Abstract
The immune system has been implicated as an important modulator of tissue regeneration. However, the mechanisms driving injury-induced immune response and tissue repair remain poorly understood. For over 200 years, planarians have been a classical model for studies on tissue regeneration, but the planarian immune system and its potential role in repair is largely unknown. We found through comparative genomic analysis and data mining that planarians contain many potential homologs of the innate immune system that are activated during injury and repair of adult tissues. These findings support the notion that the relationship between adult tissue repair and the immune system is an ancient feature of basal Bilateria. Further analysis of the planarian immune system during regeneration could potentially add to our understanding of how the innate immune system and inflammatory responses interplay with regenerative signals to induce scar-less tissue repair in the context of the adult organism.
Keywords: Planarian, regeneration, innate immunity, wound repair
1. Introduction
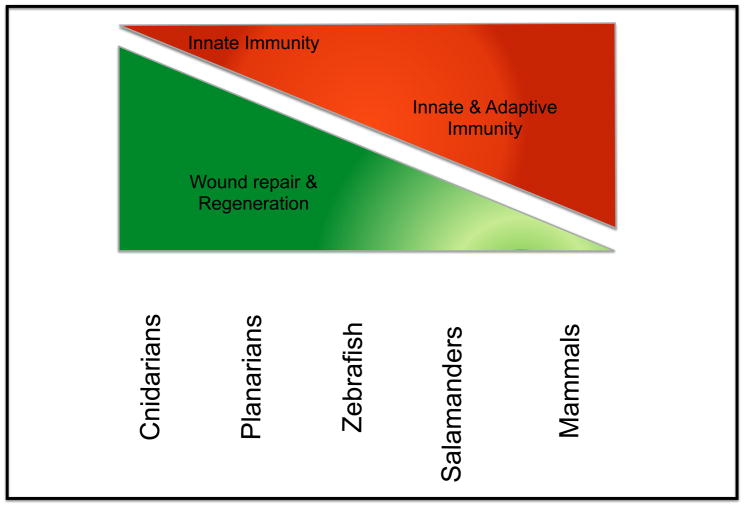
The process by which form and function is reestablished after tissue injury has puzzled the scientific community for hundreds of years [1–3]. Tissue injury triggers localized and systemic signals that orchestrate mechanical and cellular responses aimed at reducing the surface area of the wound, repair damage, and coordinating functional integration between new and pre-existing structures [4, 5]. The immune system and the inflammatory responses associated with injury have been implicated as critical modulators of wound repair and regeneration [6–10]. For instance, effective repair of wounded skin in mammals, and regeneration of complex structures in amphibians (e.g. tail and limbs) rely on the correct synchronization of the immune response upon injury [6–11]. Recent studies suggest that the difference in regenerative capacity of some species is inversely correlated with the complexity of the immune system (Figure 1) [7, 9]. However, the mechanisms integrating immune responses to injury and the process of tissue repair remain poorly understood.
Figure 1. Inverse correlation between immune system complexity and regenerative capacity.
With increasing complexity of the immune system, the regenerative capacity of the organism is decreased. In some invertebrate species without an adaptive immune system, and salamanders with a more complex immune system scar-less repair occurs. In contrast, mammals tend to have scar-forming injury repair and reduced regenerative capacity.
Upon injury the host-mediated immune response not only defends against infection, it facilitates the removal of cellular debris near the site of the wound. These functions may also modulate the cellular response that initiates repair and reestablishes tissue function. It is conceivable that both the injury-induced immune response and the process of tissue repair evolved together to promote and preserve multicellularity. The possibility of shared origin is supported by the fact that regeneration is an attribute widely distributed across multicellular organisms, and immunity is an ancient feature that emerged from the common ancestor of Cnidaria and Bilateria [12–18]. Coexistence of both regeneration and the immune system is observed in the simplest animals such as the diploblastic cnidarian Hydra, which regenerate entire body parts in presence of a primitive complement system and which have many genes associated with the mammalian immune response [16, 17, 19, 20]. Evaluating the evolution and intermingling of the immune response and process of regeneration following injury may provide a new depth to our understanding of tissue repair mechanisms across the animal kingdom.
Planarians are non-parasitic flatworms that provide compelling evolutionary reasons to analyze injury-induced immune responses and the process of regeneration in metazoans [21, 22]. Planarians display bilateral symmetry and derivatives of all three germ layers (ectoderm, mesoderm and endoderm). Adult planarians can regenerate any part of their body and constantly renew aging and damaged tissues (Figure 2), which are features missing in other commonly used invertebrate models (e.g. Drosophila and C. elegans). Regeneration and adult tissue turnover in planarians proceed through activation of somatic stem cells known as neoblasts [23–25]. Planarians have provided a classical model to study regeneration for over 200 years, but their immune system remains largely unexplored [1, 26–28]. To our knowledge, no systematic analysis of the planarian immune system has been published to date. We present a brief survey of the most salient components of the innate immune system through genomic analysis between the freshwater planarian Schmidtea mediterranea and other animal species. We analyzed two sources: the S. mediterranea genome database, SmedGD [29] and transcriptomic work of Sandman et al. [30] to evaluate genes associated with the immune system and their expression during regeneration. Our findings suggest evolutionary conservation of the triclad immune system. We identified components of the S. mediterranea immune system that may protect against infections, distinguish between commensal and pathogenic microorganisms, and that actively participate during wound repair, regeneration and maintenance of adult tissues.
Figure 2. The planarian Schmidtea mediterranea constantly renew aging and damaged tissues, and can regenerate any part of their body upon injury.
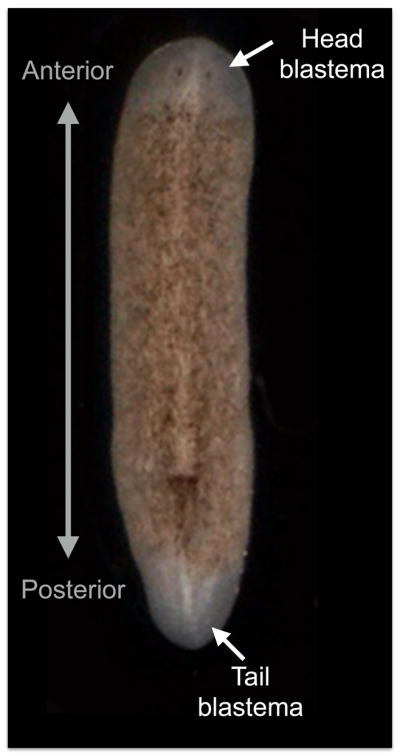
A) Adult specimen of S. mediterranea. Red dotted line describe plan of amputation. B) Regeneration of the anterior part including the entire brain, part of the digestive system, muscles, and other derivatives of the three embryonic germ layers are re-established in only seven days after decapitation. After amputation tissue contraction around the injured area is followed by the formation of the regenerative blastema, which is the unpigmented tissue where the progeny of the dividing neoblasts is instructed to recreate the missing parts. Scale bar is 200 μm.
2. The planarian model system and tissue regeneration
Planarians are known for their extraordinary capacity to regenerate adult tissues. These animals can regenerate any part of their body including their entire digestive system, brain and neural connections, muscles, and connective tissues. Planarians are easy and inexpensive to maintain under laboratory conditions and are amenable to molecular, genetic, behavioral and computational analysis [1, 26, 31–34]. During the past 20 years, planarian research has attracted attention due to the accessibility of this model organism and the opportunities to progress our understanding in long standing biomedical problems associated with regeneration, cancer and degenerative diseases. Several tools available to aid in the study of planarians, include the S. mediterranea genome, proteomic data, results from high-throughput RNAi-screens and transcriptomic analysis [1, 25, 26, 29, 32, 34–38]. Altogether with the availability of standard genetic and biochemical techniques, planarians present an attractive model to study wound repair and regeneration and the possible interplay of the immune system during these processes.
Detailed information about the biology of planarians and recent advances on research pertaining to its regenerative capacities has been reviewed elsewhere [1, 23–25, 28]. In this section, we aim at providing a brief overview of the process of regeneration in S. mediterranea, the most commonly used planarian species. Planarian regeneration is quick, taking about a week to reestablish form and function. Immediately following amputation, contraction around the injury reduces the damaged surface. In the first 24 hours local cell death peaks, followed by neoblast proliferation and H+/K+-ATPase-mediated tissue depolarization around the wounded area [39–42]. The antagonistic process of cell death and proliferation around the injured area is critical for the progression of regeneration. The H+/K+-ATPase-mediated ion flux and consequent tissue depolarization is required for anterior tissue specification and may provide signaling that guides neoblast proliferation, migration and remodeling [5, 39, 40]. Decisions regarding polarity are defined within the first few hours post-amputation and differentiated tissues may provide molecular cues and guidance during regeneration [1, 25, 28, 43]. Neoblasts, the only dividing cells in adult planarians serve as the exclusive source of new cells during the formation of the blastema (a regenerative outgrowth around the injured area).
Neoblast division gives rise to cellular progeny that migrate and differentiate within the blastema. Remodeling and integration between the new and pre-existing tissue is an active process during regeneration but little is known about the signals mediating these events. The planarian model provides the opportunity to analyze local and systemic signaling during tissue repair and regeneration, providing a unique prospective for understanding injury-immune responses in the adult body.
3. The planarian innate immune system
The immune system in adult animals is essential to maintaining body homeostasis as it distinguishes between commensal and pathogenic microorganisms, protects against infections, and serves as a signaling system during tissue repair. Most studies associating the immune system and flatworms focus on parasitic species that invade and activate host immune responses [44–46]. There is a wealth of literature associated with this aspect of host-interaction that has important practical and clinical applications and is discussed elsewhere [44–46]. We instead concentrate on the immune response against pathogens that invade non-parasitic planarians and the injury-immune activation associated with adult tissue regeneration.
The immune response is comprised of two parts: the innate and adaptive responses. Innate immune responses make up the first responders of the immune system and impart broad (often termed non-specific) protection from classes of pathogens. This response is immediate, occurring within only a few hours of infection. The innate immune system provides the first line of defense giving the adaptive (or acquired) immune response the days to weeks needed to fully develop. The adaptive immune system then has the task of clearing the microbial infection using pathogen-specific immune responses [47, 48]. There is no systematic proof of an advanced adaptive immune response in invertebrate animals [49]. On the other hand, innate immune responses are markedly similar between many invertebrates and vertebrates [50–52].
The innate immune system is the first barrier of defense against invasive microbes. It is comprised of many players, from the physical barrier and secretion of mucus, anti-microbial peptides, fatty acids and lysozymes to phagocytes that capture and digest the invading pathogens [53]. Microbes express classes of surface proteins, termed pathogen-associated molecular patterns, which are recognized by host pathogen recognition receptors (PRR) expressed on host cells. In vertebrates, recognition of pathogens through these PRRs results in inflammatory signals, differentiation of immune cells and migration of leukocytes into infected or damaged tissue [53].
The planarian immune system remains largely unexplored, but it is known that under laboratory conditions planarians suffer from infection just as other eukaryotic species, with bacterial infection a common threat in routine maintenance [54]. Treatment with antibiotics usually resolves the infection but it is unknown what types of bacteria commonly infect S. mediterranea under laboratory conditions. Nonetheless, a mechanism of pathogen recognition must exist in S. mediterranea to prevent or control these infections. Histological and functional work identified active phagocytic cells around areas of wounding; suggesting the possibility that a primitive innate immune system participates during tissue repair, cancer, and infection [55–57]. Table 1 includes a collection of candidate genes from the S. mediterranea genome that are potential components of the innate immune system [29]. Table 2 provides a sample of candidate genes of the innate immune system and their expression during head regeneration in S. mediterranea [30]. Details about these planarian candidate innate immune genes are provided below.
Table 1.
Potential candidate genes of the planarians innate immune system. Source SmedGD [29].
| Innate immune Component | Domain found | Maker ID from Planarian databse |
|---|---|---|
| Toll-Like Receptor (TLR) or MyD88 or CD14 | Leucine rich repeat | mk4.000112.15, mk4.000148.12, mk4.002858.00, mk4.007365.00, mk4.019024.00, mk4.026212.00 |
| TIR domain | mk4.000285.01, mk4.000346.04, mk4.027932.00 | |
| SARM 1 | SAM domain + TIR domain | mk4.008544.03 |
| Selectins | C-type Lectin like domain | mk4.000042.08, mk4.000389.11, mk4.020584.01 |
| Complement | CUB domain | mk4.001431.05, mk4.001438.03, mk4.000798.07, mk4.003730.02, mk4.007851.01 |
| alpha2-macroglobulin | mk4.004852.05, mk4.014934.00, mk4.043975.00, mk4.008692.00, mk4.008429.01, mk4.043975.00 | |
| C1q domain | mk4.008251.00 | |
| Complement factors or Integrins | Von Willebrand factor type A domain | mk4.007402.00 |
| Cell adhesion molecules (ICAM, VCAM) | Immunoglobulin I-set domain | mk4.006497.00 |
| Urokinase-type plasminogen activator | Kringle Domain | mk4.000652.03 |
| IRAK like kinase I | Possible IRAK similar to c elegans and Protein Kinase domain | mk4.004443.00, mk4.004443.01, mk4.027874.00 |
| TRAF-6 | zn finger domain of TRAF and TRAF MATH domain | mk4.000002.01 - mk4.000002.47 |
| TRAF related | MATH domains | mk4.000007.04 - mk4.000007.34 |
Table 2.
Expression of innate immune candidate genes during planarian regeneration. From transcriptomic analysis of head regeneration performed by Sandmann et. al., [30].
| Innate immune Component | Domain found | Maker ID | Time Post amputation | |||
|---|---|---|---|---|---|---|
| 1 hour | 6 hour | 10–18 hour | 24–72 hour | |||
| Toll Like Receptor (TLR) or MyD88 | Leucine rich repeat | mk4.007365.00 |
|
|
|
|
| Leucine rich repeat | mk4.002858.00 |
|
|
|
|
|
| TIR domain | mk4.000285.01 |
|
|
|
|
|
| Selectins | C-type Lectin like domain | mk4.000389.11 |
|
|
|
|
| C-type Lectin like domain | mk4.020584.01 |
|
|
|
|
|
| Complement | CUB domain | mk4.000798.07 |
|
|
|
|
| CUB domain | mk4.003730.02 |
|
|
|
|
|
| CUB domain | mk4.001431.05 |
|
|
|
|
|
| alpha2-macroglobulin | mk4.005639.01 |
|
|
|
|
|
| alpha2-macroglobulin | mk4.008692.00 |
|
|
|
|
|
| C1q domain | mk4.008251.00 |
|
|
|
|
|
| Complement factors or Integrins | Von Willebrand factor type A domain | mk4.007402.00 |
|
|
|
|
| IRAK like kinase I | Possible IRAK similar to c elegans and Protein Kinase domain | mk4.027874.00 |
|
|
|
|
| TRAF-6 | zn finger domain of TRAF | mk4.000002.06 |
|
|
|
|
| TRAF MATH domain | mk4.000002.07 |
|
|
|
|
|
3.1. Mucus and anti-microbial peptides
Surfaces that secrete mucus are highly susceptible to pathogens and mucosal surfaces are known to contain large amounts of bacteria that could be pathogenic or symbionts [58, 59]. Mucus containing antimicrobial activity can be generated as a front line of defense. Microbial adhesion to mucus limits colonization of the pathogens, providing a defense mechanism to infection [60]. Planarians are normally covered by a thin layer of mucus that may act as a protective barrier and allows the presence of commensal microorganisms. Stressful situations, such as wounding, in S. mediterranea induces secretion of mucus that may have a double role in blocking pathogen entry and aiding in the healing process. Recent evidence suggests that planarian derived mucus proteins display similarities with mucosal secretion from humans [35]. However it is unclear whether wound repair and regeneration in planarians require components of the mucus to repair tissue.
Anti-microbial peptides are small peptides with clusters of positively charged and hydrophobic amino acids [61]. These small peptides are evolutionarily conserved and often secreted in mucus by phagocytic cells, and are known to have immunomodulatory functions [52]. For example, anti-microbial peptides are known to be secreted upon injury in insects [62]. Altincicek et al. screened for septic wounding inducible genes by introducing bacterial lipopolysaccharide into the wound site of planaria, and identified the induction of several potential anti-microbial peptides [63]. These anti-microbial homologs are likely involved in regulation of commensal bacterial and pathogen killing in planaria. Further a sequence suggested to be an antimicrobial peptide resistance and lipid A acylation protein, PagP, was found within the planarian genome (mk4.054786.01.01). How these anti-microbial peptides are activated during injury remains to be ascertained.
3.2. Pathogen recognition receptors
Initiation of innate immune responses relies on early signals following recognition of pathogen- or damage-associated molecular patterns (PAMPs and DAMPs) [64]. These early events involve recognition of molecules such as lipopolysaccharide, flagellin or dsRNA, found on or in invading pathogens or released by damaged and dying cells [64]. Two members of the PRR family, NOD-like receptors and toll-like receptors (TLRs) are conserved from early invertebrates to mammals and are maintained with a small number of protein structures [65].
TLRs are comprised of a cytoplasmic tail, a transmembrane and an extracellular domain that contains leucine rich repeat (LRR) motifs [66]. Based on chrystallographic studies LRRs are shaped in a 3D horseshoe format that contain the leucine rich repeats. This domain is important for interaction with other molecules, pathogen detection and provides structure [66, 67]. In mammals interactions through these LRRs trigger a conformation change to the TLR that results in a signaling cascade that activates proinflammatory cytokines and type 1 interferons [68]. LRRs are highly conserved across species, and their primary function is in the formation of protein-protein interactions [67]. Vertebrate and invertebrates have various numbers of TLRs each with specific functions, mostly targeted towards pathogen recognition [69–71].
Our analysis of the planarian genome identified several gene segments that contain LRRs (mk4.007365.00, mk4.002858.00, mk4.000112.15, mk4.026212.00, mk4.000148.12). Whether these sequence similarities have TLR-like activity or are unrelated repeats remains to be investigated. CD14, also involved with innate immune function contain multiple LRR repeats, and whether the LRR repeats found in planarians belongs to TLR or CD14 functionality is unknown. Recent transcriptomic analysis performed on a head regeneration time course found upregulation of LRR homolog mk4.007365.00 and mk4.002858.00 from one to 72 hours post amputation [30].
A planarian genome wide search for TIR, the cytoplasmic domain of TLR, identified four TIR-like domain sequences (mk4.008544.03, mk4.000285.01, mk4.000346.04, mk4.027932.00). One gene segment (mk4.008544.03) contains two SAM domains suggesting that this might be a SARM1 protein. SARM1 proteins negatively regulate innate immune function by blocking signaling downstream of the PRRs [72]. None of the TIR domain containing gene segments included any LRR sequences suggesting that these might not be TLRs, or that the pattern recognition motifs in the planaria differ from traditional LRR repeats. One TIR-like sequence in planaria (mk4.000285.01) is also upregulated for 6 hours post-amputation [30]. Upregulation of these TIR and LRR motifs may correspond with detection of, or response to, infection during injury, and warrants further attention.
Mammalian TLR family members share several signaling molecules including the adaptor protein MyD88, TRAF6 (TNF receptor-associated kinase 1), the protein kinase IRAK (IL-1R-associated kinase) and TAK1 (TGF-beta-activated kinase) binding protein. MyD88 protein also contains a TIR domain and a death domain. Though we found no significant homology for death domains near the four planarian TIR domains, we cannot eliminate the existence of a MyD88-like adaptor protein in planaria. TRAF proteins contain three domains: a MATH domain, RING-type zinc finger domains and TRAF-type zinc finger domains. Analysis of the planarian genome identified MATH homologs domains (mk4.000007.00 and mk4.000002). The gene segments between mk4.000002.00.01 and mk4.000002.33.01 contains both MATH and RING-type zinc finger domains and is upregulated throughout regeneration [30]. Region mk4.000007.00 also remain upregulated 1 – 72 hours post amputation. The gene segments between mk4.000002.00.01 and mk4.000002.33.01 contains both Math and RING-type zinc finger domains and is also upregulated throughout regeneration. We identified one possible TAK1 homolog (mk4.001525.00.01) that is relatively upregulated through the regeneration time course [30]. Possible homologs for IRAK also can be found within the genome, but with no significant up/down regulation during regeneration (mk4.004443.00, mk4.004443.01). IRAK proteins contain kinase domains and mk4.027874.00 a homologous kinase domain remained upregulated up to 6 hours post amputation. Based on these findings, it seems likely that planaria have some form of pathogen-recognition and signaling to distinguish between self-tissue and pathogenic microbes.
3.3. Complement cascade
The complement system is a highly conserved component of innate immunity. In mammals, complement functions to recognize and clear invading pathogens by inducing inflammatory responses, activating phagocytic cells and directly lysing microbes [73]. In mammals the complement system is made up of a cascading series of protein interactions that begin via three independent pathogen recognition pathways [73]. These three pathways converge on one protein C3 and continue through an identical cascade to form a membrane attack complex and amplify inflammatory signals. One arm of this system uses antibodies-coated pathogens in the recognition of pathogens and initiation of the complement cascade [73]. Although in mammals complement links the innate and adaptive immune responses, the complement system appears to be very ancient, with complement-like proteins found in most invertebrates and vertebrates [17, 74, 75]. We identified several complement factors and domain homologies within the S. mediterranea genome. The CUB domain (for complement C1r/C1s, Uegf, Bmp1) is found in the C1 component of the complement cascade. We have identified several homologs for the CUB domain (mk4.001431.05, mk4.001438.03, mk4.000798.07, mk4.007851.01, mk4.003730.02) and found mk4.000798.07 to be upregulated 6 hours post amputation [30]. mk4.001431.05 and mk4.003730.02 which are found adjacent to a Sushi domain (mk4.003591.01), another domain found in the C1 complement subcomponent, remained upregulated very early during regeneration. A C1q-like domain (mk4.008251.00), the C terminal domain of the C1 enzyme that triggers classical complement activation [76] was also upregulated post amputation up to 6 hours [30]. Several C3 complement homologs for the alpha-2-macroglobulin domain were also identified in planaria. As with other early invertebrates, planarians appear to have a conserved complement system that may act to control invading pathogens, but this requires confirmation and further study.
3.4. Phagocytic cells (reticulocytes)
Macrophages are known to play an important role in the resolution of acute inflammation by clearing apoptotic and necrotic tissue due to injury and immune responses [77]. Macrophages also ingest and breakdown invading pathogens and in this way act in host defense. In mammals macrophages differentiate down two distinct pathways dependent upon the local signals encountered. M1 classical macrophage activation results in macrophages that produce inflammatory cytokines and act in host defense to control microbial infections. Alternatively activated macrophages aid in wound repair by phagocytosing damaged tissue and secreting growth factors [78]. Macrophage function is essential during tissue repair in most vertebrate animals including mice [79], zebrafish [80], and salamanders [8]. However, wound healing in PU.1 mice (no neutrophils or macrophages) occurs without scarring, establishing alternative regulatory modes for phagocytic cells during regeneration.
In planarians, a phagocytic, mesenchymal cell, termed the reticular cell has been previously described [55–57]. Within ten hours of injury in the presence of bacteria, the reticular cell migrates into the wound, phagocytoses and encapsulate bacteria. Reticular cells thus have the capability to recognize foreign particles as distinct from self, migrate and phagocytose pathogens. Phagocytic engulfment of cellular debris has also been observed during regeneration after fissioning and after amputation [55–57]. We speculate the reticular cells in planarians may represent a primitive form of macrophage.
Although it is unconfirmed whether planarian phagocytic cells express PRRs, in other organisms, macrophages are one of the major cells types that express PRRs and respond to pathogen-associated molecular patterns [53, 81]. Macrophages produce many proteins that induce inflammation, migration of other cells into the site of infection or injury, killing of pathogens and down-modulation of these responses following clearance of the infection or damage [78]. Perforin is a lytic enzyme produced and secreted by mammalian macrophages that lyses invading pathogens and infected cells. A membrane attack complex component/perforin/C9 homolog was identified during sepsis-injury in planaria [63], suggesting a role for this enzyme in host defense of planaria. Alterations in the debris removal process or inflammatory responses might alter the final outcome of the repair, thus indicating a potential role for phagocytic cells in both host defense and wound repair and regeneration.
A recently identified group of molecules secreted by macrophages, maresins (macrophage mediators in resolving inflammation) enhance macrophage phagocytosis and limit local polymorphonuclear neutrophil infiltration [82]. When exposed to human maresin MaR1, the rate of planarian anterior tissue regeneration increased [83]. Further, planaria biosynthesized MaR1 upon injury, and the formation of MaR1 was blocked by a lipoxygenase (LOX) inhibitor, eliminating the enhancement in tissue regeneration. Together these data indicate that key planarian signaling components can respond to human MaR1, and thus conservation exists between human and planarian maresins and their signal cascades. These data further support a link between immune cell activities and regenerative mechanisms.
3.5. Cell adhesion molecules
Cell adhesion molecules are typically transmembrane proteins expressed on the cell surface. These proteins mediate cell-cell interactions including, signal transduction, cellular communication, and migration. In mammals, cell adhesion molecules have been divided into four families based on protein structure: Immunoglobulin (Ig) superfamily, cadherins, selectins and integrins [84, 85].
The Ig superfamily is calcium-independent and composed of variable numbers of Ig-like repeats. Analysis of the planarian genome revealed several Ig-like domains (mk4.006497.00) that are commonly found in adhesion molecules such as the ICAMs. The cadherin family, a calcium-dependent cell adhesion molecule, usually contains 3 to 5 internal repeats of cadherin and form homodimers. Cadherins play a crucial role in mediating innate and adaptive immune functions by maintaining epithelial barrier function and regulating leukocyte migration [86, 87]. Most integrins function as receptors for proteins within the extracellular matrix and can bind to a single ligand or to multiple ligands. In mammals, selectins are adhesion molecules with a trans-membrane glycoprotein that are expressed on leukocytes and activated epithelial cells [88]. The vertebrate selectin family consists of L-selectin, E-selectin and P-selectin and these are mostly involved with leukocyte homing and migratory responds to inflammation or pathogens. They mediate leukocyte rolling and adhesion during mammalian wound repair and leukocyte extravasation [89]. Selectins belong to the C-type lectin superfamily of proteins, are calcium-dependent and are widely found in metazoans [90]. Although C-type lectin domains have been linked to a large range of functions, pathogen recognition remains a primary function amongst most of this family [90].
Fusaoka et al. recently identified several vertebrate neural cell adhesion molecule orthologs in planarian Dugesia japonica that when knocked-down using RNAi altered neural network morphology [91]. Our analysis of the planarian genome revealed homologs for several Ig-like domains typically found in adhesion molecules, integrin-related sequences, C-type lectin domains and Cys-FGFR domains. Some of these domains were upregulated post-amputation and remained mostly elevated at 30 minutes to 72 hours [30]. C-type lectin domains are associated with immune responses, cell adhesion and cell death [90]. Studies have shown that invertebrate genomes have an abundance of C-type lectin-like domains [92]. C-type lectin receptors play a crucial role in pathogen recognition and could mediate functions associated with cell adhesion. We found several homologs of C-type lectin domains (mk4.020584.01, mk4.000042.08, mk4.000389.11) that were upregulated during regeneration in planaria [30]. Analysis of the planarian genome also identified homologs for 8, 11 and 15 cadherins and for E-Cadherin. Several of these homologs were upregulated during regeneration. However, the specific function these proteins during planaria wounding and regeneration or in host defense has not been evaluated.
3.6. Nitric oxide
Nitric oxide is utilized in many animal species as a cell signaling and cytotoxic molecule as it diffuses readily through fluids and across cell membranes [93]. It is synthesized from L-arginine by the enzyme nitric oxide synthase in response to calcium-dependent and independent signals. Apart from pathogen defense nitric oxide stimulates clearing of cellular debris and the activation, growth and death of immune cells [94, 95]. A homolog for nitric oxide synthase has been discovered in several flatworm species [96]. Adh3 an evolutionarily conserved gene that encodes the enzyme alcohol dehydrogenase, is another protein involved in nitric oxide metabolism that has been identified in planaria [97]. The existence of a nitric oxide synthase homolog and an alcohol dehydrogenase suggests another host defense mechanism by which planarians might control pathogens. Nitric oxide also has a documented role in mammalian wound repair. In rats fed with an arginine-free diet (arginine suppresses nitric oxide production) wound healing was impaired [98], and in patients nitric oxide levels are low in diabetic sores [95]. Nitric oxide may represent another evolutionarily conserved example with functions in both host defense and wound repair.
3.7. Double Strand RNA (dsRNA)
RNAi technology is widely used in planarian research. dsRNA and miRNA have been implicated biologically as important transcription regulators that mediate many signaling pathways during development, repair, regeneration and maintenance of homeostasis [99]. Existence of dsRNA creates red flags within eukaryotic cells, indicating viral infection [100, 101]. Eukaryotic cells have evolved with specific rapid and successful mechanisms to remove dsRNA as foreign material. Scientists have manipulated this knowledge to successfully downregulate genes and their functions from cells. Within an eukaryotic organism this RNAi silencing pathway can be used not only for the removal of the viral bi-products, but for changes in gene expression levels and subsequent signaling pathways that might play a crucial role in immune functions. Analysis of the planarian genome revealed homologs for DICER and AGO2 (mk4.000678.05 and mk4.001736.00), however these genes were not upregulated during regeneration in the planaria [30] implying this mode of immune regulation might not be important in wound repair and regeneration.
3.8. Metalloendopeptidases
Metalloendopeptidases are highly conserved across evolution and are ubiquitously expressed [102, 103]. Matrix metalloproteinases (MMPs) in mammals play a critical role in extracellular matrix remodeling and thus wound repair. There are 24 mammalian MMPs, some that may also have a role in immune responses, activating cytokines and chemokines to induce inflammation and immunity [104, 105]. Injury induces the expression of MMPs, and these perform multiple, distinct functions in wound repair, cell migration and bacterial inhibition and killing [106]. MMP-like proteins have been identified in a diverse array of invertebrates [107, 108]. Planarians have four MMP-like genes (Smed-mmp1, Smed-mmp2, Smed-mt-mmpA and Smed-mt-mmpB) with roles in proliferation, apoptosis and cell migration [109]. Two of these, Smed-mmp-1 and Dj-mmp-1, are essential for homeostatic remodeling of extracellular matrix, but not wound repair [109]. Although gene expression was unchanged following wounding, knock-down of Smed-mt-mmpA resulted in a delay in blastema regeneration [109]. Smed-mmp1 was also found to be expressed during septic-wounding in planaria, suggesting a role for this MMP in innate immune responses in addition to structural homeostasis [63]. A detailed analysis of MMP-like gene activity in planaria should be evaluated in response to injury and infection as these proteins have potential to be important in both biological activities.
4. Concluding Remarks
The presence of molecules associated with innate immune response in planarians along with their activation during regeneration implies that the injury-mediated immune response is an ancient feature of basal Bilateria. Specific regulatory mechanisms of the immune system during planarian regeneration remain to be addressed. In addition to the potential vertebrate immune homologs that we identified, it is unknown if other non-vertebrate immune mechanisms exist in planarians for fighting pathogens. It is possible that further analysis of the injury-induced immune response may reveal invertebrate-specific responses as part of their ancient immune defense mechanisms. Thus, the planarian model provides great opportunities to evaluate evolutionary mechanisms and studies of molecular players during tissue regeneration and host defense that are difficult to interrogate in other experimental models. Planarian studies mostly focus on the adult organism, which facilitate analysis of local and systemic signals in the presence of a fully developed immune system. RNAi, transcriptomic and proteomic approaches could be used to categorize components of the immune system modulating particular phases of wound healing, blastema formation and tissue remodeling (Figure 3). Critical aspects such as the role of commensal bacteria during tissue repair and the specific role of primitive phagocytic cells (reticulocytes) could be investigated in the context of the whole organism. The absence of an adaptive immune system in planarians presents some limitations within the model for comparison to human mechanisms. However, comparative studies between planarians and salamanders evaluating scar-free repair and regeneration [9] may point toward common factors within the innate immune system with potential clinical applications.
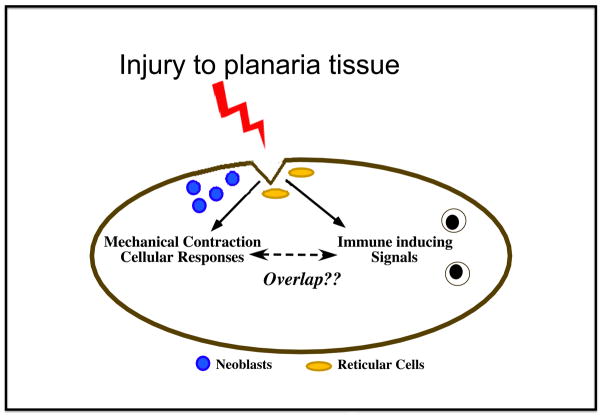
Figure 3. Injury induces immune and repair signaling in planarians.
Following injury or amputation, molecular signals induce proliferation of neoblasts and formation of the blastema. Reticular cells migrate into the injury site and can be found with phagocytosed dead tissue. Sepsis-injury induces expression of potential host defense molecules [63]. As immune-inducing and regeneration signaling events likely occur simultaneously, it is possible that some of these signals impact both biological pathways. In addition, immune components alter the rate of regeneration and potentially impact the structural outcome of the tissue reconstruction.
Evaluation of healing in autoimmune patients suggests that a dysregulated inflammatory response delays or impairs the wound repair process [110]. It thus appears that a proper balance of immune response must be maintained during wound repair. The data seem to suggest that too strong of an immune response (or perhaps a more developed immune system) may contribute to scaring during wound repair and reduced regenerative capacity, while a lower level of inflammation (or more primitive immune signaling) may be critical in ensuring that the appropriate cell types migrate into the damaged environment and contribute to the clean-up of localized debris and dead cellular tissue (Figure 3) [6, 7, 9, 11]. This information could be readily addressed by analyzing planarian regeneration and their enigmatic immune system.
Acknowledgments
We thank Daniel Ramirez for comments on the manuscript. KKH is supported by the National Institutes of Health grant R00HL090706. Research in the Oviedo Lab is supported by the University of California, Merced; the Health Sciences Research Institute at UC Merced; Jane Vilas faculty award; Hellman Fellows Fund, and the NIH-National Cancer Institute grant number R21CA176114 to NJO.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elliott SA, Sanchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:301–326. doi: 10.1002/wdev.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galliot B. Hydra, a fruitful model system for 270 years. The International journal of developmental biology. 2012;56:411–423. doi: 10.1387/ijdb.120086bg. [DOI] [PubMed] [Google Scholar]
- 3.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King RS, Newmark PA. The cell biology of regeneration. J Cell Biol. 2012;196:553–562. doi: 10.1083/jcb.201105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin M. Large-scale biophysics: ion flows and regeneration. Trends in cell biology. 2007;17:261–270. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Eming SA, Hammerschmidt M, Krieg T, Roers A. Interrelation of immunity and tissue repair or regeneration. Semin Cell Dev Biol. 2009;20:517–527. doi: 10.1016/j.semcdb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Godwin JW, Brockes JP. Regeneration, tissue injury and the immune response. J Anat. 2006;209:423–432. doi: 10.1111/j.1469-7580.2006.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godwin JW, Rosenthal N. Scar-free wound healing and regeneration in amphibians: immunological influences on regenerative success. Differentiation. 2014;87:66–75. doi: 10.1016/j.diff.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Murawala P, Tanaka EM, Currie JD. Regeneration: the ultimate example of wound healing. Semin Cell Dev Biol. 2012;23:954–962. doi: 10.1016/j.semcdb.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. The Journal of investigative dermatology. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- 12.Hemmrich G, Miller DJ, Bosch TC. The evolution of immunity: a low-life perspective. Trends Immunol. 2007;28:449–454. doi: 10.1016/j.it.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez Alvarado A. Regeneration in the metazoans: why does it happen? Bioessays. 2000;22:578–590. doi: 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 15.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 16.Nonaka M. Evolution of the complement system. Subcell Biochem. 2014;80:31–43. doi: 10.1007/978-94-017-8881-6_3. [DOI] [PubMed] [Google Scholar]
- 17.Nonaka M, Kimura A. Genomic view of the evolution of the complement system. Immunogenetics. 2006;58:701–713. doi: 10.1007/s00251-006-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Developmental cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch TC. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 20.Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Riutort M, Alvarez-Presas M, Lazaro E, Sola E, Paps J. Evolutionary history of the Tricladida and the Platyhelminthes: an up-to-date phylogenetic and systematic account. The International journal of developmental biology. 2012;56:5–17. doi: 10.1387/ijdb.113441mr. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez Alvarado A. The freshwater planarian Schmidtea mediterranea: embryogenesis, stem cells and regeneration. Curr Opin Genet Dev. 2003;13:438–444. doi: 10.1016/s0959-437x(03)00082-0. [DOI] [PubMed] [Google Scholar]
- 23.Reddien PW. Specialized progenitors and regeneration. Development. 2013;140:951–957. doi: 10.1242/dev.080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baguna J. The planarian neoblast: the rambling history of its origin and some current black boxes. The International journal of developmental biology. 2012;56:19–37. doi: 10.1387/ijdb.113463jb. [DOI] [PubMed] [Google Scholar]
- 25.Aboobaker AA. Planarian stem cells: a simple paradigm for regeneration. Trends in cell biology. 2011;21:304–311. doi: 10.1016/j.tcb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Lobo D, Beane WS, Levin M. Modeling planarian regeneration: a primer for reverse-engineering the worm. PLoS Comput Biol. 2012;8:e1002481. doi: 10.1371/journal.pcbi.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentile L, Cebria F, Bartscherer K. The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Dis Model Mech. 2011;4:12–19. doi: 10.1242/dmm.006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rink JC. Stem cell systems and regeneration in planaria. Dev Genes Evol. 2013;223:67–84. doi: 10.1007/s00427-012-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robb SM, Ross E, Sanchez Alvarado A. SmedGD: the Schmidtea mediterranea genome database. Nucleic acids research. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandmann T, Vogg MC, Owlarn S, Boutros M, Bartscherer K. The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol. 2011;12:R76. doi: 10.1186/gb-2011-12-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shomrat T, Levin M. An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. The Journal of experimental biology. 2013;216:3799–3810. doi: 10.1242/jeb.087809. [DOI] [PubMed] [Google Scholar]
- 32.Adamidi C, Wang Y, Gruen D, Mastrobuoni G, You X, Tolle D, et al. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome research. 2011;21:1193–1200. doi: 10.1101/gr.113779.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labbe RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DD, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onal P, Grun D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. The EMBO journal. 2012;31:2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bocchinfuso DG, Taylor P, Ross E, Ignatchenko A, Ignatchenko V, Kislinger T, et al. Proteomic profiling of the planarian Schmidtea mediterranea and its mucous reveals similarities with human secretions and those predicted for parasitic flatworms. Mol Cell Proteomics. 2012;11:681–691. doi: 10.1074/mcp.M112.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boser A, Drexler HC, Reuter H, Schmitz H, Wu G, Scholer HR, et al. SILAC proteomics of planarians identifies Ncoa5 as a conserved component of pluripotent stem cells. Cell reports. 2013;5:1142–1155. doi: 10.1016/j.celrep.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 37.Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Developmental cell. 2005;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beane WS, Morokuma J, Adams DS, Levin M. A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chemistry & biology. 2011;18:77–89. doi: 10.1016/j.chembiol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beane WS, Morokuma J, Lemire JM, Levin M. Bioelectric signaling regulates head and organ size during planarian regeneration. Development. 2013;140:313–322. doi: 10.1242/dev.086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sanchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Developmental biology. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Developmental biology. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends in genetics : TIG. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins JJ, 3rd, Newmark PA. It’s no fluke: the planarian as a model for understanding schistosomes. PLoS pathogens. 2013;9:e1003396. doi: 10.1371/journal.ppat.1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chuah C, Jones MK, Burke ML, McManus DP, Gobert GN. Cellular and chemokine-mediated regulation in schistosome-induced hepatic pathology. Trends in parasitology. 2014;30:141–150. doi: 10.1016/j.pt.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez JL, Leung G, McKay DM. Cestode regulation of inflammation and inflammatory diseases. International journal for parasitology. 2013;43:233–243. doi: 10.1016/j.ijpara.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14:181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 48.Ankeny DP, Popovich PG. Mechanisms and implications of adaptive immune responses after traumatic spinal cord injury. Neuroscience. 2009;158:1112–1121. doi: 10.1016/j.neuroscience.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hauton C, Smith VJ. Adaptive immunity in invertebrates: a straw house without a mechanistic foundation. Bioessays. 2007;29:1138–1146. doi: 10.1002/bies.20650. [DOI] [PubMed] [Google Scholar]
- 50.Chalmers IW, Hoffmann KF. Platyhelminth Venom Allergen-Like (VAL) proteins: revealing structural diversity, class-specific features and biological associations across the phylum. Parasitology. 2012;139:1231–1245. doi: 10.1017/S0031182012000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez RM, Lopez-Vazquez A, Lopez-Larrea C. Immune systems evolution. Adv Exp Med Biol. 2012;739:237–251. doi: 10.1007/978-1-4614-1704-0_15. [DOI] [PubMed] [Google Scholar]
- 52.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 53.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annual review of immunology. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 54.Oviedo NJ, Nicolas CL, Adams DS, Levin M. Establishing and maintaining a colony of planarians. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5053. pdb prot5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morita M, Best JB. Electron microscopic studies of planarian regeneration. II. Changes in epidermis during regeneration. The Journal of experimental zoology. 1974;187:345–373. doi: 10.1002/jez.1401870305. [DOI] [PubMed] [Google Scholar]
- 56.Morita M. Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia. 1991;227:193–199. [Google Scholar]
- 57.Morita M. Structure and function of the reticular cell in the planarian Dugesia dorotocephala. In: Cannon L, editor. Biology of turbellaria and some related flatworms. Abo/Turku, Finland: Springer; 1995. pp. 189–196. [Google Scholar]
- 58.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 59.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 62.Zasloff M. Antibiotic peptides as mediators of innate immunity. Curr Opin Immunol. 1992;4:3–7. doi: 10.1016/0952-7915(92)90115-u. [DOI] [PubMed] [Google Scholar]
- 63.Altincicek B, Vilcinskas A. Comparative analysis of septic injury-inducible genes in phylogenetically distant model organisms of regeneration and stem cell research, the planarian Schmidtea mediterranea and the cnidarian Hydra vulgaris. Front Zool. 2008;5:6. doi: 10.1186/1742-9994-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Q, Zmasek CM, Godzik A. Domain architecture evolution of pattern-recognition receptors. Immunogenetics. 2010;62:263–272. doi: 10.1007/s00251-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 68.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alcaide M, Edwards SV. Molecular evolution of the toll-like receptor multigene family in birds. Mol Biol Evol. 2011;28:1703–1715. doi: 10.1093/molbev/msq351. [DOI] [PubMed] [Google Scholar]
- 70.Song X, Jin P, Qin S, Chen L, Ma F. The evolution and origin of animal Toll-like receptor signaling pathway revealed by network-level molecular evolutionary analyses. PLoS One. 2012;7:e51657. doi: 10.1371/journal.pone.0051657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satake H, Sekiguchi T. Toll-like receptors of deuterostome invertebrates. Front Immunol. 2012;3:34. doi: 10.3389/fimmu.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 73.Carroll MC. Complement and humoral immunity. Vaccine. 2008;26 (Suppl 8):I28–33. doi: 10.1016/j.vaccine.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nonaka M, Yoshizaki F. Evolution of the complement system. Mol Immunol. 2004;40:897–902. doi: 10.1016/j.molimm.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 75.Pinto MR, Melillo D, Giacomelli S, Sfyroera G, Lambris JD. Ancient origin of the complement system: emerging invertebrate models. Adv Exp Med Biol. 2007;598:372–388. doi: 10.1007/978-0-387-71767-8_26. [DOI] [PubMed] [Google Scholar]
- 76.Kishore U, Reid KB. C1q: structure, function, and receptors. Immunopharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 77.Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 78.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goren I, Allmann N, Yogev N, Schurmann C, Linke A, Holdener M, et al. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am J Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem. 2012;287:25353–25360. doi: 10.1074/jbc.M112.349126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ulvila J, Vanha-Aho LM, Ramet M. Drosophila phagocytosis - still many unknowns under the surface. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2011;119:651–662. doi: 10.1111/j.1600-0463.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- 82.Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, Porter TF, et al. Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. The Journal of experimental medicine. 2009;206:15–23. doi: 10.1084/jem.20081880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serhan CN, Dalli J, Karamnov S, Choi A, Park CK, Xu ZZ, et al. Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:1755–1765. doi: 10.1096/fj.11-201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez-Amaro R, Sanchez-Madrid F. Cell adhesion molecules: selectins and integrins. Crit Rev Immunol. 1999;19:389–429. [PubMed] [Google Scholar]
- 85.Petruzzelli L, Takami M, Humes HD. Structure and function of cell adhesion molecules. Am J Med. 1999;106:467–476. doi: 10.1016/s0002-9343(99)00058-3. [DOI] [PubMed] [Google Scholar]
- 86.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119:1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 87.Tian X, Liu Z, Niu B, Zhang J, Tan TK, Lee SR, et al. E-cadherin/beta-catenin complex and the epithelial barrier. J Biomed Biotechnol. 2011;2011:567305. doi: 10.1155/2011/567305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ley K. The role of selectins in inflammation and disease. Trends Mol Med. 2003;9:263–268. doi: 10.1016/s1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 89.Prozialeck WC, Edwards JR. Cell adhesion molecules in chemically-induced renal injury. Pharmacol Ther. 2007;114:74–93. doi: 10.1016/j.pharmthera.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cambi A, Figdor CG. Dual function of C-type lectin-like receptors in the immune system. Curr Opin Cell Biol. 2003;15:539–546. doi: 10.1016/j.ceb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 91.Fusaoka E, Inoue T, Mineta K, Agata K, Takeuchi K. Structure and function of primitive immunoglobulin superfamily neural cell adhesion molecules: a lesson from studies on planarian. Genes Cells. 2006;11:541–555. doi: 10.1111/j.1365-2443.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- 92.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 93.Wink DA, Hanbauer I, Grisham MB, Laval F, Nims RW, Laval J, et al. Chemical biology of nitric oxide: regulation and protective and toxic mechanisms. Curr Top Cell Regul. 1996;34:159–187. doi: 10.1016/s0070-2137(96)80006-9. [DOI] [PubMed] [Google Scholar]
- 94.Schwentker A, Billiar TR. Nitric oxide and wound repair. Surg Clin North Am. 2003;83:521–530. doi: 10.1016/S0039-6109(02)00207-4. [DOI] [PubMed] [Google Scholar]
- 95.Witte MB, Barbul A. Role of nitric oxide in wound repair. Am J Surg. 2002;183:406–412. doi: 10.1016/s0002-9610(02)00815-2. [DOI] [PubMed] [Google Scholar]
- 96.Margaretha KS, Gustafsson AM, Lindholm KM, Reuter Maria, Christel A. Lundström, Terenina N. No news on the flatworm front! Nitric oxide synthase in parasitic and free-living flatworms. Hydrobiologia. 1998;383:161–166. [Google Scholar]
- 97.Godoy L, Gonzalez-Duarte R, Albalat R. Analysis of planarian Adh3 supports an intron-rich architecture and tissue-specific expression for the urbilaterian ancestral form. Comp Biochem Physiol B Biochem Mol Biol. 2007;146:489–495. doi: 10.1016/j.cbpb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 98.Seifter E, Rettura G, Barbul A, Levenson SM. Arginine: an essential amino acid for injured rats. Surgery. 1978;84:224–230. [PubMed] [Google Scholar]
- 99.Cullen BR. Derivation and function of small interfering RNAs and microRNAs. Virus research. 2004;102:3–9. doi: 10.1016/j.virusres.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 100.Lindenbach BD, Rice CM. RNAi targeting an animal virus: news from the front. Molecular cell. 2002;9:925–927. doi: 10.1016/s1097-2765(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 101.Jacobs BL, Langland JO. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 102.Massova I, Kotra LP, Fridman R, Mobashery S. Matrix metalloproteinases: structures, evolution, and diversification. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1998;12:1075–1095. [PubMed] [Google Scholar]
- 103.Fanjul-Fernandez M, Folgueras AR, Cabrera S, Lopez-Otin C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim Biophys Acta. 2010;1803:3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 104.Nissinen L, Kahari VM. Matrix metalloproteinases in inflammation. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagen.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 105.Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol. 2013;13:649–665. doi: 10.1038/nri3499. [DOI] [PubMed] [Google Scholar]
- 106.Parks WC. Matrix metalloproteinases in repair. Wound Repair Regen. 1999;7:423–432. doi: 10.1046/j.1524-475x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 107.Sarras MP, Jr, Yan L, Leontovich A, Zhang JS. Structure, expression, and developmental function of early divergent forms of metalloproteinases in hydra. Cell Res. 2002;12:163–176. doi: 10.1038/sj.cr.7290123. [DOI] [PubMed] [Google Scholar]
- 108.Quinones JL, Rosa R, Ruiz DL, Garcia-Arraras JE. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Developmental biology. 2002;250:181–197. doi: 10.1006/dbio.2002.0778. [DOI] [PubMed] [Google Scholar]
- 109.Isolani ME, Abril JF, Salo E, Deri P, Bianucci AM, Batistoni R. Planarians as a model to assess in vivo the role of matrix metalloproteinase genes during homeostasis and regeneration. PLoS One. 2013;8:e55649. doi: 10.1371/journal.pone.0055649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loots MA, Lamme EN, Zeegelaar J, Mekkes JR, Bos JD, Middelkoop E. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. The Journal of investigative dermatology. 1998;111:850–857. doi: 10.1046/j.1523-1747.1998.00381.x. [DOI] [PubMed] [Google Scholar]