Abstract
One-third of type 2 diabetes patients do not respond to metformin. Genetic variants in metformin transporters have been extensively studied as a likely contributor to this high failure rate. Here, we investigate, for the first time, the effect of genetic variants in transcription factors on metformin pharmacokinetics (PK) and response. Overall, 546 patients and healthy volunteers contributed their genome-wide, pharmacokinetic (235 subjects), and HbA1c data (440 patients) for this analysis. Five variants in specificity protein 1 (SP1), a transcription factor that modulates the expression of metformin transporters, were associated with changes in treatment HbA1c (P < 0.01) and metformin secretory clearance (P < 0.05). Population pharmacokinetic modeling further confirmed a 24% reduction in apparent clearance in homozygous carriers of one such variant, rs784888. Genetic variants in other transcription factors, peroxisome proliferator–activated receptor-α and hepatocyte nuclear factor 4-α, were significantly associated with HbA1c change only. Overall, our study highlights the importance of genetic variants in transcription factors as modulators of metformin PK and response.
Metformin is first-line therapy for type 2 diabetes, and it is also one of the most commonly prescribed drugs worldwide. 1–10 Despite 50 years of clinical use, its mechanism of action remains controversial. It has been well established that metformin activates adenine monophosphate–activated protein kinase, which may contribute to many of the pharmacological outcomes of metformin, including the inhibition of gluconeogenesis, reduction of glucose absorption, and enhancement of glucose uptake and utilization.2,6,11
There is considerable variability in the glycemic response and pharmacokinetic characteristics of metformin. In terms of pharmacokinetics (PK), metformin is not metabolized, and is excreted unchanged in the urine, with a half-life of roughly 5 h.2,5,6,10 The pharmacokinetic variability of metformin is unusually high for a renally cleared drug. In particular, mean plasma concentrations of metformin fluctuate between 0.4 and 1.3 mg/l at a dose of 1,000 mg twice daily.1,2,5,6,8,12–17 Metformin relies on facilitated transport for uptake into various tissues as well as for renal elimination. Specifically, transporters that mediate metformin elimination and tissue distribution include organic cation transporters (OCTs) and multidrug and toxin extrusion proteins (MATEs), and may contribute to the wide variation in metformin PK. Pharmacokinetic variability contributes to variation in response to metformin: various research groups have observed dose-response relationships with fasting plasma glucose and HbA1c levels. 18–20 Metformin response variability is substantial; >30% of patients receiving metformin are classified as poor responders.1,5,8,10
To date, many pharmacogenetic studies have focused on the relationship between genetic variants in transporters and metformin pharmacokinetic parameters, and there has been one genome-wide association study for metformin response. 1,5,8,12–17,21–24
For example, OCT1 is a major determinant of metformin uptake into hepatocytes, and genetic polymorphisms of OCT1 have been associated with reduced response and changes in metformin PK in healthy subjects and diabetes patients. 1,7–9 Recently, promoter variants of MATE1 and MATE2K, transporters that determine the efflux of metformin into the urine, were also shown to be associated with metformin disposition and response in healthy subjects and diabetes patients.5,15,25 Understanding genetic predictors of variability in terms of both its response and disposition is important in the rational use of metformin for the treatment of patients with type 2 diabetes.
Although genetic studies have demonstrated associations between single-nucleotide polymorphisms (SNPs) in transporters and metformin PK and pharmacodynamics (PD), each individual SNP accounts only for a small fraction of the variation in HbA1c among type 2 diabetes patients. This is not surprising given that metformin disposition is governed by multiple transporters rather than a single transporter (Figure 1). With this in mind, we proposed to study genetic variants in transcription factors that may regulate the expression levels of multiple metformin transporters and thus may have larger effects on metformin disposition and response than variants in a single transporter. A subset of transcription factors have been shown to modulate the expression levels of OCTs (SLC22) and MATEs (SLC47), which are involved in determining metformin PK.26 For example, transfection of hepatocyte nuclear factor 4-α (HNF4-a) has been shown to increase transcript levels of OCT1 in hepatocytes.3,7,9 Specificity protein 1 (SP1) has been implicated in modulating mRNA levels of MATE1.27–31 Activating enhancer binding protein (AP)2 has been shown to have a repressive effect on MATE1 gene expression.3,28,30 Other transcription factors have also been linked to modulating the expression levels of OCTs and MATEs involved in metformin disposition.7,9,27,29,31,32 To date, the impact of transcription factor polymorphisms on metformin PK and response phenotypes has not been studied. Our hypothesis is that, compared with genetic variants in transporter genes, genetic variants in transcription factors may have a stronger impact on overall metformin plasma and tissue levels. This is because transcription factors modulate expression levels of a system of transporters, leading to stronger effect sizes on pharmacological outcomes.
Figure 1.
Candidate transcription factors known to modulate gene expression levels of transporters involved in metformin disposition. (a) A cell diagram that depicts a putative network of transcription factors working in concert to modulate gene expression levels of metformin transporters. (b) A high-level gene diagram that highlights a mechanism by which a single-nucleotide polymorphism change in a transcription factor gene may modulate the pharmacological outcome of metformin. PK/PD, pharmacokinetics/pharmacodynamics.
In this study, we first investigated the effect of genetic variants in a subset of genes on metformin response, specifically HbA1c levels in type 2 diabetes patients. The genes included were relevant metformin transcription factors cited in the literature and demonstrated to play a modulatory role on key metformin transporters. Subsequently, for the most significant transcription factor variants associated with HbA1c change, we further investigated their relationship with metformin PK using two approaches: (i) In a subset of healthy subjects with abundant pharmacokinetic measurements and available urine data, we used multiple linear regression to investigate the effect of the top transcription factor variants on measured metformin secretory clearance, which is a major route of metformin elimination. (ii) Using data combined from type 2 diabetes patients and healthy subjects, we then developed a population pharmacokinetic model to investigate the effect of prioritized transcription factor variants, shown to be significantly associated with secretory clearance, on various metformin pharmacokinetic parameters.1,5,8,14
Our study suggests the importance of transcription factors and genetic variants in transcription factors with regard to the pharmacological outcomes of metformin use, namely HbA1c levels and metformin pharmacokinetic parameters. Variants in SP1 exhibited the strongest association with both metformin PK and PD.
RESULTS
Characteristics of type 2 diabetes patients and healthy subjects
Baseline characteristics of patients and healthy subjects are summarized in Table 1. Clinical data included longitudinal HbA1c measurements from 440 type 2 diabetes patients. A total of 2,382 metformin plasma samples in healthy subjects (102) and patients (133) were used to develop a population pharmacokinetic model. Of the 102 healthy subjects, 57 subjects also had available urine samples, which enabled the collection of creatinine levels and the subsequent calculation of metformin secretory clearance.
Table 1.
Baseline characteristics of patients with type 2 diabetes and healthy volunteers dosed with metformin
| Characteristic | Type 2 diabetes patients | Healthy volunteers | |||
|---|---|---|---|---|---|
| Marshfield Clinic | Kaiser Southeast | Vanderbilt | Study no. 6112/6113 | Study no. 865/767 | |
| N | 149 | 133 | 162 | 57 | 45 |
| Available pharmacokinetic data, n | 0 | 133 | 0 | 57 | 45 |
| Male, n | 65 | 44 | 78 | 21 | 23 |
| Female, n | 84 | 89 | 84 | 36 | 22 |
| European American (%) | 100 | 18 | NA | 32 | 84 |
| African American (%) | — | 76 | NA | 58 | 11 |
| Asian American (%) | — | 4 | NA | 10 | 5 |
| Other (%) | — | 2 | NA | — | — |
| Quantitative traits | |||||
| Age (years) | 57 (23–90) | 58(33–79) | 59(33–81) | 25(18–45) | 31 (18–44) |
| Average body weight (kg) | 98(51–212) | 98(58–182) | 93(34–184) | 73(49–136) | 70 (44–112) |
| BaselineHbA1c(%) | 7.5(5.5–12.8) | 7.7(5.2–11.9) | 7.6(5.8–14.9) | — | — |
| Average metformin daily dose (mg) | 990 (500–2,000) | 1,000(250–2,500) | 1,000(250–2,000) | NA | NA |
| Metformin dose (mg) (healthy volunteers) |
NA | NA | NA | 1,850 | 1,850/850 |
Quantitative data shown reflect median (range). Of the Kaiser Southeast cohort, four patients did not have reported baseline HbA1c levels.
Study 6112/6113/865: Healthy volunteers in these studies were administered 1,000 mg of metformin, followed by 850 mg of metformin after a 12-h interval. Healthy subjects in studies 6112/6113 also had urine data available to calculate metformin secretory clearance. See refs. 5,8, and 9.
Study 767: Healthy volunteers were given a single dose of metformin (850 mg). See ref. 9.
Vanderbilt ethnicity is not reported (NA). Principal components were used for analysis across all studies.
All healthy volunteers and 133 patients with sparse pharmacokinetic information were used to build the population pharmacokinetic model.
NA, not available.
Top transcription factor variants from a multivariate regression approach
A total of five transcription factors were selected (AP1, AP2, SP1, HNF4-α, and peroxisome proliferator–activated receptor (PPAR)-α) for multivariate linear regression with treatment HbA1c levels (Supplementary Table S2). Among the tested genetic variants that met our criteria (see Methods section), 40 SNPs were associated with HbA1c change at 3 months after metformin initiation, adjusted for baseline levels (Treatment HbA1c). Among the 40 SNPs, a multivariate linear regression on metformin secretory clearance was performed as one method for investigating a pharmacokinetic mechanism. A total of six SNPs in two genes were significantly associated with metformin secretory clearance and treatment HbA1c levels using a multivariate regression model. Of these six genetic variants, five were located in the SP1 region and one was located in the AP2 region (Supplementary Table S1 online).
Population pharmacokinetic model
In addition to metformin secretory clearance, a population-based modeling approach was used to investigate the effect of prioritized transcription factor variants on the systemic plasma levels of metformin in both patients and healthy subjects. The six-transcription factor SNPs (associated with metformin PK and PD) identified from the multiple linear regression analysis, along with 13 genetic variants in metformin transporters (Supplementary Table S5 online) and demographic variables, were investigated.
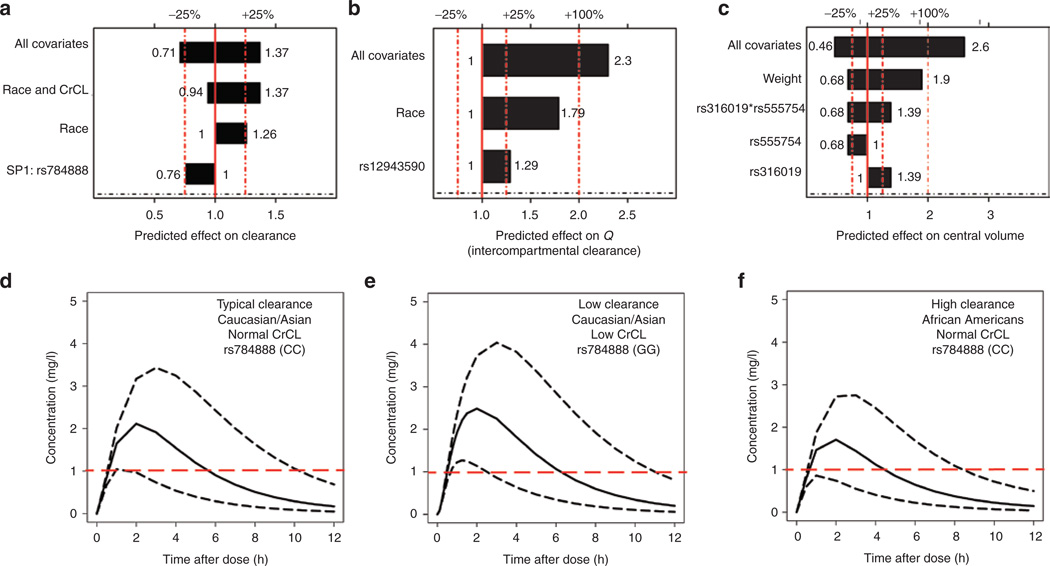
A two-compartment model with a delayed absorption (one-transit compartment) best described the data. A schematic of the final model with included covariates is shown in Figure 2. Pharmacokinetic profiles were overall quite similar between healthy subjects and patients. The final model consisted of statistically significant covariates on central compartment volume (Vc/F), apparent clearance (CL/F), and metformin peripheral flow (Q/F). Covariates that affected Vc/F were body weight, OCT3 variant rs555754, and OCT2 variant rs316019. Based on different combinations of these covariates, Vc/F may decrease by as much as 54% and increase by as much as 260% (see Figure 3). For peripheral flow (Q/F), ethnicity and the MATE2 variant rs12943590 were significant. In Figure 3, the reference ethnic population was European American to more clearly show the range of covariate effects on Q/F and CL/F. Finally, for CL/F, creatinine clearance (CrCL), ethnicity, and the SP1 variant rs784888 were significant. Different combinations of these covariates may decrease CL/F by as much as 29% and increase it by as much as 37%. Final parameter estimates are summarized in Table 2.
Figure 2.
Final metformin population pharmacokinetic model structure and visual predictive check. (a) A two-compartment model with delayed absorption best characterizes the data. All parameters are ratios over the bioavailability of metformin (F). The model structure includes the final model covariates determined by a stepwise covariate analysis (SCM). (b) Visual predictive check of the final population pharmacokinetic model. The shaded regions indicate the 95th and 5th percentiles (ends) and the range of median simulated profiles (center) of simulated predictions from the visual predictive check. Overlaid back points are combined healthy volunteer (n = 102) and type 2 diabetes patient (n = 133) data observations. The solid line indicates the median of the observed data, with red lines indicating the 95th and 5th percentiles of observations. CrCL, creatinine clearance; CL, apparent clearance of metformin; Ka, absorption rate constant; Ktr, transit rate constant, Vc, apparent central volume of distribution; Vp, apparent peripheral volume of distribution.
Figure 3.
Predicted effects of demographic and genetic factors on metformin pharmacokinetic parameters. (a) Predicted effects of covariates on apparent clearance (CL/F) with the reference ethnicity identified as European Americans. (b) Predicted effect of covariates on intercompartmental clearance (Q/F). (c) Predicted effects of covariates on central compartment volume (Vc/F). (d, e, and f) Simulations of pharmacokinetic profiles after a single 850-mg dose of metformin with variability on clearance. Simulations are based on the predicted metformin clearance estimates. Predicted clearance estimates are based on the covariates described in Figure 3a. Ethnicity, creatinine clearance (CrCL), and SP1 variant status are shown in each panel. Dashed line at 1 mg/l indicates the lower target of metformin concentration based on therapeutic range. (d) Black solid line = typical value of clearance for a Caucasian with normal creatinine clearance (80–130ml/min) who is homozygous CC for SNP rs784888. Dashed black lines indicate the 97.5 and 2.5 percentiles of the interindividual variability (ETA) distribution for CL/F for this patient. (e) Black solid line = typical value of clearance for a Caucasian with low creatinine clearance (<70ml/min) who is homozygous GG for SNP rs784888. Dashed lines indicate the 97.5 and 2.5 percentiles of the interindividual variability (ETA) distribution for CL/F for this patient. (f) Black solid line = typical value of clearance for an African American with normal creatinine clearance (80–130ml/min) who is homozygous CC for SNP rs784888. Dashed lines indicate the 97.5 and 2.5 percentiles of the interindividual variability (ETA) distribution for CL/F or this patient. CrCL, creatinine clearance; SNP, single-nucleotide polymorphism.
Table 2.
Population pharmacokinetic model-derived estimates and bootstrap results for pharmacokinetic parameters of metformin
| Final model parameter | Median (%RSE)a |
Median (90% Cl)b |
|---|---|---|
| Total clearance (CL/F; I/h) | 78.4 (6) | 77.5 (70–87) |
| Central volume of distribution (Vc/F; I) | 76.8(15) | 76.7 (53–95) |
| Peripheral flow (Q/F; I/h) | 18.1 (9) | 18.8(16–32) |
| Peripheral volume of distribution (Vp/F; I) | 413(43) | 419(286–3,293) |
| Mean transit time (h) | 0.207(24) | 0.2033(0.14–0.37) |
| Absorption rate(ka; 1/h) | 0.312(5) | 0.31 (0.28–0.34) |
| Interindividual variability (% variance) | ||
| Between-subject variability (CL) | 50(10) | 50 (40–60) |
| Between-subject variability (Vc) | 45(13) | 45 (34–64) |
| Between-subject variability (Q) | 41 (23) | 40 (27–49) |
| Covariance of parameters | ||
| Correlation between total clearance and central volumeof distribution (CL-V) |
0.05 (5) | 0.10 (−0.33–0.29) |
| Residual error model | ||
| Studies 6112/6113 | ||
| Proportional error (%) | 14(12) | 0.14(0.11–0.16) |
| Additive error | 0.02(32) | 0.02(0.01–0.02) |
| Study 865 | ||
| Proportional error (%) | 12(6) | 12(11–13) |
| Additive error | 0.01 | - |
| Study 787 | ||
| Proportional error (%) | 21(13) | 20(16–25) |
| Additive error | 0.01 (20) | 0.007(0.003–0.012) |
| Patient data | ||
| Proportional error (%) | 20(13) | 0.2(0.16–0.24) |
| Additive error | 0.01 | - |
Typical value of pharmacokinetic parameter in final model. RSE, relative standard error (%), also known as the precision of the population pharmacokinetic parameter estimate.
Confidence interval (CI) forthe population pharmacokinetic parameterfollowing bootstrap results. Reference ethnicity of model output is African Americans, due to the high proportion of African Americans in the cohort.
The final model explained 5% of the variability in CL/F, 23% of the variability in Vc/F, and 13% of the variability in Q/F. The SP1 variant rs784888 and ethnicity reduced the variance of interindividual variability in CL/F by 1.6 and 2.3%, respectively. Based on model estimates, an extreme case of high metformin clearance (low exposure) occurs in an African-American patient with normal renal function and homozygous CC for the SP1 variant rs784888. The performance of the final model based on model diagnostics was adequate (Table 2 and Figure 2). The final equations for CL/F, Vc/F, and Q/F were (Eqs. 1, 2, and 3, respectively):
| (1) |
| (2) |
| (3) |
where 78.4 l/h is the typical metformin apparent clearance for an African American with a creatinine clearance of 112 ml/min and homozygous reference (CC genotype) for the SP1 variant, rs784888. The imputed SP1 variant can take on a value between 0 and 2, which quantifies the presence of a G allele. Similarly, 76.8 l is the typical central compartment volume for an African American weighing 75 kg (WT), and is a homozygous reference for both rs555754 (SLC22A3) and rs316019 (SLC22A2). ΘCRCL, ΘEthnicity, CL, Θrs784888, Θrs555754, ΘWT, Θrs316019, Θrs12943590, and ΘEthnicity, Q, are the corresponding effect sizes for creatinine clearance and ethnicity on CL/F, and SP1 variant rs784888, OCT3 variant rs555754, body weight, OCT2 variant rs316019, MATE2 variant rs12943590, and ethnicity on Q/F, respectively. The number of subjects in the data set with an imputed value of greater than or equal to 1 for variants rs784888, rs555754, rs316019,and rs12943590 were 54, 131, 106, and 60, respectively. African Americans were used as the reference ethnic population in the model due to a large representation of African Americans in our cohort. In Figure 3, the reference ethnic population was changed to European American to more clearly show the range of covariate effects on each pharmacokinetic parameter.
Genetic variants in SP1 were critical determinants of variation in PK and PD of metformin
Our top finding was in regard to the gene encoding the transcription factor SP1, a gene previously noted to potentially modulate transcript levels of OCT3 and MATE1.28,30 Our regression results showed multiple independent SNPs in the SP1 locus associated with pharmacodynamic and pharmacokinetic phenotypes of metformin (Supplementary Table S1 online). Five of the six total variants linked to metformin PK and PD were in the SP1 gene region. Most of the SNPs were in noncoding regions, including intronic, upstream, and downstream regions of SP1. The most strongly associated SNP, rs784892, is in the intronic region of the downstream gene, AMHR2 (anti-Mullerian hormone receptor, type II). This SNP was strongly associated with both metformin PD (HbA1c) and PK (metformin secretory clearance), with β coefficients of −0.32 HbA1c per G allele (P = 0.008) and −76.9 ml/min per G allele (P = 0.02), respectively (Figure 4). A separate analysis, based on creatinine clearance, was performed for SP1 variants, and from this analysis, no statistically significant association was observed between SP1 variants and creatinine clearance. The rs784892 variant has a combined minor allele frequency of ∼11% across all ethnic groups, with African Americans (∼35%) having a higher frequency than European Americans (<1%). However, the effect of race was accounted for using principal components in our multivariate analysis, and a separate analysis in African Americans was performed to ensure that the variant has a significant effect on both treatment HbA1c levels and secretory clearance in African Americans. Statistical significance was observed for both phenotypes in the African-American cohort.
Figure 4.
Gene diagram summarizes the chromosome location of top associated single-nucleotide polymorphisms, highlighting both metformin pharmacodynamic and pharmacokinetic associated P values using multiple linear regression analysis, n = 440 patients for pharmacodynamics analysis, and n = 57 healthy subjects for the pharmacokinetic analysis. G, associated allele of rs784892; PD, pharmacodynamics, variant association with treatment HbA1c level, as defined in the text; PK, pharmacokinetics, variant association with measured metformin secretory clearance (ml/min).
Results from the final population pharmacokinetic model of metformin determined that rs784888, a SNP less than 50 kb downstream of SP1, was an important predictor of metformin apparent clearance. The rs784888 G allele led to a 12% reduction in metformin apparent clearance. This variant has a frequency of ∼9% (in African Americans = 42%, in European Americans = <1%). As with rs784892, a separate analysis confirmed this finding in African Americans.
PPAR-α and HNF4-α were major genes with polymorphisms associated with variation in PD, independent of PK
A total of 17 variants in PPAR-α and 6 variants in HNF4-α were associated with metformin PD (P < 0.01) (Figure 5; Supplementary Table S4 online). The most significant variant from our pharmacodynamic analysis was intronic SNP rs149711321 in PPAR-α (P = 1 × 10−05). This variant has a minor allele frequency of 6% reported for both European-American and African-American ethnicities. Of the 23 total variants in PPAR-α and HNF4-α, none were significantly associated with metformin PK in healthy subjects.
Figure 5.
A zoomed-in view of genetic polymorphisms in HNF4-α and PPAR-α associated with treatment-HbA1c levels. Circles represent the location and the–log10 P value of the association. Recombination rates are also overlaid on the figure, with each peak representing relatively high recombination rates for that region.
Deletion in AP2, a repressor, was associated with changes in metformin PK and PD
A total of 11 variants in AP2 were linked to changes in HbA1c levels in type 2 diabetes patients receiving metformin. Of these, 1 imputed deletion variant in the intronic region of AP2 was associated with an increase in metformin secretory clearance (P = 0.004) and an increase in treatment HbA1c levels (P = 0.003) (Supplementary Table S1 online). The minor allele frequency of this variant was 6%, with a slope (β) of 0.45% change in HbA1c per minor allele. This variant was not associated with the PK of metformin in our population pharmacokinetic model. However, it is important to note that the model did not specifically include metformin secretory clearance as a pharmacokinetic parameter due to the absence of urine data.
Significant impact on metformin pharmacokinetic parameters of other covariates in the population pharmacokinetic model
Transporter variants previously associated with metformin PK from noncompartmental approaches were also significantly associated with metformin kinetics (CL/F, Vc/F, and Q/F) in the population pharmacokinetic model. In particular, a MATE2/ SLC47A2 variant (rs12943590) was significantly associated with peripheral flow (Figure 2), potentially increasing Q/F by as much as 30%. An interaction effect between an OCT3 variant (rs555754) and an OCT2 variant (rs316019) on metformin central volume was also observed. Based on the combination of minor alleles from rs555754 and rs316019, metformin Vc/F may decrease by as much as 32% and increase by as much as 39% (Figure 3). Although significant in the final model, the downstream significance of these variants will require replication by other studies before the findings can be clinically translated.
The effect of ethnicity was investigated using our population pharmacokinetic model. Ethnicity was found to be a significant predictor of metformin flow, specifically for the pharmacokinetic parameters CL/F and Q/F. For both parameters, African Americans had significantly higher values as compared with those of European Americans (Figure 3). As compared with African Americans, European Americans had an approximately 26% lower metformin clearance and Asian Americans had a 22% lower clearance. For Q/F, European Americans were predicted to have a 46% lower peripheral flow as compared with that of African Americans. These effects were independent of creatinine clearance and body weight. Simulations in Figure 3 compare the effect of typical and extreme values of metformin clearance after an 850-mg dose (based on different covariate combinations).
DISCUSSION
Previous pharmacogenetic studies of metformin PK have focused on a few nonsynonymous variants in transporter genes.1,5,14,15 Pharmacogenetic investigation of variants in gene expression modulators of key transporters involved in metformin PK is a novel approach to understanding the variability in response to metformin. This study tested the effect of genetic variants in key transcription factor genes on metformin PD, with a focus on glycemic response to metformin in type 2 diabetes patients. Subsequently, the top hits associated with metformin response were examined for a pharmacokinetic mechanism in both type 2 diabetes patients and healthy subjects using two approaches. Four important findings emerged from our combined PK/PD analysis: (i) SNPs in SP1 were associated with metformin PD in type 2 diabetes patients, and this association had a pharmacokinetic basis. (ii) SNPs in AP2, in particular a deletion variant, were associated with the PK and PD of metformin. (iii) SNPs in PPAR-α and HNF4-α were associated with the PD of metformin but did not have a pharmacokinetic mechanism. (iv) Finally, African Americans were observed to have greater apparent clearances as compared with those of European Americans and Asian Americans.
To date, the role of SP1 in metformin pharmacology has not been investigated despite previous studies indicating that SP1 modulates the gene expression of MATE1 and OCT3, two important metformin transporters.28,30 Our findings strongly suggest an important role of SP1 in governing metformin disposition and response. Using standard regression, we observed that five genetic variants in SP1 were associated with metformin PK and PD. Multiple independent effects were observed even after accounting for linkage disequilibrium. A population-based approach demonstrated that one such SNP located downstream of SP1, rs784888, significantly affected metformin apparent clearance, therefore impacting systemic plasma levels of metformin. Comparing the effect size of this SP1 variant on CL/F with the effect sizes of transporter variants observed in previous studies revealed that the effect size for the SP1 variant (12%) was greater than those previously reported for variants in transporters and CL/F.5,8,33 Interestingly, rs784888 is in strong linkage disequilibrium with rs147778161 (r2 > 0.8),an intronic SNP of SP1 that was initially removed due to not passing our initial pharmacodynamic cutoff (P < 0.01). The observed associations for SP1 are biologically plausible, considering the expression of MATE1 on the apical membrane in the proximal tubule of the kidney and its role in metformin renal secretion. The role of OCT3 in the renal elimination of metformin is still not fully elucidated; therefore, it is not known whether SNPs in SP1 may modulate metformin PK by regulating the expression of OCT3 in addition to MATE1 and other metformin transporters. Furthermore, we performed a separate transcription factor binding analysis using a transcription factor binding tool (FIMO), and found that in addition to regulating the expression of MATE1 and OCT3, SP1 may also modulate levels of OCT2 and MATE2-K, transporters that are known to play a very important role in metformin elimination from the kidney (Supplementary Table S3 online). Although speculative, we propose that genetic variants in SP1 may affect the binding affinity or the expression level of the transcription factor, which could then have a combined effect on MATE1, OCT2, OCT3, and MATE2-K levels, globally affecting the pharmacokinetic and pharmacodynamic outcomes of metformin. Furthermore, the regression results suggest a clinically significant impact of SP1 variants on metformin PK and PD. For example, typical patients homozygous GG for rs784892 achieved on average treatment HbA1c levels that were 1.1% lower than patients homozygous AA. This finding, if replicated, would have clinical significance, given that metformin reduces HbA1c levels by 1.12% on average (i.e., from 8.0% A1c to 6.9% A1c) within the first year of therapy.27,34,35 Furthermore, in healthy subjects, there was a 98 ml/min reduction in metformin secretory clearance on average in homozygous GG carriers as compared with homozygous AA carriers. This pharmacokinetic mechanism supports our pharmacodynamic finding, in which a lower metformin secretory clearance in homozygous carriers of the variant allele is expected to increase metformin exposure and hence reduce HbA1c levels to a greater extent than in carriers of the reference allele. In our population-based approach, rs784892 trended in the same direction as observed in our regression analysis for metformin secretory clearance but was only of borderline significance in stepwise covariate modeling building and was not included in the final model. However, SP1 variant rs784888 was included. This variant was predicted to lower metformin clearance by up to 24% in patients with homozygous GG. A lower clearance of metformin was predicted to increase metformin exposure, potentially leading to a more favorable response to metformin.
Interestingly, one deletion variant in AP2 was associated with an increase in metformin secretory clearance and a reduction in the glycemic response to metformin. This finding was similar to those observed for polymorphisms in SP1, and consistent with previous studies in our laboratory showing that AP2 is an important repressor of SLC47A1 (MATE1) transcription.3,5 This finding is biologically plausible, with high expression of AP2 and MATE1 found in the kidney.36 Furthermore, we performed additional gene expression analysis using data from the Cancer Genome Atlas to investigate correlations between high-priority transcription factors and transporter genes (Supplementary Figure S1 online). Gene expression data, along with our supplemental motif binding analysis, suggested a regulatory role of AP2 in SLC47A1 and SLC47A2. In addition to SP1, AP2 may also play an important role in modulating metformin PK and PD via its role as a global regulator of multiple transporter genes.
PPAR-α and HNF4-a are multifunctional transcription factors that are primarily expressed in the liver.7,27 PPAR-α is a known regulator of lipid metabolism in the liver. In a study by the Diabetes Prevention Program, one SNP in PPAR-α was associated with diabetes incidence and another variant showed a significant interaction with metformin intervention.37 Mutations in HNF4-a have also been linked to type 2 diabetes.38 Moreover, literature evidence suggests that both PPAR-α and HNF4-α are important regulators of SLC22A1 (OCT1), a key transporter mediating the uptake of metformin into the liver, the primary site of action.5,27 Our regression results suggest that PPAR-α and HNF4-a are important modulators of metformin PD, perhaps independent of PK. A total of 23 genetic variants in PPAR-α and HNF4-a were associated with treatment HbA1c levels, none of which were explained by a pharmacokinetic mechanism. Furthermore, gene expression levels for both transcription factors were strongly correlated with OCT1 expression, as well as with other metformin transporters in the liver. PPAR-α and HNF4-a may be involved in regulating OCT1 and other genes in the liver that play a role in metformin disposition and glycemic response.
In addition to genetics, ethnicity was a significant predictor of metformin PK, affecting intercompartmental clearance (Q/F) and apparent clearance (CL/F). African Americans had significantly higher mean CL/F and Q/F estimates as compared with those of European Americans and Asian Americans. The effects of creatinine clearance and body weight did not confound this effect observed in our model. Furthermore, based on our simulations, for African Americans to achieve similar metformin exposure to that of European Americans, a 26% increase in dose should be considered. This takes into account similar creatinine clearances and SP1 genotype status for the two ethnicities. This means that if, for example, a European American individual would start with an 850-mg dose of metformin, an African American would require at least a 1,000-mg dose of the drug to attain similar exposure levels. The impact of ethnicity on metformin response still needs to be investigated. In addition, further studies and different approaches are required to validate this observation for metformin PK.
Overall, this study demonstrates that genetic variants in key transcription factor genes, along with transporters and ethnicity, are important determinants of metformin PK and PD. Transcription factors may regulate gene expression levels through either enhancer or repressor activity. In some cases, for example, SP1 and AP2, the association of genetic variants with PD may be mediated through pharmacokinetic mechanisms, which ultimately control systemic blood levels of the drug. For example, SP1 may regulate the expression of a system of transporters in the kidney that is involved in metformin elimination. In other cases, PPAR-α, for example, the observed mechanisms for the effects of SNPs are unclear. PPAR-α SNPs may modulate metformin PD independently of the effects on systemic levels of metformin. Clearly, future studies are needed to further clarify the biological roles of SP1, AP2, HNF4-α, and PPAR-α in the disposition and action of metformin.
METHODS
Healthy human subjects
Data from four healthy volunteer studies from the University of California, San Francisco, were pooled for this study, as previously described.5,39 Studies 6112, 6113, and 865 followed similar protocols. Healthy subjects were dosed with 1,000 mg of metformin, followed by an 850-mg dose of metformin on the second day of the study. Participants from study 767 were given a single 850-mg dose of metformin. During the short duration of the study in healthy volunteers, metformin levels in the liver were not expected to have reached steady state.
Patients with type 2 diabetes
Diabetes patients of European-American, African-American, or Asian-American ancestry were recruited into a multicenter retrospective study as previously described.5,15 All patients were metformin naive, had HbA1c levels measured before and after initiation of metformin therapy (between 3 and 18 months), and had a medication possession ratio of >80%.
Selection of transcription factor genes and variants
The candidate gene study consisted of genes and SNPs from transcription factors known to modulate levels of metformin-related transporters. PharmGKB was the resource used to determine the list of transporters used in this study. 2,40 Transcription factors were selected based on evidence from previous publications linking the transcription factor to one of the metformin-related transporters.7,28,41 A total of five transcription factors were selected: SP1, AP2, AP1, HNF4-α, and PPAR-α. After filtering out SNPs with low minor allele frequencies (<5%) in the type 2 diabetes cohort, we selected SNPs in transcription factors within 50,000 base pairs upstream and downstream of each transcription factor gene.
Phenotype selection for multivariate regression analysis
For the pharmacodynamic analysis, a minimum treatment HbA1c value between 3 and 18 months postinitiation of metformin, referred to as the treat-ment-HbA1c, was selected as the phenotype of interest. Top SNPs associated with treatment HbA1c were selected and subsequently tested on metformin secretory clearance. Pharmacokinetic parameters from a healthy subject study were previously determined using noncompartmental analysis.5,42,43 Secretory clearance was the primary parameter of interest, as this parameter is assumed to be the most sensitive to changes in transporter function and expression level; it was calculated using the following formula:
Linear regression analysis
Linear regression was performed using PLINK (v1.07), assuming an additive genetic model.39,44 Imputation was performed using IMPUTE2 software (version 2).42,43,45 Variants with <5% minor allele frequency were excluded from the regression analysis. For the regression analysis with HbA1c, a statistical base model (Supplementary Methods online) was established before testing for the effect of transcription factor variants. Top variants associated with treatment-HbA1c with an adjusted P value <0.01 were filtered. These SNPs were then tested against metformin secretory clearance (P < 0.05). In healthy subjects, the statistical model was corrected for principal components and age.
Population pharmacokinetic modeling of metformin and final model selection
Data from five studies (patient study, and healthy volunteer studies 6112, 6113, 865, and 767), which include healthy volunteers and type 2 diabetes patients, were analyzed using nonlinear mixed effect modeling (NONMEM 7) with the first-order conditional estimation method with interaction (FOCE-I). Model selection was informed by using the objective function value (−2log likelihood) and visual inspection of diagnostic plots. The final structural model was parameterized in terms of CL/F, apparent central and peripheral volumes of distribution (Vc/F and Vp/F), apparent intercompartmental clearance (Q/F), mean transit time, and first-order absorption (ka). Interindividual variability was estimated for CL/F, Vc/F, and Q/F. The Stepwise Covariate Model tool in Perl Speaks NONMEM was used to develop the final model with statistically significant covariates on metformin pharmacokinetic parameters. Finally, a bootstrap was performed with 1,000 samples to obtain 95% confidence intervals for all pharmacokinetic parameters used to characterize the final model.
Supplementary Material
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
-
✓
Previous studies have focused on the effects of genetic polymorphisms in membrane transporters on metformin PK and response.
WHAT QUESTION DID THIS STUDY ADDRESS?
-
✓
In this study, we investigated the effects of genetic variants in transcription factor genes on the PK and PD of metformin in healthy volunteers and in patients with type 2 diabetes.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
-
✓
This study provides evidence that genetic polymorphisms in transcription factors SP1 and AP2 may have a significant impact on the PK and PD of metformin. This study also shows that HNF4-α and PPAR-α are linked to metformin PD, independent of PK. Finally, this study provides model-based evidence to suggest that African Americans have higher flow rates than European Americans and Asians.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY AND THERAPEUTICS
-
✓
To date, this is the first study to explore the effect of transcription factors on metformin PK and PD. In the future, gen-otyping of transcription factor genes along with transporter genes may be used to inform metformin therapy.
ACKNOWLEDGMENTS
This research was supported by a grant from the National Institutes of Health (NIH) National Institute of General Medical Sciences (GM61390), NIH P30 DK063720, and the NIH Pharmacogenomics Research Network (PGRN)-RIKEN Strategic Alliance.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
AUTHOR CONTRIBUTIONS
R.M.S., S.G., and K.M.G. wrote the manuscript. R.M.S., S.G., and K.M.G. designed the research. R.M.S., S.G., S.W.Y., R.C., and K.M.G. performed the research. R.M.S., S.G., J.D.M, M.K., J.A.M., C.W., X.L., J.W., and K.M.G. analyzed the data. R.M.S., S.G., S.S., C.B., S.M., M.D.S., M.M.H, R.L.D., D.M.R., and K.M.G. contributed new reagents/analytical tools.
CONFLICTOF INTEREST
The authors declared no conflict of interest.
References
- 1.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 2008;83:273–280. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways. Pharmacogenet. Genomics. 2012;22:820–827. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha Choi J, et al. Identification and characterization of novel polymorphisms in the basal promoter of the human transporter, MATE1. Pharmacogenet. Genomics. 2009;19:770–780. doi: 10.1097/FPC.0b013e328330eeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander GC, Sehgal NL, Moloney RM, Stafford RS. National trends in treatment of type 2 diabetes mellitus, 1994–2007. Arch Intern Med. 2008;168:2088–2094. doi: 10.1001/archinte.168.19.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacker SL, et al. The effect of novel promoter variants in MATE1 and MATE2 on the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacol Ther. 2013;93:186–194. doi: 10.1038/clpt.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham G, Punt J. Clinical pharmacokinetics of metformin. Clin. Pharmacokinet. 2011;50:81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 7.Saborowski M, Kullak-Ublick GA, Eloranta JJ. The human organic cation transporter-1 gene is transactivated by hepatocyte nuclear factorial pha. J. Pharmacol. Exp. Ther. 2006;317:778–785. doi: 10.1124/jpet.105.099929. [DOI] [PubMed] [Google Scholar]
- 8.Shu Y, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. HNF4A in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab. Pharmacokinet. 2007;22:287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
- 10.Cook MN, Girman CJ, Stein PP, Alexander CM. Initial monotherapy with either metformin or sulphonylureas often fails to achieve or maintain current glycaemic goals in patients with Type 2 diabetes in UK primary care. Diabet. Med. 2007;24:350–358. doi: 10.1111/j.1464-5491.2007.02078.x. [DOI] [PubMed] [Google Scholar]
- 11.Owen M, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348:607–614. [PMC free article] [PubMed] [Google Scholar]
- 12.GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group. Zhou K, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2010;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker ML, Visser LE, van Schaik RHN, Hofman A, Uitterlinden AG, Strieker BHC. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2008;58:745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, et al. Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet. Genomics. 2009;19:497–504. doi: 10.1097/FPC.0b013e32832cc7e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi JH, et al. A common 5’-UTR variant in MATE2-K is associated with poor response to metformin. Clin. Pharmacol. Ther. 2009;90:674–684. doi: 10.1038/clpt.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzvetkov MV, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin. Pharmacol. Ther. 2009;86:299–306. doi: 10.1038/clpt.2009.92. [DOI] [PubMed] [Google Scholar]
- 17.Takane H, Shikata E, Otsubo K, Higuchi S, leiri I. Polymorphism in human organic cation transporters and metformin action. Pharmacogenomics. 2008;9:415–422. doi: 10.2217/14622416.9.4.415. [DOI] [PubMed] [Google Scholar]
- 18.Scarpello JH, Howlett HC. Metformin therapy and clinical uses. Diab. Vase. Dis. Res. 2008;5:157–167. doi: 10.3132/dvdr.2008.027. [DOI] [PubMed] [Google Scholar]
- 19.Fujioka K, et al. Efficacy, dose-response relationship and safety of once-daily extended-release metformin (Glucophage® XR) in type 2 diabetic patients with inadequate glycaemic control despite prior treatment with diet and exercise: results from two double-blind, placebo-controlled studies. Diabetes Obes. Metab. 2005;7:28–39. doi: 10.1111/j.1463-1326.2004.00369.x. [DOI] [PubMed] [Google Scholar]
- 20.Garbar A, Duncan TG, Goodman AM, Mills DJ, Rohlf JL. Efficacy of metformin in type II diabetes: results of a double-blind, placebo-controlled, dose-response trial. Am. J. Med. 1997;103:491–497. doi: 10.1016/s0002-9343(97)00254-4. [DOI] [PubMed] [Google Scholar]
- 21.Becker Ml, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, Strieker BH. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 2009;9:242–247. doi: 10.1038/tpj.2009.15. [DOI] [PubMed] [Google Scholar]
- 22.Song IS, et al. Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin. Pharmacol. Ther. 2008;84:559–562. doi: 10.1038/clpt.2008.61. [DOI] [PubMed] [Google Scholar]
- 23.Wang ZJ, Yin OQ, Tomlinson B, Chow MS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet. Genomics. 2008;18:637–645. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
- 24.Kim DH, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin. Cancer Res. 2009;15:4750–4758. doi: 10.1158/1078-0432.CCR-09-0145. [DOI] [PubMed] [Google Scholar]
- 25.Chung JY, et al. Functional characterization of MATE2-K genetic variants and their effects on metformin pharmacokinetics. Pharmacogenet. Genomics. 2013;23:365–373. doi: 10.1097/FPC.0b013e3283622037. [DOI] [PubMed] [Google Scholar]
- 26.Shu Y, Bello CL, Mangravite LM, Feng B, Giacomini KM. Functional characteristics and steroid hormone-mediated regulation of an organic cation transporter in Madin-Darby canine kidney cells. Pharmacogenet. Genomics. 2001;299:392–398. [PubMed] [Google Scholar]
- 27.Nie W, Sweetser S, Rinella M, Green RM. Transcriptional regulation of murine Slc22a1 (Oct1) by peroxisome proliferator agonist receptor-alpha and -gamma. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G207–G212. doi: 10.1152/ajpgi.00057.2004. [DOI] [PubMed] [Google Scholar]
- 28.Kajiwara M, et al. Critical roles of Sp1 in gene expression of human and rat H+/ organic cation antiporter MATE1. Am. J. Physiol. Renal Physiol. 2007;293:F1564–F1570. doi: 10.1152/ajprenal.00322.2007. [DOI] [PubMed] [Google Scholar]
- 29.Cho SK, et al. Rifampin enhances the glucose-lowering effect of metformin and increases OCT1 mRNA levels in healthy participants. Clin. Pharmacol. Ther. 2011;89:416–421. doi: 10.1038/clpt.2010.266. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, et al. Genetic and epigenetic regulation of the organic cation transporter 3, SLC22A3. Pharmacogenomics J. 2013;13:110–120. doi: 10.1038/tpj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asaka J, Terada T, Ogasawara K, Katsura T, Inui K. Characterization of the Basal promoter element of human organic cation transporter 2 gene. J. Pharmacol. Exp. Ther. 2007;321:684–689. doi: 10.1124/jpet.106.118695. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien VP, et al. Hepatocyte nuclear factor 1 regulates the expression of the organic cation transporter 1 via binding to an evolutionary conserved region inintron 1 of the OCT1 gene. J. Pharmacol. Exp. Ther. 2013;347:181–192. doi: 10.1124/jpet.113.206359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group. Zhou K, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. War. Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little RR, Rohlfing CL. Analytical goals for HbAI c are HbAI c results good enough for optimal use? J. Diabetes. 2011;3:3–6. doi: 10.1111/j.1753-0407.2010.00109.x. [DOI] [PubMed] [Google Scholar]
- 35.Hirst JA, Farmer AJ, Ali R, Roberts NW, Stevens RJ. Quantifying the effect of metformin treatment and dose on glycemic control. Diabetes Care. 2012;35:446–454. doi: 10.2337/dc11-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suda S, Rai T, Sohara E, Sasaki S, Uchida S. Postnatal expression of KLF12 in the inner medullary collecting ducts of kidney and its trans-activation of UT-A1 urea transporter promoter. Biochem. Biophys. Res. Commun. 2006;344:246–252. doi: 10.1016/j.bbrc.2006.03.138. [DOI] [PubMed] [Google Scholar]
- 37.Jablonski K, et al. Diabetes Prevention Program Research Group Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohlke KL, Boehnke M. The role of HNF4A in the riskof type 2 diabetes. Curr. Diab. Rep. 2005;5:149–156. doi: 10.1007/s11892-005-0043-y. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whirl-Carrillo M, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet. Genomics. 2010;20:687–699. doi: 10.1097/FPC.0b013e32833fe789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3:Genes, Genomes, Genetics. 2011;1:6457–6470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Cancer Institute. [Accessed 28 February 2012];The Cancer Genome Atlas. < http://cancergenome.nih.gov>.
- 45.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.