Abstract
Apomyoglobin folding proceeds through a molten globule intermediate (low-salt form; I1) that has been characterized by equilibrium (pH 4) and kinetic (pH 6) folding experiments. Of the eight alpha-helices in myoglobin, three (A, G, and H) are structured in I1, while the rest appear to be unfolded. Here we report on the structure and stability of a second intermediate, the trichloroacetate form of the molten globule intermediate (I2), which is induced either from the acid-unfolded protein or from I1 by > or = 5 mM sodium trichloroacetate. Circular dichroism measurements monitoring urea- and acid-induced unfolding indicate that I2 is more highly structured and more stable than I1. Although I2 exhibits properties closer to those of the native protein, one-dimensional NMR spectra show that it maintains the lack of fixed side-chain structure that is the hallmark of a molten globule. Amide proton exchange and 1H-15N two-dimensional NMR experiments are used to identify the source of the extra helicity observed in I2. The results reveal that the existing A, G, and H helices present in I1 have become more stable in I2 and that a fourth helix--the B helix--has been incorporated into the molten globule. Available evidence is consistent with I2 being an on-pathway intermediate. The data support the view that apomyoglobin folds in a sequential fashion through a single pathway populated by intermediates of increasing structure and stability.
Full text
PDF
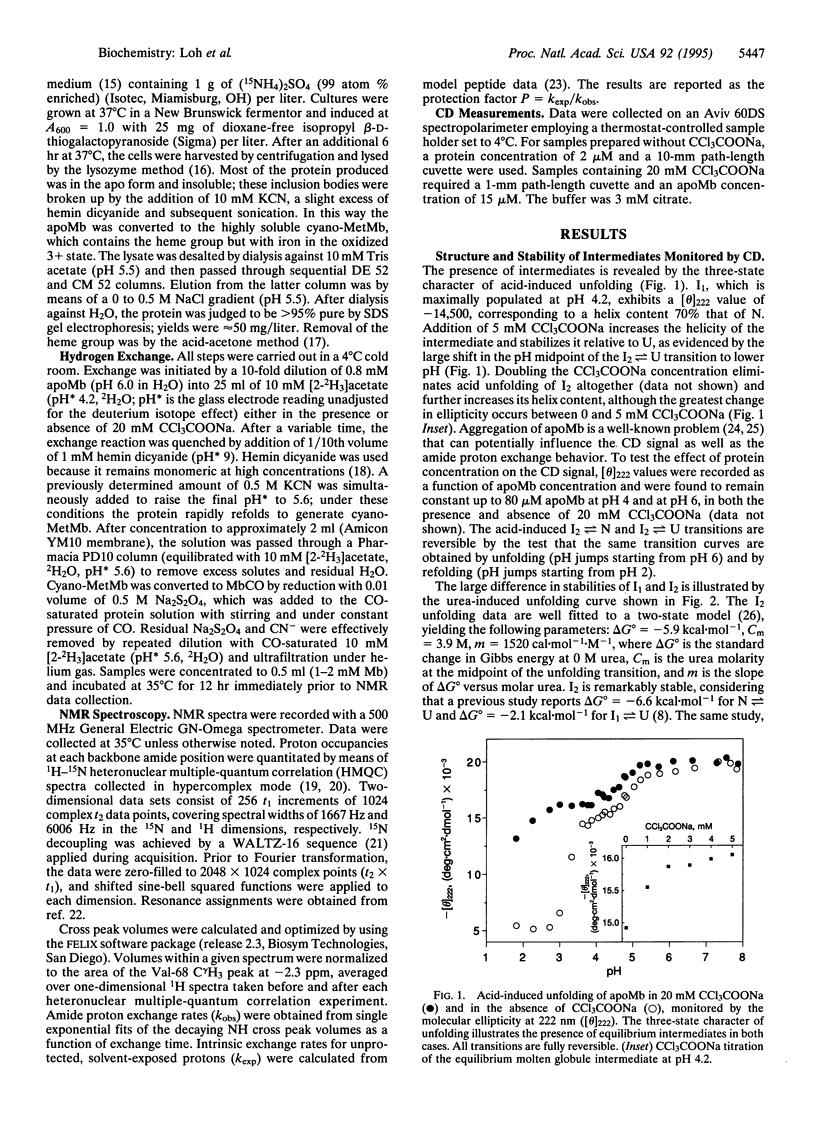
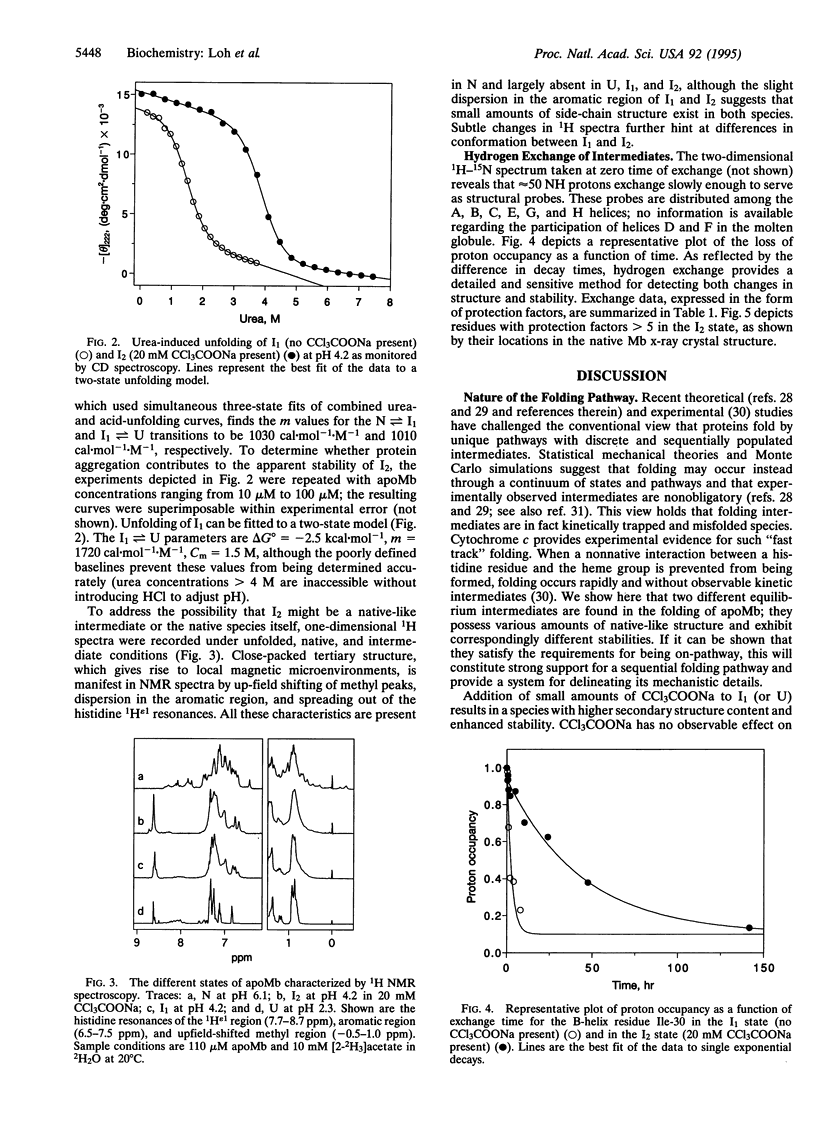


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bai Y., Milne J. S., Mayne L., Englander S. W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R. L. The nature of protein folding pathways: the classical versus the new view. J Biomol NMR. 1995 Feb;5(2):103–109. doi: 10.1007/BF00208801. [DOI] [PubMed] [Google Scholar]
- Barrick D., Baldwin R. L. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993 Apr 13;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Nicholson H., Schellman J. A. Folding kinetics of T4 lysozyme and nine mutants at 12 degrees C. Biochemistry. 1992 Feb 11;31(5):1464–1476. doi: 10.1021/bi00120a025. [DOI] [PubMed] [Google Scholar]
- Cull M., McHenry C. S. Preparation of extracts from prokaryotes. Methods Enzymol. 1990;182:147–153. doi: 10.1016/0076-6879(90)82014-s. [DOI] [PubMed] [Google Scholar]
- De Young L. R., Dill K. A., Fink A. L. Aggregation and denaturation of apomyoglobin in aqueous urea solutions. Biochemistry. 1993 Apr 20;32(15):3877–3886. doi: 10.1021/bi00066a006. [DOI] [PubMed] [Google Scholar]
- FANELLI A. R., ANTONINI E., CAPUTO A. Studies on the structure of hemoglobin. I. Physicochemical properties of human globin. Biochim Biophys Acta. 1958 Dec;30(3):608–615. doi: 10.1016/0006-3002(58)90108-2. [DOI] [PubMed] [Google Scholar]
- Goto Y., Calciano L. J., Fink A. L. Acid-induced folding of proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Nishikiori S. Role of electrostatic repulsion in the acidic molten globule of cytochrome c. J Mol Biol. 1991 Dec 5;222(3):679–686. doi: 10.1016/0022-2836(91)90504-y. [DOI] [PubMed] [Google Scholar]
- Goto Y., Takahashi N., Fink A. L. Mechanism of acid-induced folding of proteins. Biochemistry. 1990 Apr 10;29(14):3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- Griko Y. V., Privalov P. L. Thermodynamic puzzle of apomyoglobin unfolding. J Mol Biol. 1994 Jan 28;235(4):1318–1325. doi: 10.1006/jmbi.1994.1085. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Barrick D., Baldwin R. L. Probing the stability of a partly folded apomyoglobin intermediate by site-directed mutagenesis. Biochemistry. 1991 Apr 30;30(17):4113–4118. doi: 10.1021/bi00231a001. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Wright P. E., Baldwin R. L. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990 Sep 28;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- Jennings P. A., Wright P. E. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993 Nov 5;262(5135):892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Wilz S., Karplus M., Petsko G. A. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 A resolution. J Mol Biol. 1986 Nov 5;192(1):133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. A folding model of alpha-lactalbumin deduced from the three-state denaturation mechanism. J Mol Biol. 1977 Aug 5;114(2):241–258. doi: 10.1016/0022-2836(77)90208-x. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Nishii I., Kataoka M., Tokunaga F., Goto Y. Cold denaturation of the molten globule states of apomyoglobin and a profile for protein folding. Biochemistry. 1994 Apr 26;33(16):4903–4909. doi: 10.1021/bi00182a019. [DOI] [PubMed] [Google Scholar]
- Ogasahara K., Yutani K. Unfolding-refolding kinetics of the tryptophan synthase alpha subunit by CD and fluorescence measurements. J Mol Biol. 1994 Mar 4;236(4):1227–1240. doi: 10.1016/0022-2836(94)90023-x. [DOI] [PubMed] [Google Scholar]
- Onuchic J. N., Wolynes P. G., Luthey-Schulten Z., Socci N. D. Toward an outline of the topography of a realistic protein-folding funnel. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3626–3630. doi: 10.1073/pnas.92.8.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUMEN N. M., APPELLA E. Molecular association behavior of apomyoglobin I from the seal (Phoca vitulina). Arch Biochem Biophys. 1962 Apr;97:128–133. doi: 10.1016/0003-9861(62)90053-x. [DOI] [PubMed] [Google Scholar]
- Robertson A. D., Baldwin R. L. Hydrogen exchange in thermally denatured ribonuclease A. Biochemistry. 1991 Oct 15;30(41):9907–9914. doi: 10.1021/bi00105a014. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994 May 19;369(6477):248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- Santoro M. M., Bolen D. W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988 Oct 18;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- Sanz J. M., Johnson C. M., Fersht A. R. The A-state of barnase. Biochemistry. 1994 Sep 20;33(37):11189–11199. doi: 10.1021/bi00203a015. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Hiller R., Englander S. W. The barriers in protein folding. Nat Struct Biol. 1994 Mar;1(3):149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Springer B. A., Sligar S. G. High-level expression of sperm whale myoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault Y., Pochapsky T. C., Dalvit C., Chiu M. L., Sligar S. G., Wright P. E. 1H and 15N resonance assignments and secondary structure of the carbon monoxide complex of sperm whale myoglobin. J Biomol NMR. 1994 Jul;4(4):491–504. doi: 10.1007/BF00156616. [DOI] [PubMed] [Google Scholar]
- Yee S., Peyton D. H. Proton NMR investigation of the reconstitution of equine myoglobin with hemin dicyanide. Evidence for late formation of the proximal His93F8-iron bond. FEBS Lett. 1991 Sep 23;290(1-2):119–122. doi: 10.1016/0014-5793(91)81240-9. [DOI] [PubMed] [Google Scholar]