van der Heijden et al., 2008). Microbial biomass nitrogen (N) was measured using the chloroform-fumigation extraction technique of Vance et al. (1987).
Kinetic parameters (Vmax and Km) of potential nitrification were measured following a protocol adapted from Koper et al. (2010) and Dassonville et al. (2011). Potential denitrification enzyme activity (DEA) was measured according to Attard et al. (2011). Abundance of nitrifiers [ammonium-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB)] as well as nitrite oxidizers [Nitrospira (NIP) and Nitrobacter (NIB)] were quantified based on the gene copy numbers present in soil of the ammonium monooxigenase gene amoA (amoA-AOA amoA-AOB) and 16S rRNA gene of NIP and nrxA gene of NIB, respectively. Abundances of denitrifiers were measured based on the abundance of the nitrite reductase genes [nirK (NRK) and nirS (NRS)]. Quantitative real-time PCR (qPCR) was done following Grigulis et al. (2013). Briefly, soil samples stored at –20°C were used for DNA analyses. Soil DNA was extracted from 0·5 g of fresh soil using the FastDNA SPIN Kit for soil (MP Biomedicals, Irvine, CA, USA) and a Precellys24 Instrument (Bertin Technologies, Montigny-le-Bretonneux, France). Quantity and quality of extracted DNA were tested by spectrophotometry (Nanodrop; PeqLab, Erlangen, Germany). qPCR was carried out using SYBR green as the fluorescent dye. Efficiencies of all performed qPCRs were in the range 85–99 %. R2 values were always above 0·95. Possible inhibition of PCR was tested in advance and appropriate dilutions were chosen (data not shown).
Plant trait analysis
Vegetation composition was quantified in each replicate field using the Botanal method (Lavorel et al., 2008), and above-ground plant traits [SLA, LNC, leaf C concentration (LCC), leaf C/N ratio, leaf dry matter content (LDMC), height] were measured using standard protocols (Cornelissen et al., 2003). Community-weighted mean traits were calculated following Casanoves et al. (2011). A more detailed protocol of vegetation sampling and above-ground trait measurements is given by Grigulis et al. (2013).
Roots were sampled from a dedicated soil core in each of four quadrats per field, which was weighed before careful washing in tepid water to allow separation of roots by floatation using sieve stacks with different mesh, namely 4 mm, 2 mm and 0·2 mm, and separation of live and dead roots was based on visual clues (Bahn et al., 2006). Roots were placed into an alcohol solution (ethanol 10 %, acetic acid 5 %, v/v) and stored at 4°C to maintain freshness until root morphology measurements using digital scanning. Before analysis, roots were suspended in 1 cm of demineralized water in a 29 × 42-cm clear acrylic tray and scanned at 300 d.p.i. with an Epson Expression 10000XL flatbed scanner. Each digital root image was processed using WINRHIZO software (Regent Instruments Inc., Sainte-Foy-Sillery-Cap-Rouge, Canada) to determine total root length and average root diameter. Roots were then weighed, dried at 70°C and reweighed to calculate root dry matter content (RDMC) and specific root length (SRL). Finally, dry roots were ground to a fine powder for analysis of N and C concentrations. Being obtained from community-level soil cores, these root trait measures represented community-weighted means.
Two additional root properties were calculated, namely total root length and root mass per mass of soil (see the list of abbreviations for plant, soil and microbial variables, as well as their respective category in Table 1).
Table 1.
Plant traits, and microbial and soil variables measured and their abbreviations
| Variable | Abbreviation | Units |
|---|---|---|
| Leaf functional traits | ||
| Leaf carbon content | LCC | mg of C g−1 leaf dry mass |
| Leaf carbon/nitrogen ratio | Leaf C/N ratio | |
| Leaf dry matter content | LDMC | mg dry mass g−1 leaf fresh mass |
| Leaf nitrogen content | LNC | mg of N g−1 leaf dry mass |
| Specific leaf area | SLA | m2 of leaf kg−1 leaf dry mass |
| Vegetative height | Vegetative height | cm |
| Root functional traits | ||
| Root carbon content | RCC | mg of C g−1 root dry mass |
| Root carbon/nitrogen ratio | Root C/N ratio | |
| Root diameter | Root Diameter | mm |
| Root dry matter content | RDMC | mg dry mass g−1 root fresh mass |
| Root length soil mass | RLSM | m of root g−1 dry soil |
| Root mass soil mass | RMSM | g of dry root g−1 dry soil |
| Root nitrogen content | RNC | mg of N.g-1 root dry mass |
| Specific root length | SRL | m of root g−1 root dry mass |
| Microbial response variables | ||
| Microbial biomass N | MBN | μg−1 N g−1 dry soil |
| Fungal to bacterial PLFA ratio | F/B ratio | |
| Abundance of ammonia-oxidizing Archaea | AOA | no. of gene copies g−1 dry soil |
| Abundance of ammonia-oxidizing bacteria | AOB | no. of gene copies g−1 dry soil |
| AOA to AOB ratio | AOA/AOB ratio | |
| Abundance of nitrite oxidizers (Nitrospira) | NIP | no. of gene copies g−1 dry soil |
| Abundance of nitrite oxidizers (Nitrobacter) | NIB | no. of gene copies g−1 dry soil |
| NIP to NIB ratio | NIP/NIB ratio | |
| Abundance of the nitrite reductase gene (nirK) | NRK | no. of gene copies g−1 dry soil |
| Abundance of the nitrite reductase gene (nirS) | NRS | no. of gene copies g−1 dry soil |
| NRK to NRS ratio | NRK/NRS ratio | |
| Kinetic parameters of potential nitrification (maximal nitrification rate) | Vmax | μg N-NH4+ g−1 dry soil h−1 |
| Kinetic parameters of potential nitrification (ammonium affinity = 1/Km) | Km | μg N-NH4+ ml−1 |
| Potential denitrification enzyme activity | DEA | μg N-N2O g−1 dry soil h−1 |
| Soil properties | ||
| Dissolved organic nitrogen | DON | μg N g−1 dry soil |
| Soil nitrate | Soil NO3− | μg N-NO3− g−1 dry soil |
| Soil ammonium | Soil NH4+ | μg N-NH4+ g−1 dry soil |
| In situ nitrate absorbed in resin | NO3− sorption | μg N-NO3− g−1 resin d−1 |
| In situ ammonium absorbed in resin | NH4+ sorption | μg N-NH4+ g−1 resin d−2 |
| Soil ammonium to soil nitrate ratio | Soil NH4+/NO3− ratio | |
| Soil organic matter | SOM | % |
| Soil water content | SWC | % |
| Total dissolved nitrogen | TDN | μg N g−1 dry soil |
| Water filled pore space | WFPS | % |
| Water holding capacity | WHC | % |
Community-weighted mean traits were calculated only for leaf functional traits which were measured at the species level whereas below-ground plant traits were measured directly at the community level.
Data analysis
We used correlative modelling to quantify the respective contributions of soil properties and plant traits to variation in microbial functional variables. Data were analysed using a hypothesis-driven process, using linear mixed models with restricted maximum likelihood (REML) estimates (Grigulis et al., 2013). Briefly, in our model, grasslands were considered as replicates grouped within each of the three experimental sites (Stubai, Lautaret, Yorkshire Dales). The REML algorithm allows for the specification of the covariance structure induced by such grouping of the data and provides estimations of parameter effects and variance components for both fixed (soil properties and plant traits) and random (site) effects in the model of microbial functional variables. Analyses were carried out using the average information (AI) algorithm to estimate variance parameters using the software package JMP 7·0 (SAS Institute, Cary, NC, USA). All variables were tested for normality, and log transformations applied, as required, prior to analysis. To provide an objective methodology for the selection of the most parsimonious model, analyses were conducted following Díaz et al. (2007).
Prior to the REML analyses, the number of variables was reduced using principal component analysis to avoid possible multiple correlations among explanatory variables. Then, as described by Grigulis et al. (2013), each response variable was added to the model in a stepwise manner, with those variables significant in the presence of previously fitted variables being retained in the model, and variables no longer significant in the presence of other variables, due to colinearity, being removed from the model. This was especially the case for multiple correlated plant traits that form the ‘plant economics spectrum’ (Reich, 2014), but which we decided not to select a priori, or to combine into a single multivariate proxy, to preserve interpretability based on individual traits. Assuming that soil abiotic properties have a larger influence on microbial traits than plant traits (Fierer and Jackson, 2006; de Vries et al., 2012), multi-variable models for each microbial trait were developed by first fitting those soil properties significant in single variable models, and then fitting those plant effects significant in single variable models. Hence, residual plant effects were considered once impacts of soil properties had been accounted for. Final combined models reported variance explained by sites, and once this variance was accounted for, the variance explained by soil properties and by plant traits, respectively. In addition, within plant traits we examined the relative contributions to variance of above- and below-ground traits.
RESULTS
Effect of soil abiotic properties on microbial community structure, abundances and functioning
Five microbial functional variables linked to potential microbial activities (Km, Vmax), to gene abundance of nitrifiers (NIP, AOA/AOB ratio) and denitrifier groups (NRS) were related only to soil properties (Table 2). Once site variation was removed, models explained between 25 % (Km) and 93 % (Vmax) of their variation. Specifically, except for the AOA/AOB ratio, these microbial functional variables were positively correlated to soil pH. Microbial gene abundances and activities related to nitrification (i.e. AOA/AOB ratio, NIP and Vmax) were linked to the substrate or products (i.e. NH4+ or NO3−) of nitrification. Soil pH and total soil C content explained a large proportion of the variation in microbial biomass N (MBN; 97 %) (Table 2).
Table 2.
Fixed effects plant trait and soil property variables retained within the multiple variable REML models for each of the microbial properties; also presented is the percentage variation in each microbial property explained by the site, the retained fixed effects, the proportion of explanation afforded by the fixed effects due to plant traits and soil parameters, respectively, and the significance (P), Wald statistic and the direction and magnitude of the standardized effect for each of the retained fixed effects
| Response variable | Retained fixed effect | % variation explained by site | % variation explained after site effect was removed |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % variation explained by fixed effect | Proportion of % variation explained by plant traits | Proportion of % variation explained by soil parameters | P | Wald effect | Effect | |||||
| Microbial abundances | Total microbes | Microbial biomass N | pH | 59·5 | 89·8 | 2·9 | 97·1 | <0·0001 | 41·01 | −0·190 |
| Total Soil Carbon | <0·0001 | 69·22 | 0·085 | |||||||
| Specific Leaf Area | 0·0012 | 15·39 | −0·020 | |||||||
| Fungi and bacteria | F/B ratio | Total Soil Carbon | 97·6 | 57·1 | 47·9 | 52·1 | 0·0039 | 10·95 | −0·003 | |
| Soil DON | 0·0375 | 5·04 | 0·017 | |||||||
| Root Diameter | <0·0001 | 15·29 | −0·190 | |||||||
| Nitrifiers | AOA | pH | 63·1 | 78·8 | 14·6 | 85·4 | <0·001 | 30·04 | 0·500 | |
| Soil NO3– | 0·002 | 12·99 | 0·470 | |||||||
| RDMC | 0·0048 | 10·29 | −5·690 | |||||||
| AOB | NH4+ sorption | 17·7 | 74·9 | 28·8 | 71·2 | <0·001 | 18·88 | 0·900 | ||
| RDMC | 0·0265 | 5·95 | −3·350 | |||||||
| RLSM | 0·0325 | 5·43 | −0·530 | |||||||
| AOA/AOB ratio | Soil total Nitrogen | 51·5 | 74·4 | 0·0 | 100·0 | <0·001 | 23·91 | −1·270 | ||
| NH4+ sorption | <0·001 | 37·36 | 0·950 | |||||||
| NO3– sorption | 0·0013 | 17·67 | −0·120 | |||||||
| Soil NH4+ | 0·0164 | 7·88 | 0·880 | |||||||
| NIP | pH | 24·3 | 68·7 | 0·0 | 100·0 | <0·001 | 20·55 | 0·428 | ||
| Soil NH4+/NO3– ratio | <0·001 | 26·63 | −0·600 | |||||||
| NIB | NO3– sorption | 6·9 | 62·1 | 11·4 | 88·6 | 0·023 | 6·57 | 0·400 | ||
| RDMC | 0·042 | 4·92 | −4·930 | |||||||
| NIP/NIB ratio | NH4+ sorption | 42·1 | 58·4 | 24·3 | 75·7 | <0·001 | 19·04 | −1·060 | ||
| Leaf C/N ratio | 0·0248 | 6·22 | 0·036 | |||||||
| Denitrifiers | NRK | NH4+ sorption | 55·1 | 74·4 | 52·7 | 47·3 | 0·0112 | 8·18 | 0·258 | |
| Root C/N ratio | <0·001 | 28·86 | 0·017 | |||||||
| NRS | pH | 41·7 | 56·6 | 0·0 | 100·0 | <0·001 | 25·59 | 0·385 | ||
| NRK/NRS ratio | Root C/N ratio | 31·3 | 79·2 | 100·0 | 0·0 | <0·001 | 47·37 | 94·480 | ||
| Vegetative Height | 0·0074 | 18·93 | 87·410 | |||||||
| Microbial activities | Nitrification | Vmax | pH | 13·3 | 93·3 | 0·0 | 100·0 | <0·001 | 63·28 | 0·300 |
| NO3– sorption | 0·0197 | 6·93 | 0·081 | |||||||
| Soil NH4+/NO3– ratio | <0·001 | 49·75 | −0·457 | |||||||
| Km | pH | 25·1 | 25·0 | 0·0 | 100·0 | 0·0231 | 5·97 | 0·248 | ||
| Denitrification | DEA | Total Soil Carbon | 30·2 | 85·0 | 13·0 | 87·0 | <0·001 | 78·44 | 0·006 | |
| Soil NH4+/NO3– ratio | <0·001 | 26·28 | −0·160 | |||||||
| Leaf Carbon Content | 0·0008 | 15·94 | −0·006 | |||||||
RLSM (root length per gram of dried soil), C/N ratio (carbon/nitrogen content ration), RDMC (root dry matter content), DON (dissolved organic nitrogen), NO3– and NH4+ sorption (in situ nitrate and ammonium absorbed in resin), F/B ratio (fungal to bacterial PLFA ratio), AOA and AOB (ammonia-oxidizing archaea and bacteria, respectively), NIP and NIB for nitrite oxidizers (genera Nitrospira and Nitrobacter, respectively), NRK and NRS (nitrite reductase genes), Vmax and Km (kinetic response variables of potential nitrification), DEA (potential denitrification enzyme activity).
Effect of plant traits on microbial community structure, abundances and functioning
The addition of plant traits, particularly below-ground traits, increased the explanatory power of models for nine microbial functional variables, namely F/B ratio, AOA, AOB, NIB, NIP/NIB ratio, NRK, NRK/NRS ratio and DEA. The NRK/NRS ratio was the sole microbial property for which only plant traits (root C/N and vegetative height) and no soil properties were retained. All other potential microbial activities and gene abundances were related to combined effects of soil properties and plant traits. The variation explained by this set of parameters varied from 57 % (F/B ratio) to 90 % (microbial biomass N) (once site variation was removed), with 3–100 % of that variation explained by plant traits (Table 2). The role of plant traits in explaining the variation of microbial functional variables was present in models for both the nitrification and the denitrification processes, as well as for the F/B ratio. In the following section, we focus on the most relevant models having a strong contribution of plant traits.
Plant traits explained about half of the within-site variation in the F/B ratio, which was strongly and negatively related to the single below-ground trait root diameter (48 % of the 57 % explained by fixed effects). Total soil C concentration and soil DON accounted for the other half of explained within-site variation, showing negative and positive correlations, respectively, with the F/B ratio (Table 2, Fig. 2).
Fig. 2.
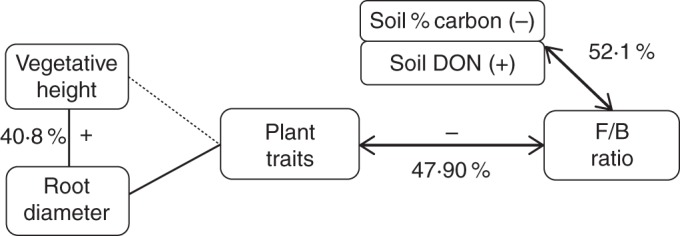
Schematic diagram showing potential links and percentages of explanation of soil properties and plant traits on the F/B ratio (fungal to bacterial PLFA ratio). Positive and negative correlations are represented by + and – symbols next to the arrows when only one variable was involved in the relationship, and by (+) and (–) next to each variable when two variables were involved in the relationships.
Overall, four of the six models for microbial gene abundances involved in nitrification processes retained plant traits (Table 2). For example, 71 % of within-site variation in the gene abundance of amoA from AOB were explained by a model combining a positive relationship with one soil parameter (in situ available soil NH4+) and negative relationships with two plant traits (RDMC and root length by soil mass) (Table 2). In this model, the addition of plant traits improved the model fit, leading to a total of 75 % of the variance explained compared with 58·5 % with soil properties alone. Likewise, the abundance of NRK, a gene involved in denitrification, showed a strong positive relationship with the root C/N ratio, which contributed 52·7 % of the 74 % of explained within-site variance. In addition, NH4+ availability was positively related to NRK and improved model fit from 39 to 74 % (Fig. 3, Table 2). The NRK/NRS ratio, by contrast, was exclusively explained by two plant traits (root C/N ratio and vegetative height) (Table 2). The potential DEA was strongly linked to soil parameters, with total soil C concentration and NH4+/NO3– explaining most (87 %) of its within-site variation, with LCC explaining the remaining 13 %.
Fig. 3.
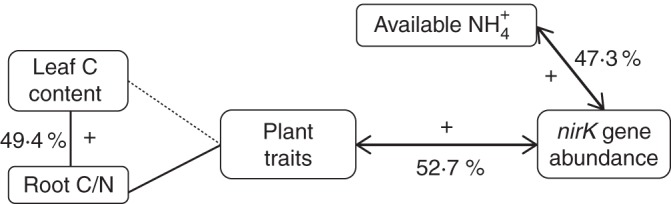
Schematic diagram showing potential links and percentages of explanation of soil properties and plant traits on NRK gene abundance. Positive and negative correlations are represented by + and – symbols.
Relative effects of different plant traits
Above-ground plant traits explained more than 50 % of the variation in below-ground properties and traits, once site variation had been removed. Our results showed that, although above- and below-ground plant traits were both retained in models, they differed in their contributions to variation in microbial functional features. In fact, when retained in models root C/N ratio and RDMC captured a greater proportion of variation of microbial characteristics than above-ground plant traits (Table 2). Hence, the contribution of above-ground plant traits to explained variation ranged from 2·9 to 25 % and was never the best explaining variables. In contrast, the contribution of below-ground plant traits ranged from 11·4 to 75 % and represented the best category of explaining variables in two models as compared with above-ground plant traits or soil properties (Table 2).
DISCUSSION
Overall, our findings indicate the importance of incorporating both above- and below-ground plant traits, in addition to soil abiotic properties, into models explaining variations in soil microbial functional variables associated with N cycling (Fig. 4, size of dashed black arrows). Indeed, both plant compartments explained significant and independent components of variation in soil microbial community composition and functional variables across the broad range of grasslands studied, pointing to their significance as drivers of microbial communities. Also, although several above- and below-ground plant traits were correlated (Table 3), as expected from previous studies (Freschet et al., 2010; Birouste et al., 2012), below-ground traits retained in models captured a greater proportion of variation in measured microbial properties than did leaf traits (Table 2).
Fig. 4.
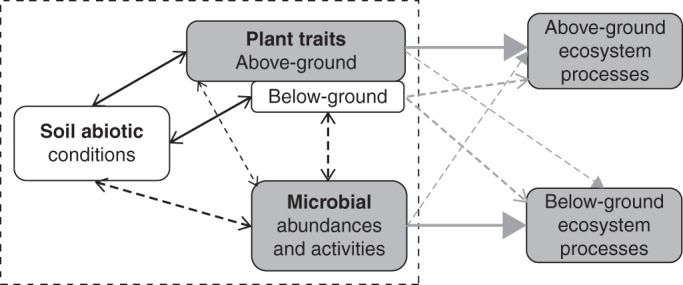
Schematic diagram showing the potential mechanisms by which soil and plants are related to microbial communities and ecosystem processes. The grey boxes and arrows depict the links in Grigulis et al. (2013) and the dashed box represents the links modelled in the present study. The size of the dashed black arrows is proportional to the relative contribution of soil abiotic properties, and above- and below-ground plant traits on variance of microbial functional variables. The dashed grey arrows extrapolate the potential changes that the present study could bring to the contribution of plant and microbial communities on ecosystem processes described by Grigulis et al. (2013).
Table 3.
Fixed effects leaf trait variables retained within the multiple variable REML models for each of the root properties; also presented is the percentage variation in each root property explained by the site, the retained fixed effects, and the significance (P), Wald statistic and the direction and magnitude of the standardized effect for each of the retained fixed effects
| Response variable | Retained fixed effect | % variation explained by site | % variation explained by model (after site effect removed) | P | Effect | Wald |
|---|---|---|---|---|---|---|
| RLSM | LDMC | 56·5 | 34·0 | 0·025 | 2·63 | 5·88 |
| Leaf N Content | 0·043 | −0·02 | 4·71 | |||
| RMSM | Leaf C Content | 66·7 | 59·0 | <0·001 | 0·54 | 29·44 |
| Vegetative Height | 0·011 | −0·76 | 14·20 | |||
| Root Diameter | Vegetative Height | 89·1 | 40·7 | <0·001 | 0·01 | 14·91 |
| SRL | Leaf C Content | 87·0 | 35·2 | 0·0023 | −0·02 | 12·14 |
| RDMC | Leaf C Content | 30·5 | 39·9 | 0·003 | 0·00 | 14·03 |
| Leaf N Content | 0·045 | 0·00 | 4·54 | |||
| Root C Content | Leaf C Content | 10·6 | 50·6 | < 0·001 | 1·08 | 17·25 |
| Vegetative Height | 0·005 | −1·66 | 9·85 | |||
| Root N Content | Leaf C Content | 4·5 | 37·3 | 0·004 | −0·08 | 10·16 |
| Root C/N ratio | Leaf C Content | 2·7 | 49·3 | <0·001 | 0·48 | 17·60 |
RLSM and RMSM (root length and root mass per gram of dried soil), RDMC (root dry matter content), LDMC (leaf dry matter content), C content and N content were the content of carbon and nitrogen in leaves or roots.
Effects of soil abiotic properties on microbial response variables
Abiotic soil properties (pH and nutrient availability) constituted major drivers of soil microbial activities and abundances, as demonstrated in previous studies (e.g. Fierer and Jackson, 2006; Leininger et al., 2006; de Vries et al., 2012). Soil pH was strongly related to many microbial variables (e.g. NEA, AOA, NIP, MBN) and, when retained in models, this parameter had the largest contribution to total explained variation, which is consistent with the findings of previous studies (Fierer and Jackson, 2006; Eskelinen et al., 2009; Laughlin, 2011). Logically, soil nutrient availability (NH4+ and NO3−), which influences microbial growth and activity, also strongly contributed to variation in microbial functional characteristics (Henry et al., 2008; Attard et al., 2010; Verhamme et al., 2011; Le Roux et al., 2013). In contrast, although soil water availability was tested in single or multiple REML models among other explanatory variables for each of the microbial measurements (Supplementary Data Table S2), it was never retained in final models, which was presumably due to its correlation with soil N or ammonium concentrations (data not shown).
Effects of plant traits on microbial processes
To the best of our knowledge, no other study has linked both above- and below-ground plant traits to both soil microbial activities and microbial gene abundances associated with N cycling in field conditions. The inclusion of plant traits significantly improved statistical models for response variables relevant to nitrification and denitrification. Moreover, for one measure, the NRK/NRS ratio, plant traits were the only parameters retained in the final model, and for two measures, F/B ratio and NRK, root traits predicted a large proportion of the variation.
A few studies have highlighted significant effects of plant traits on soil microbial biomass and activities at the individual plant, ecosystem and landscape scale (Orwin et al., 2010; Laughlin, 2011; de Vries et al., 2012). We found that fine roots (i.e. high SRL), which tend to be less developed in slow-growing plants typical of nutrient-poor environments (Craine et al., 2003), were positively associated with the F/B ratio. This suggests that plant traits of slow-growing species could mediate their effects on the composition of microbial communities in nutrient-poor and/or carbon-rich environments. This is consistent with findings of associations of a higher abundance of fungi relative to bacteria with low community-level SLA (de Vries et al., 2012) or high rooting density of individual species (Orwin et al., 2010). Nevertheless, the link observed between root traits and the relative abundance of fungi and bacteria should be considered with caution given possible over-estimation of fungal biomass due to the potential contribution of root cells to the abundance of polyunsaturated PLFAs such as 18:2ω6 (Zelles, 1997; Kaiser et al., 2010), even after sieving and manual removal of root fragments. We also observed a negative relationship between total soil C concentrations and ratio of fungal to bacterial PLFAs, which contrasts with recent findings (de Vries et al., 2012). A plausible explanation for this difference is that a decrease in total soil C could reflect a drop in labile soil C content, which usually favours bacterial communities at the expense of fungal communities (Fontaine and Barot, 2005).
Plant traits contributed to explaining the variation in microbial functional gene abundances, but had low contribution to variation in potential microbial activities (0 % for nitrification and 13 % for denitrification). In fact, all variation in N-related microbial processes was predicted by soil parameters, as they controlled the availability of N-NH4+ and N-NO3− processed by nitrification and denitrification, and thereby outweighed any direct plant trait effects. The most influential plant trait predicting variation in the abundance of genes involved in nitrification (e.g. AOA, AOB and NIB) was RDMC (Fig. 5), with only root length and leaf C/N also being retained in models of the gene abundance of nitrifiers. Such relationships could be mediated by N availability because both RDMC (Craine et al., 2001) and nitrifier gene abundance (Attard et al., 2010; Verhamme et al., 2011) are known to be strongly linked to soil nutrient availability. Grasslands with high N availability are often characterized by exploitative plant communities (e.g. low RDMC) and high N turnover rates (e.g. high nitrification rate and nitrifier abundance) (Robson et al., 2007). Consequently, the negative relationship between RDMC and nitrifier gene abundances may simply result from their simultaneous responses to a change of soil N availability.
Fig. 5.
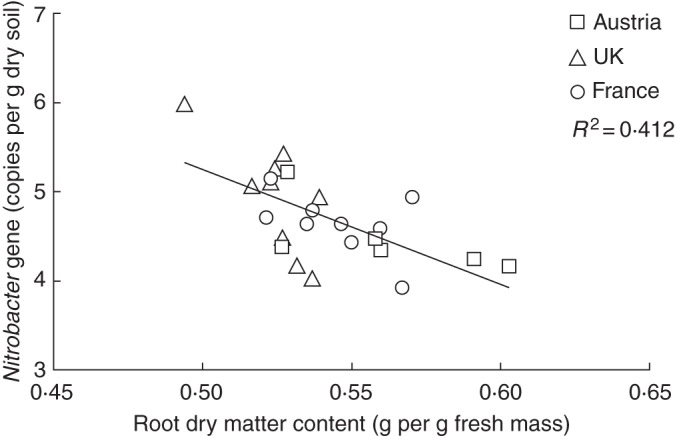
Relationship between root dry matter content and gene copy numbers of Nitrobacter.
Concerning denitrifier gene abundances, two of the three response variables studied (NRK and consequently NRK/NRS ratio) were primarily explained by plant traits in the statistical models (Table 2). In both cases, the root C/N ratio was the most significant variable with an increase of this ratio being related to increased NRK. As root exudate quality impacts on microbial communities (Griffiths et al., 1999), we suggest that root C/N ratio is a proxy of exudate quality and production (Valé et al., 2005), and notably a proxy of the exudate C/N ratio which directly influences microbial communities (Bremer et al., 2007; Henry et al., 2008). Nevertheless, more studies are needed concerning the quality of root exudates and their assimilation by microbial communities to support this hypothesis. Additionally, root C/N ratio may influence the C/N ratio of labile soil fractions (Fornara et al., 2011), with possible indirect effects on denitrifier abundance (Yin et al., 2002). Hence, these two potential mechanisms suggest that high root C/N ratio might be related to an exudation of high C/N compounds, which could change C/N ratios of labile soil fractions and impact the NRK community (Henry et al., 2008).
Although unlikely because sampling was done in early summer, we cannot exclude an indirect effect of root decomposition on soil C availability and then on denitrifiers. It is also possible that a high root C/N ratio could be either related to low N uptake by plants (Craine et al., 2003), or the result of competition with denitrifiers which could limit plant N uptake and lead to low N in roots. These characteristics are typical plant traits in nutrient-poor environments where soil NO3− availability is low, which may also impact the gene abundance of some denitrifiers. Finally, leaf traits had significant effects in models of variation in NRK/NRS ratio and DEA, improving model fit by 25 % (with vegetative height) and 13 % (with LCC), respectively (Table 2). This suggests an indirect effect of leaf traits on denitrifiers linked to litter quality, as a correlation was shown between leaf litter quality and LCC (Fortunel et al., 2009) but also with vegetative height in one of our study sites (Lavorel and Grigulis, 2012). Litter quality, by driving partly the nature of C sources available for microbes, may impact the gene abundance of microbial communities and their ability to use N resources (Wardle, 1992; Falcão Salles et al., 2012).
Relative importance of plant above- and below-ground traits for variation in microbial functional variables
The relative abundance of microbial genes related to key N-processes were in some cases significantly related to both above- and below-ground plant traits. As detailed above, a small set of below-ground plant traits (root diameter, RDMC, root C/N ratio) were highly influential in predicting variation in soil microbial community structure across the grassland sites. Likewise, Orwin et al. (2010) found that RNC and root C concentration (RCC), along with root biomass (not a functional trait per se), showed stronger relationships to soil community structure (microbial biomass and F/B ratio) than did leaf traits. Consequently, although root and leaf traits are dependent on each other (Craine et al., 2001; Freschet et al., 2010), they probably ensure distinct effects or relationships with N-related microbial communities and processes. In the present study, five root traits were retained in the models describing the structure, abundances and activities of microbial communities, and it is difficult to identify a specific root trait for studying plant–microbial relationships. However, all of the selected root traits were linked to plant nutrient uptake capacity (Craine et al., 2003), and were likely to be highly influenced by soil nutrient availability (Craine et al., 2001), and have been suggested to impact root exudate quantity and quality (Valé et al., 2005).
The strong contribution of root traits to statistical models of microbial response variables could be related to two potential mechanisms. On the one hand, root traits reflect plant responses to soil parameters including pH and nutrient conditions, with effects on microbial activities and community structure being additional to direct impacts of these soil parameters. On the other hand, root traits determine the amount and the quality of plant C and N supply to soil microbial communities (mainly denitrifiers), which depend on both soil nutrient status and plant nutrient uptake (Van der Krift et al., 2001). However, the amount of root exudates may also be linked to the strength of the C sink in microbial communities (Bahn et al., 2013). Therefore, such feedback effects from microbial communities to plant traits should be incorporated for a more complete understanding of complex inter-relationships among soil abiotic properties, plant traits and microbial community structure and activities, especially using experimental approaches (Harrison and Bardgett, 2010). In the same way, the mycorhizal status of roots could be included in our analysis as a key root trait, especially given the known roles of mycorrhizal fungi in soil N cycling (Read and Perez-Moreno, 2003; Veresoglou et al., 2011).
Finally, the contribution of below-ground plant traits to microbial functional variables could change the estimation of the relative contributions of plant and microbial parameters to N-cycling processes (Grigulis et al., 2013). This assumption is supported by recent studies showing the role of root traits on N-cycling through their link with root litter decomposability (Birouste et al., 2012) or microbial community composition (Orwin et al., 2010). By accounting for these effects, the contribution of microbial variables to variations in soil ecosystem properties across grasslands, as studied by Grigulis et al. (2013), could decrease in favour of root traits (Fig. 4, dashed grey arrows).
CONCLUSIONS
Our findings highlight the importance of incorporating plant traits, and especially root traits, in analyses of variation in soil N-related microbial activities and community structure and abundances across a broad range of European mountain grasslands. As expected, we found that soil abiotic properties, including soil pH and nutrient availability, were the primary drivers of soil microbial functional variables. However, consistent with our first hypothesis, indirect pathways via above- and below-ground plant functional traits explained additional variation in soil microbial community (microbial gene abundances of N-related processes and F/B ratio) across grassland communities. Also, consistent with our second hypothesis, below-ground traits were more closely linked to microbial properties than were leaf traits, suggesting that consideration of root traits, in addition to leaf traits, increases our ability to explain variation in the structural and functional characteristics of microbial communities in grassland soils. Finally, we suggest that future experiments are needed to advance our understanding of the mechanisms involved in those indirect effects from soil abiotic properties to microbial functional variables that operate via plant functional traits.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This study was conducted as part of ERA-Net BiodivERsA project VITAL, ANR-08-BDVA-008, with the national funders ANR, FWF, NERC, BMBF and Austrian Science Fund (FWF project number I 242-B17). The Lautaret site is part of the long-term research site Zone Atelier Alpes, a member of the ILTER-Europe and LTSER networks. The sites at Stubai are part of the LTSER platform Tyrolean Alps. We acknowledge the Joseph Fourier Alpine research station (CNRS UMS 3370) for infrastructure support, and Edith Primat, Hanna Secher-Frommell and Marie-Pascale Colace for help with field and laboratory measurements at Lautaret. We thank Nadine Guillaumaud for her support in the field campaign and her help for the measurements of microbial activities on the AME platform (UMR5557-USC1364). qPCR to estimate abundances of Nitrobacter and Nitrospira were performed at the DTAMB platform (FR41, University Lyon1). We also thank Dario Fornara and an anonymous reviewer for their considerable input in improving the manuscript.
LITERATURE CITED
- Attard E, Poly F, Commeaux C, et al. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environmental Microbiology. 2010;12:315–326. doi: 10.1111/j.1462-2920.2009.02070.x. [DOI] [PubMed] [Google Scholar]
- Attard E, Recous S, Chabbi A, et al. Soil environmental conditions rather than denitrifier abundance and diversity drive potential denitrification after changes in land uses. Global Change Biology. 2011;17:1975–1989. [Google Scholar]
- Bahn M, Knapp M, Garajova Z, Pfahringer N, Cernusca A. Root respiration in temperate mountain grasslands differing in land use. Global Change Biology. 2006;12:995–1006. [Google Scholar]
- Bahn M, Lattanzi FA, Hasibeder R, et al. Responses of belowground carbon allocation dynamics to extended shading in mountain grassland. New Phytologist. 2013;198:116–126. doi: 10.1111/nph.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert A, Kleineidam K, Wissing L, et al. Changes in diversity and functional gene abundances of microbial communities involved in nitrogen fixation, nitrification, and denitrification in a tidal wetland versus paddy soils cultivated for different time periods. Applied and Environmental Microbiology. 2011;77:6109–6116. doi: 10.1128/AEM.01751-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett RD, Hobbs PJ, Frostegard A. Changes in soil fungal:bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biology and Fertility of Soils. 1996;22:261–264. [Google Scholar]
- Birouste M, Kazakou E, Blanchard A, Roumet C. Plant traits and decomposition: are the relationships for roots comparable to those for leaves? Annals of Botany. 2012;109:463–472. doi: 10.1093/aob/mcr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bremer C, Braker G, Matthies D, Reuter A, Engels C, Conrad R. Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil. Applied and Environmental Microbiology. 2007;73:6876–6884. doi: 10.1128/AEM.01536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanoves F, Pla L, Di Rienzo JA, Diaz S. FDiversity: a software package for the integrated analysis of functional diversity. Methods in Ecology and Evolution. 2011;2:233–237. [Google Scholar]
- Clément JC, Robson TM, Guillemin R, et al. The effects of snow-N deposition and snowmelt dynamics on soil-N cycling in marginal terraced grasslands in the French Alps. Biogeochemistry. 2012;108:297–315. [Google Scholar]
- Cornelissen JHC, Lavorel S, Garnier E, et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Australian Journal of Botany. 2003;51:335–380. [Google Scholar]
- Craine JM, Froehle J, Tilman GD, Wedin DA, Chapin FS. The relationships among root and leaf traits of 76 grassland species and relative abundance along fertility and disturbance gradients. Oikos. 2001;93:274–285. [Google Scholar]
- Craine JM, Wedin DA, Chapin FS, Reich PB. Relationship between the structure of root systems and resource use for 11 North American grassland plants. Plant Ecology. 2003;165:85–100. [Google Scholar]
- Dassonville N, Guillaumaud N, Piola F, Meerts P, Poly F. Niche construction by the invasive Asian knotweeds (species complex Fallopia): impact on activity, abundance and community structure of denitrifiers and nitrifiers. Biological Invasions. 2011;13:1115–1133. [Google Scholar]
- De Deyn GB, Cornelissen JHC, Bardgett RD. Plant functional traits and soil carbon sequestration in contrasting biomes. Ecology Letters. 2008;11:516–531. doi: 10.1111/j.1461-0248.2008.01164.x. [DOI] [PubMed] [Google Scholar]
- de Vries FT, Manning P, Tallowin JRB, et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecology Letters. 2012;15:1230–1239. doi: 10.1111/j.1461-0248.2012.01844.x. [DOI] [PubMed] [Google Scholar]
- Díaz S, Lavorel S, de Bello F, Quetier F, Grigulis K, Robson M. Incorporating plant functional diversity effects in ecosystem service assessments; Proceedings of the National Academy of Sciences of the United States of America; 2007. pp. 20684–20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen A, Stark S, Mannisto M. Links between plant community composition, soil organic matter quality and microbial communities in contrasting tundra habitats. Oecologia. 2009;161:113–123. doi: 10.1007/s00442-009-1362-5. [DOI] [PubMed] [Google Scholar]
- Falcão Salles J, Le Roux X, Poly F. Relating phylogenetic and functional diversity among denitrifiers and quantifying their capacity to predict community functioning. Frontiers in Microbiology. 2012;3:209. doi: 10.3389/fmicb.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities.; Proceedings of the National Academy of Sciences of the United States of America; 2006. pp. 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine S, Barot S. Size and functional diversity of microbe populations control plant persistence and long-term soil carbon accumulation. Ecology Letters. 2005;8:1075–1087. [Google Scholar]
- Fornara DA, Bardgett R, Steinbeiss S, Zak DR, Gleixner G, Tilman D. Plant effects on soil N mineralization are mediated by the composition of multiple soil organic fractions. Ecological Research. 2011;26:201–208. [Google Scholar]
- Fortunel C, Garnier E, Joffre R, et al. Leaf traits capture the effects of land use changes and climate on litter decomposability of grasslands across Europe. Ecology. 2009;90:598–611. doi: 10.1890/08-0418.1. [DOI] [PubMed] [Google Scholar]
- Freschet GT, Cornelissen JHC, van Logtestijn RSP, Aerts R. Evidence of the ‘plant economics spectrum’ in a subarctic flora. Journal of Ecology. 2010;98:362–373. [Google Scholar]
- Garnier E, Lavorel S, Ansquer P, et al. Assessing the effects of land-use change on plant traits, communities and ecosystem functioning in grasslands: a standardized methodology and lessons from an application to 11 European sites. Annals Of Botany. 2007;99:967–985. doi: 10.1093/aob/mcl215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths BS, Ritz K, Ebblewhite N, Dobson G. Soil microbial community structure: effects of substrate loading rates. Soil Biology & Biochemistry. 1999;31:145–153. [Google Scholar]
- Grigulis K, Lavorel S, Krainer U, et al. Relative contributions of plant traits and soil microbial properties to mountain grassland ecosystem services. Journal of Ecology. 2013;101:47–57. [Google Scholar]
- Grime JP. Evidence for existence of 3 primary strategies in plants and its relevance to ecological and evolutionary theory. American Naturalist. 1977;111:1169–1194. [Google Scholar]
- Harrison KA, Bardgett RD. Influence of plant species and soil conditions on plant-soil feedback in mixed grassland communities. Journal of Ecology. 2010;98:384–395. [Google Scholar]
- Henry S, Texier S, Hallet S, et al. Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environmental Microbiology. 2008;10:3082–3092. doi: 10.1111/j.1462-2920.2008.01599.x. [DOI] [PubMed] [Google Scholar]
- Jones DL, Willett VB. Experimental evaluation of methods to quantify dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) in soil. Soil Biology & Biochemistry. 2006;38:991–999. [Google Scholar]
- Kaiser C, Frank A, Wild B, Koranda M, Richter A. Negligible contribution from roots to soil-borne phospholipid fatty acid fungal biomarkers 18:2 omega 6,9 and 18:1 omega 9. Soil Biology & Biochemistry. 2010;42:1650–1652. doi: 10.1016/j.soilbio.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koper TE, Stark JM, Habteselassie MY, Norton JM. Nitrification exhibits Haldane kinetics in an agricultural soil treated with ammonium sulfate or dairy-waste compost. FEMS Microbiology Ecology. 2010;74:316–322. doi: 10.1111/j.1574-6941.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- Kuzyakov Y. Sources of CO2 efflux from soil and review of partitioning methods. Soil Biology & Biochemistry. 2006;38:425–448. [Google Scholar]
- Kuzyakov Y. Priming effects: interactions between living and dead organic matter. Soil Biology & Biochemistry. 2010;42:1363–1371. [Google Scholar]
- Laughlin DC. Nitrification is linked to dominant leaf traits rather than functional diversity. Journal of Ecology. 2011;99:1091–1099. [Google Scholar]
- Lavorel S, Garnier E. Predicting changes in community composition and ecosystem functioning from plant traits: revisiting the Holy Grail. Functional Ecology. 2002;16:545–556. [Google Scholar]
- Lavorel S, Grigulis K. How fundamental plant functional trait relationships scale-up to trade-offs and synergies in ecosystem services. Journal of Ecology. 2012;100:128–140. [Google Scholar]
- Lavorel S, Grigulis K, McIntyre S, et al. Assessing functional diversity in the field – methodology matters! Functional Ecology. 2008;22:134–147. [Google Scholar]
- Le Roux X, Poly F, Currey P, et al. Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. Isme Journal. 2008;2:221–232. doi: 10.1038/ismej.2007.109. [DOI] [PubMed] [Google Scholar]
- Le Roux X, Schmid B, Poly F, et al. Soil environmental conditions and microbial build-up mediate the effect of plant diversity on soil nitrifying and denitrifying enzyme activities in temperate grasslands. Plos One. 2013;8:e61069. doi: 10.1371/journal.pone.0061069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay N, Personeni E, Slezack-Deschaumes S, Piutti S, Cliquet JB. Grassland species show similar strategies for sulphur and nitrogen acquisition. Plant and Soil. 2014;375:113–126. [Google Scholar]
- Leininger S, Urich T, Schloter M, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Ordoñez JC, van Bodegom PM, Witte JPM, Wright IJ, Reich PB, Aerts R. A global study of relationships between leaf traits, climate and soil measures of nutrient fertility. Global Ecology and Biogeography. 2009;18:137–149. [Google Scholar]
- Orwin KH, Buckland SM, Johnson D, et al. Linkages of plant traits to soil properties and the functioning of temperate grassland. Journal of Ecology. 2010;98:1074–1083. [Google Scholar]
- Pohl M, Stroude R, Buttler A, Rixen C. Functional traits and root morphology of alpine plants. Annals of Botany. 2011;108:537–545. doi: 10.1093/aob/mcr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read DJ, Perez-Moreno J. Mycorrhizas and nutrient cycling in ecosystems – a journey towards relevance? New Phytologist. 2003;157:475–492. doi: 10.1046/j.1469-8137.2003.00704.x. [DOI] [PubMed] [Google Scholar]
- Reich PB. The world-wide ‘fast-slow’ plant economics spectrum: a traits manifesto. Journal of Ecology. 2014;102:275–301. [Google Scholar]
- Robson TM, Lavorel S, Clement JC, Le Roux X. Neglect of mowing and manuring leads to slower nitrogen cycling in subalpine grasslands. Soil Biology & Biochemistry. 2007;39:930–941. [Google Scholar]
- Roumet C, Urcelay C, Diaz S. Suites of root traits differ between annual and perennial species growing in the field. New Phytologist. 2006;170:357–368. doi: 10.1111/j.1469-8137.2006.01667.x. [DOI] [PubMed] [Google Scholar]
- Valé M, Nguyen C, Dambrine E, Dupouey JL. Microbial activity in the rhizosphere soil of six herbaceous species cultivated in a greenhouse is correlated with shoot biomass and root C concentrations. Soil Biology & Biochemistry. 2005;37:2329–2333. [Google Scholar]
- van der Heijden MGA, Bardgett RD, van Straalen NM. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecology Letters. 2008;11:296–310. doi: 10.1111/j.1461-0248.2007.01139.x. [DOI] [PubMed] [Google Scholar]
- Van der Krift TAJ, Kuikman PJ, Moller F, Berendse F. Plant species and nutritional-mediated control over rhizodeposition and root decomposition. Plant and Soil. 2001;228:191–200. [Google Scholar]
- van der Putten WH, Bardgett RD, Bever JD, et al. Plant-soil feedbacks: the past, the present and future challenges. Journal of Ecology. 2013;101:265–276. [Google Scholar]
- Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass-C. Soil Biology & Biochemistry. 1987;19:703–707. [Google Scholar]
- Veresoglou SD, Sen R, Mamolos AP, Veresoglou DS. Plant species identity and arbuscular mycorrhizal status modulate potential nitrification rates in nitrogen-limited grassland soils. Journal of Ecology. 2011;99:1339–1349. [Google Scholar]
- Verhamme DT, Prosser JI, Nicol GW. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. Isme Journal. 2011;5:1067–1071. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle DA. A comparative-assessment of factors which influence microbial biomass carbon and nitrogen levels in soil. Biological Reviews of the Cambridge Philosophical Society. 1992;67:321–358. [Google Scholar]
- White DC, Davis WM, Nickels JS, King JD, Bobbie RJ. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- Yin SX, Chen D, Chen LM, Edis R. Dissimilatory nitrate reduction to ammonium and responsible microorganisms in two Chinese and Australian paddy soils. Soil Biology & Biochemistry. 2002;34:1131–1137. [Google Scholar]
- Zelles L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 1997;35:275–294. doi: 10.1016/s0045-6535(97)00155-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


