Abstract
Outer membrane vesicles (OMVs) are nanoscale proteoliposomes that are ubiquitously secreted by Gram-negative bacteria. Interest in these bioparticles has escalated over the years, leading to discoveries regarding their composition, production, and vaccine potential. Given that many steps in vesicle biogenesis are ‘engineerable,’ it is now possible to tailor OMVs for specific applications. Such tailoring involves modifying the OMV-producing bacterium through protein, pathway, or genome engineering in a manner that specifically alters the final OMV product. For example, targeted deletion or upregulation of genes associated with the cell envelope can modulate vesicle production or remodel the composition of vesicle components such as lipopolysaccharide. Likewise, bacteria can be reprogrammed to incorporate heterologously expressed proteins into either the membrane or lumenal compartment of OMVs. We anticipate that further research in the field of OMV engineering will enable continued design and biosynthesis of specialized vesicles for numerous biotechnological purposes ranging from the delivery of vaccines to the deconstruction of cellulosic substrates.
Introduction
Outer membrane vesicles (OMVs) are produced by vesiculation, a secretory process ubiquitous to all Gram-negative bacteria [1]. These particles form during growth as the outer membrane blebs outwards and pinches off, resulting in nanoscale (~20–250 nm) spheres of the outer membrane containing soluble periplasmic components trapped in their lumens (Fig. 1). Hence, the composition of OMVs reflects components of the outer membrane and periplasm, for example, soluble proteins, integral membrane proteins, lipoproteins, and glycolipids. While it has not yet been established whether OMV production is a stochastic or regulated process [2], vesiculation has been linked to bacterial stress, with increased vesicle production occurring during conditions of high membrane stress [3,4].
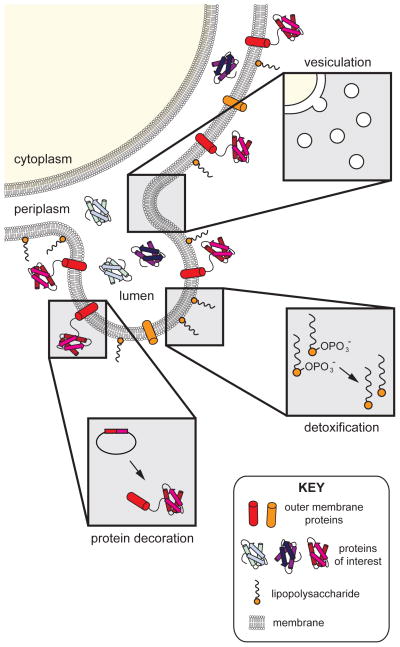
Figure 1. Engineering of bacterial OMVs.
Efforts to engineer bacterial OMVs are focused in three main areas: (1) vesiculation, or the process of membrane blebbing to produce vesicles; (2) protein decoration, or the targeting of natural or engineered biomolecules to specific subcompartments (i.e., lumen, membrane, exterior) of vesicles; and (3) detoxification, or the modification of the lipid A portion of bacterial LPS. These areas are visualized in the context of OMV structure and biogenesis. Biomolecules relevant to this review are depicted and labeled in the key.
Natural OMVs have several important roles. For example, they can contribute to bacterial survival by reducing levels of toxic compounds such as toluene [5], neutralizing environmental agents that target the OM such as antimicrobial peptides [6], aiding in the release of attacking phage [7,8], removing stress products from the cell such as misfolded periplasmic proteins [3], or nucleating the formation of bacterial communities (biofilms) [9]. OMVs released from the envelope of pathogenic bacteria play key roles in host-pathogen interactions including establishment of a colonization niche, transmission of virulence factors into host cells, and modulation of host defense and response [10–15]. These, and other characteristics of OMVs, are more fully reviewed elsewhere [1,16–18].
Because OMVs retain the physiochemical characteristics of the bacteria from which they are derived, they have been of particular interest in the context of vaccine development [19]. OMVs carry many of the same immunogenic components as the pathogens while lacking any genetic material, making them a safe vaccine platform [20]. Indeed, immunization using pathogen-derived OMVs results in the stimulation of a strong immune response and protection in murine models [21–23]. In the most notable case, Neisseria meningitidis-derived OMVs have been used in the formulation of the vaccine Bexsero (Novartis), which is approved for use in Europe [24]. Unfortunately, widespread development of OMVs in the context of vaccines is hindered by several obstacles: scale of production, versatility, and toxicity. This review will highlight progress towards enhancing bacterial vesiculation through host strain engineering, expanding the functionality of OMVs through protein decoration, and tailoring the immunogenicity and toxicity of OMVs via lipopolysaccharide (LPS) modifications. Advances in these three areas are important in showcasing OMVs as not only an effective and safe vaccine platform, but also as genetically programmable proteoliposomes with untapped potential for diverse biotechnological applications.
Engineered Vesiculation
OMVs are naturally shed at low concentration but their production can be augmented by a variety of factors, such as quorum sensing, perceived bacterial population size, and presence of a potentially hostile environment [1,3,16,17]. However, even under these circumstances, OMV production by wild-type bacteria is rarely efficient enough to achieve a reasonable scale of production for pharmaceutical and/or biotechnology applications. Additionally, the natural circumstances promoting OMV production in wild-type bacteria often require some degree of non-ideality in growth conditions, such as high temperatures, altered nutrient availability, or envelope stress [1]. This would intrinsically limit the utility of OMVs in a variety of engineered roles, such as vaccine or nucleic acid delivery systems that rely on recombinant antigen trafficking and loading [2,25].
Early efforts to stimulate OMV generation relied on mechanical or chemical disruption of Gram-negative bacteria [25]. However, more recent reports have revealed an array of factors that effectively affect vesiculation in the host bacterium. For instance, flagella proteins such as FliC [26] and “nanopods” comprised of the surface layer protein NpdA [27] participate in the production of OMVs. The deletion or overexpression of certain membrane or membrane-associated proteins can also cause a vesicle-overproducing phenotype, which results from alteration of envelope structure and/or decreased membrane stability (Table 1). Some of the earliest examples involve modifications of the Tol-Pal system, a transmembrane multiprotein complex that forms bridges between the outer membrane, peptidoglycan, and inner membrane and is generally required for outer membrane integrity [28,29]. Tol-Pal is comprised of five cell envelope proteins encoded by tolA, tolQ, tolR, tolB, and pal. A mutation in any of these genes confers a defect in outer membrane integrity (i.e., leakage of periplasmic content, detergent intolerance) and a concomitant increase in OMV production [29,30]. Likewise, overproduction of Tol proteins or proteins that interact with Tol-Pal, such as the minor coat g3p protein of filamentous phage and the translocation domains of colicins, was found to specifically destabilize the cell envelope and induce production of significant amounts of vesicles [31].
Table 1.
Genes known to alter vesiculation and/or lipid A structure
| Gene or protein name(s) | Purpose | Citation(s) |
|---|---|---|
| degP | Deletion leads to increased vesiculation | [30] |
| degS | Deletion leads to increased vesiculation | [30] |
| fliC | Deletion leads to decreased vesiculation | [26] |
| flgK | Deletion leads to decreased vesiculation | [26] |
| glnA | Deletion leads to decreased vesiculation | [30] |
| lysS/herC | Deletion leads to decreased vesiculation | [30] |
| nlpA | Deletion leads to decreased vesiculation | [30] |
| nlpI | Deletion leads to increased vesiculation | [3,30] |
| ompC | Deletion leads to increased vesiculation | [30] |
| ompR | Deletion leads to increased vesiculation | [30] |
| pepP | Deletion leads to decreased vesiculation | [30] |
| pnp | Deletion leads to increased vesiculation | [30] |
| ponB | Deletion leads to increased vesiculation | [30] |
| rmpM | Deletion leads to increased vesiculation | [54] |
| rseA | Deletion leads to increased vesiculation | [30] |
| tatC | Deletion leads to increased vesiculation | [30] |
| Tol-Pal system (tolA, tolQ, tolR, tolB, pal) | Deletion of any one gene leads to increased vesiculation | [29–33,35] |
| waaG/rfaG | Deletion leads to increased vesiculation | [30] |
| wzxE | Deletion leads to increased vesiculation | [30] |
| yieM | Deletion leads to increased vesiculation | [30] |
| ypjM | Deletion leads to decreased vesiculation | [30] |
|
| ||
| N-terminal domain of g3p phage protein | Expression leads to increased vesiculation | [31] |
| Translocation domains of colicins A and E3 | Expression leads to increased vesiculation | [31] |
|
| ||
| lpxL2 | Deletion leads to predominately tetraacylated lipid A | [56] |
| lpxM, htrB, waaN | Deletion leads to predominately tetraacylated lipid A | [57,66] |
|
| ||
| lpxL, msbB, waaM | Deletion leads to predominately pentaacylated lipid A | [57,60] |
| lpxL1 | Deletion leads to predominately pentaacylated lipid A | [54,56,67,68] |
| lpxR | Expression leads to predominately pentaacylated lipid A | [69] |
| pagL | Expression leads to predominately pentaacylated lipid A | [68] |
| msbB and pagP | Deletion of both genes leads to strictly pentaacylated lipid A | [58] |
|
| ||
| lpxO | Expression causes hydroxylation of the 3′-acyloxyacyl chain of lipid A | [65] |
|
| ||
| Hp0021 | Expression causes lipid A to lack its 1′-phosphate group | [63] |
| lpxE | Deletion causes lipid A to lack its 1′-phosphate group | [62,66] |
| lpxF | Deletion causes lipid A to lack its 4′-phosphate group | [62] |
|
| ||
| lpxT and eptA and pagP | Deletion of all three genes leads to uniform, hexaacylated, bis-phosphorylated lipid A | [65] |
Of particular interest is the tolR mutation, which has been shown to increase vesicle overproduction in different Gram-negative bacteria including Escherichia coli and Shigella sonnei to levels that are sufficient to support vaccine trials [32,33]. Despite the compromised membrane integrity, cells lacking tolR have become a valuable tool for OMV applications because they produce uniform OMVs composed of both outer membrane-associated and periplasmic proteins while excluding artificially “leaked” inner-membrane associated and cytoplasmic proteins. In addition, they can be cultivated to high densities and can even be genetically engineered for further OMV processing, such as designed antigen presentation as discussed below [29,30,32,34,35]. Given that homologs of the Tol-Pal proteins have been found in the genomes of all Gram-negative bacteria sequenced so far, it is likely that Tol-Pal manipulations, including tolR deletion, may be a universal strategy for engineering vesicle overproduction. Indeed, modification of the Tol-Pal pathway has been shown to increase the level of OMV shedding in non-pathogenic E. coli, extraintestinal pathogenic E. coli IHE3034, Shigella flexneri, S. sonnei and Salmonella enterica serovar Typhimurium [29,31,32,35].
A more systematic approach for discovering factors that affect vesiculation has been carried out using a dot blot screen to detect OMV levels in low-volume cultures [30]. This screen involved adsorbing individual cell-free supernatants to a nitrocellulose filter, after which vesicles were detected by immunoblotting with antibodies to OM components. Using this screen, 20 unique gene disruptions in E. coli were identified that caused differences in vesicle production ranging from a 5-fold decrease to a 200-fold increase relative to wild-type levels. Not surprisingly, several of the gene disruptions that increased vesiculation mapped to the Tol-Pal pathway (e.g., tolA, pal). A majority of the other identified genes were similarly linked to the cell envelope, affecting outer membrane protein expression and localization (e.g., nlpI, ompR), LPS and peptidoglycan biosynthesis (e.g., waaG, ponB) and the σE envelope stress response pathway (e.g., degP, rseA). It is conceivable that certain mutations that reliably force hypervesiculation might have unexpected benefits for different OMV applications. For example, the inner membrane integrity of strains lacking tolA and tolB was compromised [30]. Hence, these strains might be used to produce OMVs that are loaded with traditionally excluded cytoplasmic components, which are often potent immunostimulators, or recombinant deliverables, such as nucleic acids or cytoplasmic protein antigens. Strains lacking nlpI, a mutation that triggered the highest level of OMV overproduction [30], were also markedly more resistant to lethal envelope stress [3], suggesting that these strains may be useful hosts for large-scale OMV production, recombinant engineering of OMVs, or any other process that might be particularly stressful to cells. Moreover, the absence of NlpIin vesicles could reduce or eliminate the high levels of bacteria in the blood that have been attributed to this protein [36].
For many of the isolated mutants, hypervesiculation was not the result of bacterial lysis or disintegration of the bacterial envelope, nor did it simply correlate with defects in cell envelope stability or growth characteristics. Hence, despite the connections between envelope components and OMV production, a compromised envelope is not the only factor influencing vesiculation levels. The way in which these factors promote OMV production remains unclear, but they suggest the exciting possibility that vesiculation may be a regulated process, at least under certain conditions [2]. In support of a possible regulated mechanism are studies showing the selective enrichment of certain proteins in, and exclusion of others from, OMVs compared to their abundance in the outer membrane or periplasmic space. Examples of proteins preferentially enriched in OMVs are the oligomeric pore-forming cytotoxin ClyA of E. coli and other enterobacteria [37], the heat-labile enterotoxin of enterotoxigenic E. coli (ETEC) [38], the leukotoxin from Actinobacillus actinomcetemcomitans [13], the gingipain proteases of Porphyromonas gingivalis [39], and the alkaline phosphatases of Myxococcus xanthus [40]. Proteins that are reportedly excluded from OMVs include the periplasmic disulfide bond oxidase DsbA of E. coli [37] and the abundant outer membrane proteins of P. gingivalis [39].
We exploited some of these previous findings on vesicle overproduction and cargo enrichment to generate large quantities of green fluorescent protein (GFP)-enriched OMVs by expressing a ClyA-GFP fusion in cells lacking either tolR or nlpI (see the next section for more details) [34]. The biotechnological significance of producing GFP-containing OMVs to such high levels was then demonstrated by tracking the adherence and intracellular localization of fluorescent OMVs in mammalian cells [34] or characterizing the immune response of vaccinated animals [33]. We anticipate that an improved understanding of the regulatory mechanisms and physiological basis of OMV biogenesis will enable even more new applications that leverage key aspects of vesiculation amplification and control.
Interior and Exterior Protein Decoration
Naturally produced OMVs share similar characteristics and biomolecular components as the envelopes of their bacterial sources [41,42]. Building on this observation, many groups have applied genetic and biomolecular techniques to specifically target heterologous proteins to different subcompartments of OMVs, including the lumen, the membrane, and the outer surface. Early work on this front showed that E. coli cells expressing recombinant outer membrane protein Ail from Yersinia entercolitica are capable of producing OMVs that are adorned with the Ail protein on their outer surface [43]. Similarly, E. coli cells expressing heterologous GFP in the periplasmic space were observed to shed OMVs that carried the fluorescent protein in their lumens [43]. New functionality, such as enzymatic activity and binding specificity, can be imparted onto OMVs through the expression and surface localization of enzymes, targeting receptors or ligands, and protein scaffolds [34,44]. Furthermore, OMV-associated recombinant proteins can be internalized by eukaryotic cells [34,43], can stimulate a strong and specific immune response in mice [33,45–47], and can efficiently break down cellulosic materials [44]. Hence, decorating the surface and interior of OMVs with a variety of recombinant proteins results in nanoscale delivery vehicles that are capable of presenting their cargo to specific cells, tissues, whole organisms, and even biomaterials.
Engineering OMVs with recombinant proteins can be as simple as expressing the protein of interest in a vesicle overproducing host, as is the case for proteins that naturally traffic to the outer membrane [43,47] or periplasm [23]. However, many proteins of interest do not naturally partition to these locations or partition inefficiently and, therefore, must be modified accordingly. For example, fusing an export signal peptide derived from a substrate of the general secretory (Sec) or twin-arginine translocation (Tat) pathway to the N-terminus of a heterologous protein of interest targets that protein to the periplasm, and eventually into the lumen of OMVs [43,45,46] (Fig. 2A and B). However, while periplasmic targeting is useful for certain applications, many others require the protein of interest to be “displayed” on the exterior of OMVs to achieve the desired functionality. Current approaches involve fusing a protein of interest to a membrane protein anchor that co-localizes its fusion partner to the outer membrane, and hence, the membrane of OMVs (Fig. 2C and D). One notable example is cytolysin A (ClyA), a pore-forming cytotoxin expressed by E. coli and some other enterobacteria. ClyA is exported to the periplasm by an unknown mechanism (i.e., it carries no discernable signal peptide), integrated into the outer membrane, and finally enriched in OMVs [37]. Taking advantage of this preferential enrichment, chimeric fusion of a target protein to the N- or C-terminus of ClyA results in display of the target protein on the exterior of OMVs [34]. Importantly, OMV-displayed proteins, such as GFP, β-lactamase, organophosphorus hydrolase, and a single-chain Fv (scFv) antibody retained their biological activity. Bacterial autotransporters, proteins that integrate into the outer membrane and surface display their N-terminal passenger domains via self-transport through pores formed by their C-terminal beta-barrel domains [48], can also ferry their fusion partner to the exterior of OMVs [48,49]. In fact, any surface display system should work for this purpose, for instance, ice nucleation protein (INP), Lpp-OmpA, and eCPX [50].
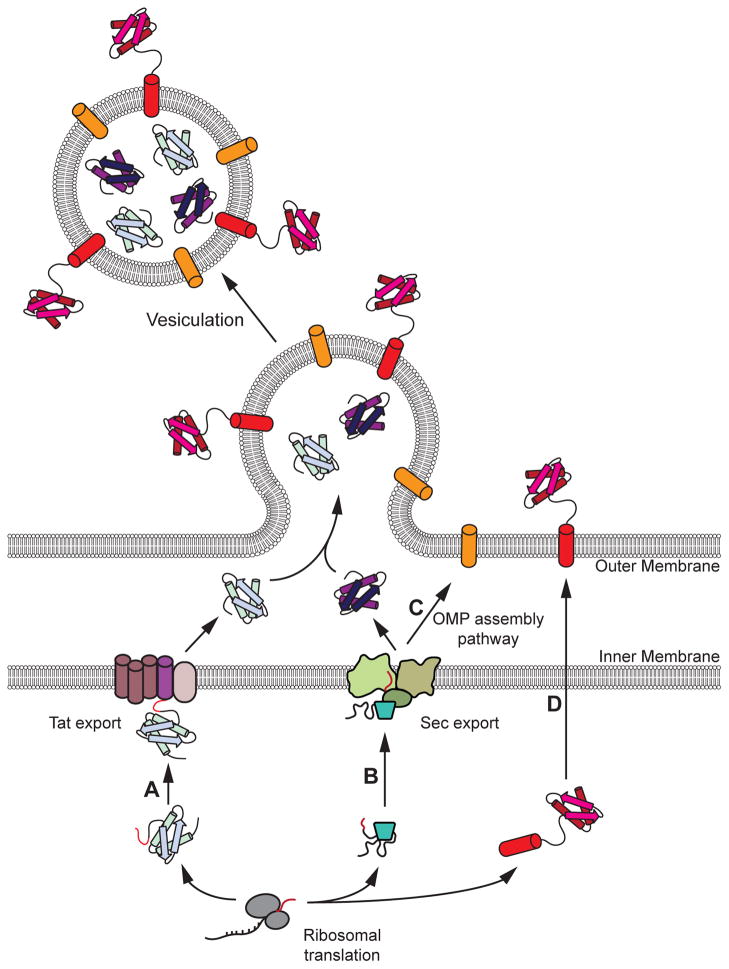
Figure 2. Decorating the interior and exterior of bacterial OMVs.
Recombinant proteins can be targeted to the lumen, the membrane, and the outer surface of OMVs. Localizing proteins in the lumen is accomplished using signal peptides that target proteins to either the (A) Tat (for folded proteins; post-translational) or (B) Sec (for unfolded proteins; post- or co-translational) export pathways. Incorporation of proteins in the membrane is accomplished using proteins that natively transit the (B) Sec pathway followed by the (C) outer membrane protein (OMP) assembly pathway. Display of proteins on the exterior is accomplished by fusion to a canonical outer membrane protein that transits the (B) Sec and (C) OMP assembly pathways, or to a non-canonical outer membrane protein (e.g., ClyA) whose export pathways (D) are undetermined.
While the above examples focus on single-gene modification strategies, incorporating multiple recombinant proteins into engineered OMVs has also been demonstrated. For example, a strain of N.meningitidis carrying several variants of the porA gene yielded multivalent OMVs that were used as a broad-spectrum vaccine capable of protecting against multiple strains of the N. meningitidis serogroup B pathogen [51]. More recently, functional assembly of multiple enzymes on OMVs was accomplished using INP to display a trivalent scaffold containing three orthogonal cohesin domains [44]. By sequentially adding three dockerin-tagged cellulases to scaffold-displaying OMVs, a multienzyme “mini-cellulosome” was assembled on the exterior of OMVs. The resulting cellulosome-displaying OMVs were observed to enhance cellulose hydrolysis by more than 20-fold over that of non-complexed enzymes. A similar trivalent mini-cellulosome structure displayed on yeast cells only improved cellulose hydrolysis by 2.5-fold over that of free enzymes [52]. The substantially higher level of enzyme synergy using OMVs is likely the result of their nanoscale dimension (~50 nm) as opposed to the microscale dimension of yeast (~5 μM), which offers a much higher enzyme to volume ratio and improved substrate accessibility.
Molecular Detoxification
The application of OMV technology to vaccine development has received the greatest attention thus far. For use in humans, however, detoxification of these liposomal particles is required. This is because the outer membrane of Gram-negative bacteria contains LPS [17], which consists of lipid A (or endotoxin), a nonrepeating “core” oligosaccharide, and a distal polysaccharide (or O-antigen) [53]. Early research towards detoxification used detergent extraction to reduce the LPS content [33,54] since it is the primary toxic component of OMVs. However, detergent extraction is laborious, cost intensive, and reduces the adjuvant activity of these vesicles, which are all serious problems for vaccine development. Recently, more attractive methods of OMV detoxification have emerged to address these issues. At the forefront of this research is the ability to genetically modify vesicle-producing bacteria in a manner that alters the physical properties of the LPS during their growth.
Detergent extraction of OMVs is the traditional method used to lower LPS content and improve vesicle yields [54]. It has been shown to result in OMV preparations that are safe for vaccine administration [20]; however, there is evidence that detergent extraction leads to the removal of other important vesicle components. These include phospholipid and lipoproteins, molecules that contribute to OMV immunogenicity [54,55]. In addition, detergent treatment can promote aggregation of vesicles or lead to changes in the OMV structure by removing essential molecules [56]. Nevertheless, this technique is still widely used for detoxifying OMVs. In fact, detergent-extracted native OMVs from N. meningitidis were still in vaccine clinical trials as recently as 2010 [20]. Unsurprisingly, although detergent extraction decreases the amount of toxic LPS, this comes at the cost of lowering the adjuvant activity of OMVs [56]. As a result, recent efforts to improve detoxification have focused on novel techniques that emphasize retention of adjuvancy.
Genetic modifications of bacterial LPS for the purposes of reducing toxicity are born from studies of natural bacterial enzymes. The endotoxic LPS of commonly used OMV-producing strains, such as E. coli, N. meningitidis, and S. enterica, naturally has six acyl chains. Enzymes that lead to the addition of the fifth, and in some cases the fourth, acyl chain of lipid A were discovered and characterized for several bacteria (Table 1). Knockout of these genes has been shown to alter the LPS structure and result in reduced toxic effects. For instance, mutant strains with an insertional inactivation of lpxL produce LPS with penta- instead of hexa-acylated lipid A, which is less toxic while still retaining the desirable adjuvant activity [57]. Similar strain engineering has been used to alter the toxicity of OMVs produced by E. coli [58,59], N. meningitidis [54,56], S. enterica [60], and Bordetella pertussis [61]. Importantly, attenuation of toxicity can be achieved in concert with enhanced vesiculation as a strain carrying both a detoxifying mutation (lpxL) and a vesicle over-producing mutation (rmpM) yielded a large quantity of less toxic, protective OMVs [54]. There are even single-gene knockouts that affect both properties, for instance, the nlpI mutation, which increases vesiculation while eliminating bacteremia [30,36].
Another approach to remodeling of the lipid A structure is to supplement bacteria with exogenous genes that encode LPS-modifying enzymes. One good source of such genes is pathogenic bacteria, such as Helicobacter pylori, that modify their lipid A by removal of phosphate groups from the 1- and 4-positions of the lipid A backbone to evade host immune systems [62]. Indeed, heterologous expression in E. coli of H. pylori Hp0021, an enzyme that removes the 1-phosphate from lipid A, resulted in biosynthesis of monophosphorylated lipid A instead of the natural diphosphorylated form [63]. As monophosphoryl lipid A, or MPL, is an FDA-approved adjuvant [64], engineering vesicle over-producing strains that carry one of these lipid A remodeling enzymes represents an important step for vaccine development. Recently, further structural diversification of lipid A was demonstrated using a combinatorial gene expression strategy in E. coli [65]. This strategy resulted in a spectrum of bioactive LPS variants with distinct immunological activities, some of which might be useful candidates for vaccine and therapeutic development.
Conclusions
As interest in OMVs for biotechnological applications has grown, so too has interest in the ability to engineer the molecular and structural properties of these nanoscale vesicle systems. The continued application of gene, pathway, and genome engineering will ultimately enable more sophisticated optimization of OMV production, leading to the creation of finely tuned bacterial “factories” required for the realistic application of OMV technology. Likewise, the same strategies will enable the creation of remodeled lipid A structures that alter the interactions of OMVs with the immune system, inducing a tailored immune response while simultaneously eliminating unwanted toxicity. The protein cargo of OMVs can be readily reprogrammed via the deployment of existing and new methods for expressing, targeting, and engineering proteins. By decorating the interior and exterior of OMVs with unique combinations of antigens, antibodies, receptors, receptor ligands and/or enzymes, the functionality of OMVs will be extended to include a wide range of biotechnologically relevant activities. Hence, while vaccine applications have drawn the most attention, it is foreseeable that additional uses will begin to emerge, including, but not limited to, bioremediation, conversion of lingo-cellulosic materials, diagnostics, and therapeutic protein and nucleic acid delivery.
Highlights.
Bacterial outer membrane vesicles (OMVs) can be rationally engineered for biotechnological applications
OMV production can be increased by manipulating cell envelope-related genes
Recombinant proteins can be controllably targeted to OMV subcompartments
Remodeling glycolipid structure alters immunological activity and toxicity of OMVs
Acknowledgments
JAR gratefully acknowledges Hertz Foundation and NSF graduate fellowship support. This work was supported by NSF CBET Award # 1264701 and NSF DGE Award # 1011509 (both to MPD), and NIH Award R21EB005669-01 (to DP and MPD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review have been highlighted as: * of special interest
** of outstanding interest
- 1.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Schwechheimer C, Sullivan CJ, Kuehn MJ. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry. 2013;52:3031–3040. doi: 10.1021/bi400164t. The authors discuss mechanistic aspects of vesiculation, in particular, the advantages of this process for non-pathogenic bacteria and the steps involved in vesicle biogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macdonald IA, Kuehn MJ. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J Bacteriol. 2013;195:2971–2981. doi: 10.1128/JB.02267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi H, Uematsu K, Hirayama H, Horikoshi K. Novel toluene elimination system in a toluene-tolerant microorganism. J Bacteriol. 2000;182:6451–6455. doi: 10.1128/jb.182.22.6451-6455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. doi: 10.1186/1471-2180-11-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeb MR. Bacteriophage T4-mediated release of envelope components from Escherichia coli. J Virol. 1974;13:631–641. doi: 10.1128/jvi.13.3.631-641.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb MR, Kilner J. Release of a special fraction of the outer membrane from both growing and phage T4-infected Escherichia coli B. Biochim Biophys Acta. 1978;514:117–127. doi: 10.1016/0005-2736(78)90081-0. [DOI] [PubMed] [Google Scholar]
- 9.Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. 2009;77:4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gankema H, Wensink J, Guinee PA, Jansen WH, Witholt B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect Immun. 1980;29:704–713. doi: 10.1128/iai.29.2.704-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 14.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 17.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 19.Rosenthal JA, Chen L, Baker JL, Putnam D, DeLisa MP. Pathogen-like particles: biomimetic vaccine carriers engineered at the nanoscale. Curr Opin Biotechnol. 2013;28:51–58. doi: 10.1016/j.copbio.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 20*.Collins BS. Gram-negative outer membrane vesicles in vaccine development. Discov Med. 2011;12:7–15. This short review highlights the use of OMVs in vaccine development, describing their production and purification as well as providing examples of their use against N. meningitidis serogroup B in several countries. [PubMed] [Google Scholar]
- 21.Camacho AI, de Souza J, Sanchez-Gomez S, Pardo-Ros M, Irache JM, Gamazo C. Mucosal immunization with Shigella flexneri outer membrane vesicles induced protection in mice. Vaccine. 2011;29:8222–8229. doi: 10.1016/j.vaccine.2011.08.121. [DOI] [PubMed] [Google Scholar]
- 22.Pierson T, Matrakas D, Taylor YU, Manyam G, Morozov VN, Zhou W, van Hoek ML. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J Proteome Res. 2011;10:954–967. doi: 10.1021/pr1009756. [DOI] [PubMed] [Google Scholar]
- 23.Schild S, Nelson EJ, Bishop AL, Camilli A. Characterization of Vibrio cholerae outer membrane vesicles as a candidate vaccine for cholera. Infect Immun. 2009;77:472–484. doi: 10.1128/IAI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorringe AR, Pajon R. Bexsero: a multicomponent vaccine for prevention of meningococcal disease. Hum Vaccin Immunother. 2012;8:174–183. doi: 10.4161/hv.18500. [DOI] [PubMed] [Google Scholar]
- 25.Chatterjee S, Chaudhuri K. Outer membrane vesicles of bacteria. Springer; Berlin Heidelberg: 2012. [Google Scholar]
- 26.Manabe T, Kato M, Ueno T, Kawasaki K. Flagella proteins contribute to the production of outer membrane vesicles from Escherichia coli W3110. Biochem Biophys Res Commun. 2013;441:151–156. doi: 10.1016/j.bbrc.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One. 2011;6:e20725. doi: 10.1371/journal.pone.0020725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol. 2006;188:5385–5392. doi: 10.1128/JB.00498-06. This study describes the application of a genetic screen in E. coli to identify gene disruptions that caused differences in vesicle production ranging from a 5-fold decrease to a 200-fold increase relative to wild-type levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry T, Pommier S, Journet L, Bernadac A, Gorvel JP, Lloubes R. Improved methods for producing outer membrane vesicles in Gram-negative bacteria. Res Microbiol. 2004;155:437–446. doi: 10.1016/j.resmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 32**.Berlanda Scorza F, Colucci AM, Maggiore L, Sanzone S, Rossi O, Ferlenghi I, Pesce I, Caboni M, Norais N, Di Cioccio V, et al. High yield production process for Shigella outer membrane particles. PLoS One. 2012;7:e35616. doi: 10.1371/journal.pone.0035616. The authors describe the successful scale-up of OMV production using S. sonnei. Yields of around 100 mg of membrane-associated proteins per liter of fermentation were reached. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, Putnam D. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci U S A. 2010;107:3099–3104. doi: 10.1073/pnas.0805532107. This study demonstrates that engineered E. coli OMVs are an easily purified vaccine-delivery system capable of greatly enhancing the immunogenicity of a low-immunogenicity protein antigen without added adjuvants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Kim JY, Doody AM, Chen DJ, Cremona GH, Shuler ML, Putnam D, DeLisa MP. Engineered bacterial outer membrane vesicles with enhanced functionality. J Mol Biol. 2008;380:51–66. doi: 10.1016/j.jmb.2008.03.076. This study describes a strategy for remodeling the outer surface of OMVs with non-native functionality by fusing several heterologous proteins to the vesicle-associated toxin ClyA of E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berlanda Scorza F, Doro F, Rodriguez-Ortega MJ, Stella M, Liberatori S, Taddei AR, Serino L, Gomes Moriel D, Nesta B, Fontana MR, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol Cell Proteomics. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Tseng YT, Wang SW, Kim KS, Wang YH, Yao Y, Chen CC, Chiang CW, Hsieh PC, Teng CH. NlpI facilitates deposition of C4bp on Escherichia coli by blocking classical complement-mediated killing, which results in high-level bacteremia. Infect Immun. 2012;80:3669–3678. doi: 10.1128/IAI.00320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wai SN, Lindmark B, Soderblom T, Takade A, Westermark M, Oscarsson J, Jass J, Richter-Dahlfors A, Mizunoe Y, Uhlin BE. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/s0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 38.Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haurat MF, Aduse-Opoku J, Rangarajan M, Dorobantu L, Gray MR, Curtis MA, Feldman MF. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 41.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 43*.Kesty NC, Kuehn MJ. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J Biol Chem. 2004;279:2069–2076. doi: 10.1074/jbc.M307628200. One of the first reports that heterologously expressed proteins can be incorporated into the membrane and lumen of OMVs derived from non-pathogenic E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park M, Sun Q, Liu F, DeLisa MP, Chen W. Positional assembly of enzymes on bacterial outer membrane vesicles for cascade reactions. Chem Commun (Camb) 2014 doi: 10.1371/journal.pone.0097103. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Muralinath M, Kuehn MJ, Roland KL, Curtiss R., 3rd Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun. 2011;79:887–894. doi: 10.1128/IAI.00950-10. These authors report that a recombinant antigen, pneumococcal PspA, can be packaged in the lumen of S. enterica OMVs. Mice intranasally immunized with these OMVs developed serum antibody responses that completely protected against a lethal challenge of Streptococcus pneumoniae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartolini E, Ianni E, Frigimelica E, Petracca R, Galli G, Berlanda Scorza F, Norais N, Laera D, Giusti F, Pierleoni A, et al. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J Extracell Vesicles. 2013;2:20181. doi: 10.3402/jev.v2i0.20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Dwyer CA, Reddin K, Martin D, Taylor SC, Gorringe AR, Hudson MJ, Brodeur BR, Langford PR, Kroll JS. Expression of heterologous antigens in commensal Neisseria spp: preservation of conformational epitopes with vaccine potential. Infect Immun. 2004;72:6511–6518. doi: 10.1128/IAI.72.11.6511-6518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jose J, Meyer TF. The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol Mol Biol Rev. 2007;71:600–619. doi: 10.1128/MMBR.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroeder J, Aebischer T. Recombinant outer membrane vesicles to augment antigen-specific live vaccine responses. Vaccine. 2009;27:6748–6754. doi: 10.1016/j.vaccine.2009.08.106. [DOI] [PubMed] [Google Scholar]
- 50.van Bloois E, Winter RT, Kolmar H, Fraaije MW. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 2011;29:79–86. doi: 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 51.van der Ley P, van der Biezen J, Poolman JT. Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine. 1995;13:401–407. doi: 10.1016/0264-410x(95)98264-b. [DOI] [PubMed] [Google Scholar]
- 52.Tsai SL, Oh J, Singh S, Chen R, Chen W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2009;75:6087–6093. doi: 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van de Waterbeemd B, Streefland M, van der Ley P, Zomer B, van Dijken H, Martens D, Wijffels R, van der Pol L. Improved OMV vaccine against Neisseria meningitidis using genetically engineered strains and a detergent-free purification process. Vaccine. 2010;28:4810–4816. doi: 10.1016/j.vaccine.2010.04.082. [DOI] [PubMed] [Google Scholar]
- 55.Ferrari G, Garaguso I, Adu-Bobie J, Doro F, Taddei AR, Biolchi A, Brunelli B, Giuliani MM, Pizza M, Norais N, et al. Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics. 2006;6:1856–1866. doi: 10.1002/pmic.200500164. [DOI] [PubMed] [Google Scholar]
- 56.van der Ley P, van den Dobbelsteen G. Next-generation outer membrane vesicle vaccines against Neisseria meningitidis based on nontoxic LPS mutants. Hum Vaccin. 2011;7:886–890. doi: 10.4161/hv.7.8.16086. [DOI] [PubMed] [Google Scholar]
- 57.van der Ley P, Steeghs L, Hamstra HJ, ten Hove J, Zomer B, van Alphen L. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect Immun. 2001;69:5981–5990. doi: 10.1128/IAI.69.10.5981-5990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Lee DH, Kim SH, Kang W, Choi YS, Lee SH, Lee SR, You S, Lee HK, Chang KT, Shin EC. Adjuvant effect of bacterial outer membrane vesicles with penta-acylated lipopolysaccharide on antigen-specific T cell priming. Vaccine. 2011;29:8293–8301. doi: 10.1016/j.vaccine.2011.08.102. The authors demonstrate that native OMVs from non-pathogenic E. coli with penta-acylated LPS are a safe, effective design for use in vaccine development. [DOI] [PubMed] [Google Scholar]
- 59.Kim SH, Kim KS, Lee SR, Kim E, Kim MS, Lee EY, Gho YS, Kim JW, Bishop RE, Chang KT. Structural modifications of outer membrane vesicles to refine them as vaccine delivery vehicles. Biochim Biophys Acta. 2009;1788:2150–2159. doi: 10.1016/j.bbamem.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee SR, Kim SH, Jeong KJ, Kim KS, Kim YH, Kim SJ, Kim E, Kim JW, Chang KT. Multi-immunogenic outer membrane vesicles derived from an MsbB-deficient Salmonella enterica serovar typhimurium mutant. J Microbiol Biotechnol. 2009;19:1271–1279. [PubMed] [Google Scholar]
- 61.Asensio CJ, Gaillard ME, Moreno G, Bottero D, Zurita E, Rumbo M, van der Ley P, van der Ark A, Hozbor D. Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine. 2011;29:1649–1656. doi: 10.1016/j.vaccine.2010.12.068. [DOI] [PubMed] [Google Scholar]
- 62.Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran AX, Karbarz MJ, Wang X, Raetz CR, McGrath SC, Cotter RJ, Trent MS. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubensky TW, Jr, Reed SG. Adjuvants for cancer vaccines. Semin Immunol. 2010;22:155–161. doi: 10.1016/j.smim.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 65**.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci U S A. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. The authors used a combinatorial gene expression strategy in E. coli to create the first library of bioactive LPS variants with diverse immunological activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han Y, Li Y, Chen J, Tan Y, Guan F, Wang X. Construction of monophosphoryl lipid A producing Escherichia coli mutants and comparison of immuno-stimulatory activities of their lipopolysaccharides. Mar Drugs. 2013;11:363–376. doi: 10.3390/md11020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders H, Feavers IM. Adjuvant properties of meningococcal outer membrane vesicles and the use of adjuvants in Neisseria meningitidis protein vaccines. Expert Rev Vaccines. 2011;10:323–334. doi: 10.1586/erv.11.10. [DOI] [PubMed] [Google Scholar]
- 68.Arenas J, van Dijken H, Kuipers B, Hamstra HJ, Tommassen J, van der Ley P. Coincorporation of LpxL1 and PagL mutant lipopolysaccharides into liposomes with Neisseria meningitidis opacity protein: influence on endotoxic and adjuvant activity. Clin Vaccine Immunol. 2010;17:487–495. doi: 10.1128/CVI.00423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CR, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–21987. doi: 10.1074/jbc.M603527200. [DOI] [PMC free article] [PubMed] [Google Scholar]