Abstract
Regulation of gene expression during cardiac development and remodeling is very complicated, involving epigenetic, transcriptional, post-transcriptional and translational regulation. Our understanding of the molecular mechanisms underlying cardiac remodeling is still far from complete. MicroRNAs are a class of small non-coding RNAs that have been shown to play critical roles in gene regulation in cardiovascular biology and disease. microRNA-22 (miR-22) is an evolutionally conserved miRNA that is highly expressed in the heart. Recent studies uncovered miR-22 as an important regulator for cardiac remodeling. MiR-22 modulates the expression and function of genes involved in hypertrophic response, sarcomere reorganization and metabolic program shift during cardiac remodeling. In this review, we will focus on the recent findings of miR-22 in cardiac remodeling and the therapeutic potential of this miRNA in the treatment of related defects resulting from adverse cardiac remodeling.
Introduction
Cardiac remodeling, which refers to the alteration in the structure (dimensions, mass, shape) and physiological function of the heart as well as changes of cardiac cells, is one of the major responses of the heart to biomechanical stresses and pathological stimuli. Cardiac hypertrophy, which is characterized by an increase in the thickness of ventricular wall, owing to the enlargement of myocyte size, is one of the major responses to stresses and/or myocardial injury. Sustained cardiac hypertrophy often leads to cardiac dilation and heart failure [1].
MicroRNA (miRNA), which were first discovered in C. elegans two decades ago, are a class of small noncoding RNAs containing ∼22 nucleotides and widely present in plants [2] and animals [3-5]. miRNAs are transcribed by RNA polymerase II as primary transcripts (pri-miRNAs) [6], which contain a signature stem-loop hairpin structure, and then are subsequently processed by RNaseIII enzymes Drosha and Dicer to generate ∼22nt long miRNA duplexes (mature miRNAs) with 2nt overhangs on the 3′ end [7]. Dicer also contributes to the loading of mature miRNAs into the RNA-induced silencing complex (RISC), where miRNAs guide the RISC to target genes by binding to imperfect complementary sites within the 3′ untranslated regions (3′UTRs) [8]. Presently, 1872 precursors and 2578 mature miRNAs have been documented in human; many of them are conserved across species. Studies have shown that miRNAs play important roles in animal development, function and diseases, including that of the cardiovascular system [9-12].
Extensive studies have demonstrated that many miRNAs are dys-regulated during cardiac hypertrophy [9, 13]. Furthermore, gain- and loss-of function studies of miRNAs have linked their function to cardiac hypertrophy and other cardiac pathologies, such as cardiac dilation, ischemic damage, in vivo and in vitro [10] [14]. In this review, we will focus on miR-22, one of the cardiac-enriched miRNAs. We will summarize the recent findings on the regulation of miR-22 in cardiac remodeling and discuss the future directions of studying this miRNA in cardiac disease.
Genomics information of miR-22 and its tissue distribution
Unlike numerous other miRNAs, many of which belong to miRNA families with multiple miRNA members, miR-22 belongs to a single member miRNA family. MiR-22 is encoded by an exon of a long non-coding RNA gene, MIR-22HG (the miR-22 host gene), in the mammalian genome. The long non-coding host gene resides in the intergenic region between two protein-coding genes, TLCD2 (TLC domain containing 2) and WDR81 (WD repeat domain 81). TLCD2 is a gene with unknown function and mutations in WDR81 are reported to be associated with autosomal recessive cerebellar ataxia, mental retardation, and dysequilibrium syndrome-2 [15]. MiR-22 is an evolutionally conserved miRNA with an identical seed sequence from fruit fly to human. Although miR-22 is ubiquitously expressed in mammals, the enriched expression of this miRNA in striated muscle tissues (cardiac and skeletal muscles) is conserved between fruit fly and mammals [16]. Recent deep sequencing analysis shown that miR-22 is the most abundant miRNA in the heart [17]. More specifically, we demonstrated that cardiomyocytes contribute most of the miR-22 expression in the heart by comparing miR-22 expression in isolated cardiomyocytes vs. non-cardiomyocytes of adult mouse hearts [18]. Consistently, the expression of miR-22 is reduced by over 90% in mutant hearts harboring cardiomyocyte-specific miR-22 deletion using alpha-MHC-Cre. In addition, we found that the expression of cardiac miR-22 is much higher in matured cardiomyocytes when compared with that of embryonic and neonatal hearts, suggesting its potential function in adult hearts [18].
Expression pattern of miR-22 in response to cardiac insult
The expression of many miRNAs is altered when the heart is under cardiac insult/stress [19]. When the heart is stressed (by transverse aortic constriction, or TAC) and undergoes remodeling, the expression of miR-22 is slightly elevated during the initial stage of hypertrophy and then gradually goes back to normal levels when the cardiac stress is prolonged [18, 20]. In another cardiac hypertrophy model, isoproterenol is able to significantly elevate the expression of cardiac miR-22 [21], suggesting the expression of miR-22 may be regulated by beta-adrenergic receptor activated signaling pathways during cardiac hypertrophy. In isolated neonatal rat cardiomyocytes, the expression of miR-22 is modestly increased when cells are stimulated by different hypertrophic stimuli, such as phenylephrine, Angiotensin II and serum [18] [22]. In addition, the circulating miR-22 was found elevated in the serum of heart failure patients and it was suggested that the elevated circulating miR-22 is caused by the increased expression of miR-22 in patients' hearts [23]. All these observations indicate that miR-22 could play important roles in cardiac response to stresses.
MiR-22 regulates cardiac remodeling
Although miR-22 is abundantly expressed in the heart, and there is no evidence of a second copy of miR-22 within the genome, it is quite a surprise that this miRNA is dispensable for animal development or cardiac morphogenesis. Global or cardiac-specific deletion of miR-22 did not cause premature death in mouse or abnormality in the heart. miR-22 null mice are fertile and viable [18]. However, miR-22 is required for normal cardiac remodeling in response to stresses (Figure 1). MiR-22 deficient mice are unable to properly develop cardiac hypertrophy in response to isoproterenol administration [18]. Instead of developing compensative cardiac hypertrophy at the early stage of remodeling, miR-22 mutant hearts quickly progress to adverse cardiac remodeling and develop cardiac dilation accompanied with increased death of cardiomyocytes and deposition of fibrotic tissue, characteristics of dilated cardiomyopathy and heart failure. Consistent with our observations, another study also found that miR-22 null mice quickly develop dilated cardiomyopathy, displaying dysregulation of hypertrophic genes in response to a pressure overload [20].
Figure 1.
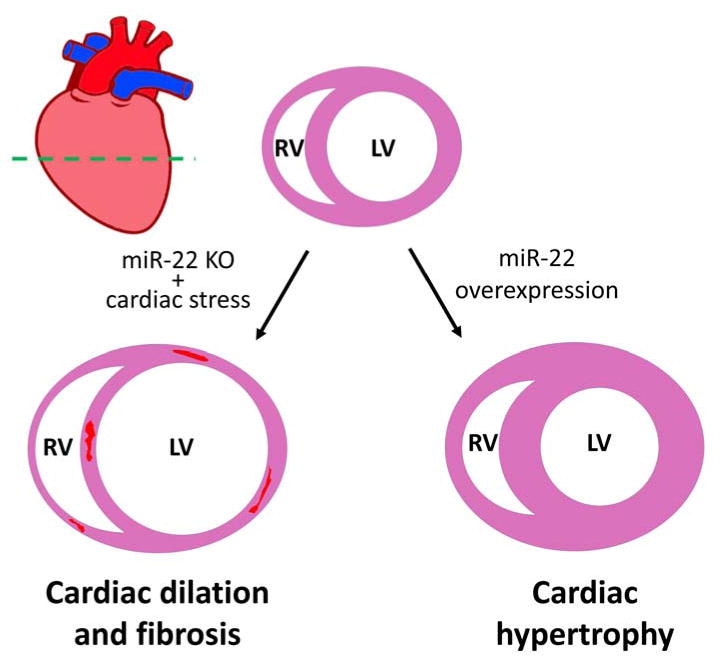
Schematic depiction of miR-22 function in the regulation of cardiac remodeling. A transverse view of heart, indicated by the green dashed line in the heart model, is used to show the functional role of miR-22 in cardiac remodeling, including cardiac hypertrophy, dilated cardiomyopathy and fibrosis. The red areas in the ventricle wall of the dilated heart designate fibrosis lesions.
In gain-of-function studies, forced expression of miR-22 in neonatal cardiomyocytes is sufficient to induce hypertrophy, which is evidenced by the increased cell surface area and elevated expression of hypertrophic marker genes, ANP and BNP [18] [24]. Consistently, ectopic overexpression of miR-22 by transgene in cardiomyocytes in vivo induced cardiac hypertrophy accompanied with an increased cross-sectional cell area of cardiomyocytes [25]. Interestingly, miR-22-induced cardiac hypertrophy is associated with a shift of transcriptome of metabolism, implying that the development of cardiac hypertrophy and the change of cardiac metabolism could be coupled. This observation is intriguing given that many of the miR-22 targets are involved in cell metabolism (Figure 2; Table 1).
Figure 2.
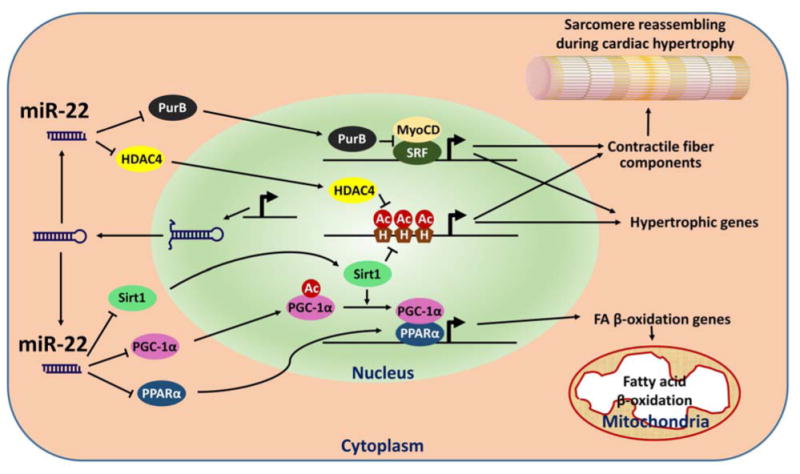
Schematic depiction of miR-22 function in cardiomyocytes. Several miR-22 targets (PurB, Sirt1, etc) and regulatory biological processes (Contraction and metabolism) are highlighted. Ac: acetyl groups; H: histone; HDAC4: histone deacetylase 4; MyoCD: myocardin; PGC-1〈: peroxisome proliferator-activated receptor gamma, coactivator 1 alpha; PPAR〈: peroxisome proliferator activated receptor alpha; PurB: purine rich element binding protein B; Sirt1: sirtuin 1; SRF: serum response factor.
Table 1.
Partial list of reported miR-22 target genes.
| Target genes | Tissue or cell content | References |
|---|---|---|
| Sirt1 | Heart, Cancer cell | [18], [25], [43] |
| HDAC4 | Heart, Cancer cell, Neuron | [18], [44], [45] |
| PPARα | Heart, Chondrocyte | [18], [25], [46] |
| PGC1-α | Heart | [25] |
| PurB | Heart | [20] |
| BMP7 | Chondrocyte, Kidney | [46], [47] |
| ER-alpha | Cancer cell | [48] |
| pTEN | Cardiomyocyte, Cancer cell line | [49], [22] |
| MAX | Cancer cell | [50] |
| EVI-1 | Cancer cell | [51] |
| CDK6 | Cancer cell | [43] |
| Sp1 | Cancer cell | [43] |
| P21 | Cancer cell | [52] |
| HIF-1α | Cancer cell | [53] |
| HDAC6 | Human adipose tissue-derived mesenchymal stem cells | [54] |
| ErbB3 | Cancer cell | [34] |
| OGN | Cardiac fibroblast | [55] |
| Irf8 | Dendritic cell | [56] |
| Rcor1 | Neuron | [45] |
| Rgs2 | Neuron | [45] |
| YWHAZ | Dendritic cell | [57] |
| TET2 | Cancer cell, hematopoietic stem cell | [29], [28] |
Genes targeted by miR-22
MiRNAs are reported to regulate the expression of protein-coding genes post-transcriptionally. To execute their biological function, the guiding strands of mature miRNAs are loaded into the RISC complex and guide the complex to the 3′ UTRs of their target mRNAs via the imperfect binding between miRNAs and the 3′ UTRs [8]. Computational algorithms have been designed to predict miRNA target genes [26, 27] and successfully predicted hundreds of miR-22 targets, many of them verified experimentally (Table 1). In our recent study, we identified two histone deacetylases, Sirt1 and HDAC4 [18], as targets of miR-22, which imply that miR-22 plays a role in epigenetic regulation of gene expression during cardiac hypertrophy (Figure 2). Indeed, miR-22 was found as an epigenetic modifier to control hematopoietic stem cell self-renewal and metastasis of breast cancer cells in hematopoietic malignancies and breast cancer, at least in part, by targeting TET (Ten Eleven Translocation) family proteins [28, 29].
Gurha and colleagues have reported that miR-22 targets purine-rich element binding protein B (PURB) in cardiomyocytes. Since PURB is a repressor of transactivation of SRF, knockout of miR-22 was believed to repress the expression of SRF-depended hypertrophic genes by de-repressing PURB [20]. PTEN, a tumor suppressor, has also been reported as a target of miR-22 in the regulation of cardiac hypertrophy, which is consistent with a previous report demonstrating inactivation of pTEN in cardiomyocytes caused cardiac hypertrophy [22]. In addition, we and others have verified the regulation of PPAR〈 and pGC1〈 by miR-22 [18] [25]. Together with Sirt1, these three master transcriptional factors/co-factors are well known to control fatty acid metabolism [30, 31]. Indeed, fatty acid metabolism signaling was down-regulated in hearts of miR-22 transgenic mice [25]. Since the heart is a major energy-consuming organ and the metabolic program often shifts during cardiac hypertrophy and diabetic cardiomyopathy, it is tempting to speculate that miR-22 plays a key role in the regulation of molecular pathways involving in the shift of metabolic program during the development of cardiomyopathies (Figure 2). miR-22 has also been reported to target CBP/p300. Overexpression of miR-22 in ischemia-reperfused hearts was shown to decrease the protein levels of CBP/p300 and acetylated p53, therefore protecting cardiomyocytes from apoptosis [32]. In addition to its critical roles in cardiac remodeling, miR-22 is also a key player in tumorigenesis, in part due to its function in negatively regulating cell proliferation. miR-22 targets many genes related to cell proliferation, including MYCBP [33], a positive regulator of oncogene c-Myc; ERBB3 [34], a member of epidermal growth factor receptor (EGFR) family and controls proliferation; MAX [35], a basic helix-loop-helix leucine zipper (bHLHZ) transcription factor and associated with Myc; and oncogene EVI-1 [36] (Table 1). It will be interesting to determine whether the expression and function of these miR-22 targets are also altered in hearts of miR-22 null mice.
Clinical aspects and therapeutic potential of miR-22 in cardiac hypertrophy
There is growing evidence that mutations in miRNA genes or key components of the miRNA machinery, Drosha, DGCR8, Dicer, are associated with human disorders [19, 37]. More recently, Harper and his colleagues tested ten SNPs near the miR-22 coding locus and found one of them was closely linked to the variability of left ventricle mass in a large cohort of human patients [38]. However, it remains to be determined whether this SNP regulates the expression and function of miR-22. Most importantly, future studies should be conducted to screen and identify miR-22 mutations in human patients with hypertrophic cardiomyopathy, dilated cardiomyopathy and heart failure in order to establish causative roles of miR-22 in cardiac diseases.
miRNAs possess great potential for the treatment of human cardiovascular diseases [39, 40]. MiR-22 has been demonstrated to regulate cardiac hypertrophy by several independent studies. Although loss-of-function of miR-22 cause premature cardiac dilation in stressed mouse hearts [18, 20], transient repression of miR-22 using antagomir was able to repress cardiac hypertrophy in isoproterenol-treated hearts without sign of dilation and without any adverse effects on cardiac function [41]. In addition, systemic administration of miR-22 antagomirs is able to reduce the blood pressure in spontaneously hypertensive rat model [42]. These results highlight that miR-22 could be a great potential target in therapeutic treatment of cardiac hypertrophy. Tu and colleagues further reported that atorvastatin attenuated angiotensin II-induced hypertrophic growth of cardiomyocyte by repressing the up-regulation of miR-22 in vitro. Pharmacological intervention with atorvastatin inhibited the cardiac hypertrophy induced by systematic administration of miR-22 mimic in vivo. However, particular attention should be paid to carefully address the issue of specificity of delivery of miR-22 inhibitors, since knocking down of miR-22 in other tissues/organs might increase the risk of promoting cell proliferation and tumorigenesis. Therefore, the future of miR-22 in clinical treatment of cardiovascular diseases is bright, but technical hurdles need to be overcome. We are confident that investigation and understanding how miR-22 regulates cardiac physiological function will contribute to the development of novel therapeutics to treat heart diseases.
Acknowledgments
We thank Ronald L. Neppl for careful reading of the manuscript and stimulating discussion.
Funding: Work in the Wang lab was supported by the March of Dimes Foundation and the NIH (HL085635, HL116919). ZP Huang is supported by the NIH training grant T32 HL007572.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, et al. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, et al. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Wang DZ. microRNAs in cardiovascular development. J Mol Cell Cardiol. 2012;52:949–957. doi: 10.1016/j.yjmcc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol. 2012;100:279–317. doi: 10.1016/B978-0-12-387786-4.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatsuguchi M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarac O, et al. Neuro-ophthalmologic findings in humans with quadrupedal locomotion. Ophthalmic genetics. 2012;33:249–252. doi: 10.3109/13816810.2012.689412. [DOI] [PubMed] [Google Scholar]
- 16.Christodoulou F, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463:1084–1088. doi: 10.1038/nature08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, et al. Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac microRNAs. Proc Natl Acad Sci U S A. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang ZP, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res. 2013;112:1234–1243. doi: 10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurha P, et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 2012;125:2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu Y, et al. In vitro and in vivo direct monitoring of miRNA-22 expression in isoproterenol-induced cardiac hypertrophy by bioluminescence imaging. European journal of nuclear medicine and molecular imaging. 2014;41:972–984. doi: 10.1007/s00259-013-2596-3. [DOI] [PubMed] [Google Scholar]
- 22.Xu XD, et al. Attenuation of microRNA-22 derepressed PTEN to effectively protect rat cardiomyocytes from hypertrophy. J Cell Physiol. 2012;227:1391–1398. doi: 10.1002/jcp.22852. [DOI] [PubMed] [Google Scholar]
- 23.Goren Y, et al. Serum levels of microRNAs in patients with heart failure. European journal of heart failure. 2012;14:147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 24.Jentzsch C, et al. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Gurha P, et al. microRNA-22 promotes heart failure through coordinate suppression of PPAR/ERR-nuclear hormone receptor transcription. PLoS One. 2013;8:e75882. doi: 10.1371/journal.pone.0075882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 28.Song SJ, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song SJ, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planavila A, et al. Sirt1 acts in association with PPARalpha to protect the heart from hypertrophy, metabolic dysregulation, and inflammation. Cardiovasc Res. 2011;90:276–284. doi: 10.1093/cvr/cvq376. [DOI] [PubMed] [Google Scholar]
- 31.Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95:568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, et al. MicroRNA-22 targeting CBP protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats. Molecular biology reports. 2014;41:555–561. doi: 10.1007/s11033-013-2891-x. [DOI] [PubMed] [Google Scholar]
- 33.Xiong J, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 34.Ling B, et al. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. Journal of cancer research and clinical oncology. 2012;138:1355–1361. doi: 10.1007/s00432-012-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ting Y, et al. Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem Biophys Res Commun. 2010;394:606–611. doi: 10.1016/j.bbrc.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Patel JB, et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene. 2011;30:1290–1301. doi: 10.1038/onc.2010.510. [DOI] [PubMed] [Google Scholar]
- 37.Amiel J, et al. miRNA, development and disease. Advances in genetics. 2012;80:1–36. doi: 10.1016/B978-0-12-404742-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 38.Harper AR, et al. Common variation neighbouring micro-RNA 22 is associated with increased left ventricular mass. PLoS One. 2013;8:e55061. doi: 10.1371/journal.pone.0055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seok HY, Wang DZ. The emerging role of microRNAs as a therapeutic target for cardiovascular disease. BioDrugs. 2010;24:147–155. doi: 10.2165/11535860-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 41.Tu Y, et al. MicroRNA-22 downregulation by atorvastatin in a mouse model of cardiac hypertrophy: a new mechanism for antihypertrophic intervention. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2013;31:997–1008. doi: 10.1159/000350117. [DOI] [PubMed] [Google Scholar]
- 42.Friese RS, et al. MicroRNA-22 and promoter motif polymorphisms at the Chga locus in genetic hypertension: functional and therapeutic implications for gene expression and the pathogenesis of hypertension. Hum Mol Genet. 2013;22:3624–3640. doi: 10.1093/hmg/ddt213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu D, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovicic A, et al. MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington's disease-related mechanisms. PLoS One. 2013;8:e54222. doi: 10.1371/journal.pone.0054222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iliopoulos D, et al. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One. 2008;3:e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long J, et al. MicroRNA-22 is a master regulator of bone morphogenetic protein-7/6 homeostasis in the kidney. J Biol Chem. 2013;288:36202–36214. doi: 10.1074/jbc.M113.498634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol. 2009;29:3783–3790. doi: 10.1128/MCB.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS One. 2010;5:e10859. doi: 10.1371/journal.pone.0010859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong J, et al. Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene. 2010;29:4980–4988. doi: 10.1038/onc.2010.241. [DOI] [PubMed] [Google Scholar]
- 51.Nagaraja AK, et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol. 2010;24:447–463. doi: 10.1210/me.2009-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuchiya N, et al. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 2011;71:4628–4639. doi: 10.1158/0008-5472.CAN-10-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamakuchi M, et al. MicroRNA-22 regulates hypoxia signaling in colon cancer cells. PLoS One. 2011;6:e20291. doi: 10.1371/journal.pone.0020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang S, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem cells and development. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jazbutyte V, et al. MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr) 2013;35:747–762. doi: 10.1007/s11357-012-9407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li HS, et al. miR-22 controls Irf8 mRNA abundance and murine dendritic cell development. PLoS One. 2012;7:e52341. doi: 10.1371/journal.pone.0052341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min S, et al. Multiple tumor-associated microRNAs modulate the survival and longevity of dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J Immunol. 2013;190:2437–2446. doi: 10.4049/jimmunol.1202282. [DOI] [PubMed] [Google Scholar]


