Abstract
The solution dependence of gas-phase unfolding for ubiquitin [M+7H]7+ ions has been studied by ion mobility spectrometry-mass spectrometry (IMS-MS). Different acidic water:methanol solutions are used to favor the native (N), more helical (A), or unfolded (U) solution states of ubiquitin. Unfolding of gas-phase ubiquitin ions is achieved by collisional heating and newly formed structures are examined by IMS. With an activation voltage of 100 V, a selected distribution of compact structures unfolds, forming three resolvable elongated states (E1-E3). The relative populations of these elongated structures depend strongly on the solution composition. Activation of compact ions from aqueous solutions known to favor N-state ubiquitin produces mostly the E1 type elongated state, whereas, activation of compact ions from methanol containing solutions that populate A-state ubiquitin favors the E3 elongated state. Presumably, this difference arises because of differences in precursor ion structures emerging from solution. Thus, it appears that information about solution populations can be retained after ionization, selection, and activation to produce the elongated states. These data as well as others are discussed.
Introduction
The mechanism through which amino acid chains fold into their native three-dimensional protein structures has been studied extensively; however, a detailed understanding is still lacking.1–4 One barrier to a more complete understanding is that protein intermediates along folding pathways often exist transiently, and at only relatively low populations.2,5, 6 Despite considerable developments in experimental techniques such as nuclear magnetic resonance (NMR) spectrometry and single-molecule Förster resonance energy transfer, detecting and characterizing protein folding intermediates in solution remains challenging.7–10 In addition to those traditional solution-based analytical methods, mass spectrometry (MS) has been used to provide insight about solvent-free protein structures, dynamics, and folding. 11 – 16 Hybrid techniques such as hydrogen/deuterium exchange,11 electron capture dissociation (ECD),12,13 fast photochemical oxidation of proteins (FPOP)14 and ion mobility spectrometry (IMS)15,16 use gas-phase protein ions as a means of elucidating how intramolecular interactions influence protein conformations and folding.
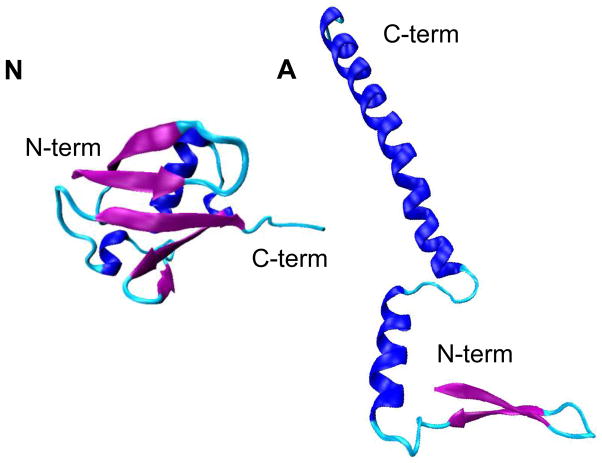
In this paper, we use hybrid IMS-MS techniques to investigate the conformations of collisionally activated ubiquitin [M+7H]7+ ions that are produced by electrospraying four acidic water:methanol solutions (100:0, 70:30, 40:60, and 10:90, pH = 2). In aqueous solutions, across a range of pH values of 1.2 to 8.4, ubiquitin favors the native (N) state.17 A more helical state of ubiquitin, the A-state, is stabilized in the 40:60 water:methanol solution at pH 2.18,19 Figure 1 illustrates the structure of the N-state ubiquitin that was characterized by X-ray crystallography20 as well as a structural model for the A state proposed previously based on NMR analyses.21 Similar to the structure of the N state, the N-terminal portion of the A state persists as the native-like β-sheet structure; however, the C-terminal side of the protein is often depicted as more elongated and studies with circular dichroism and NMR spectroscopies show that it is high in α-helical propensity.18,19,21–26 It is noted that the static picture of the A state shown in Figure 1 is oversimplified, as the A state is highly conformational flexible and is involved in dynamic equilibrium in solution.21
Figure 1.
Structures of the native (N) and A states of ubiquitin. (N) shows the native structure characterized by X-ray crystallography in ref. 17. (A) illustrates the structural model of the A state constructed based on the structure proposed in ref. 18.
In the work presented below, we probe the effect of solution structures on gas-phase unfolding for ubiquitin. The IMS approach records the time that ions take to travel through a drift tube containing a gas, and the ion’s mobility can yield insight about the overall shapes of the proteins.15,16,27 –31 Several studies have shown evidence that gas-phase protein conformations retain some aspects of their solution structures.32–37 In general, ions of low charge states show conformations of similar size to their solution states. In the case of ubiquitin, the collision cross section (1010 Å2) 38 of the compact conformation of [M+7H]7+ ions is slightly smaller than the 1090 Å2 value39 calculated from coordinates of the crystal structure by the rigorous trajectory method.28 For ions of higher charge states, the structures become more extended to reduce the repulsive Coulombic interactions. 40, 41 Based on cross section comparisons, those partially folded and elongated conformers are often significantly different from the solution structures. Despite variances in conformations between the gas phase ion and the solution state, solution structures can be correlated to their counterparts in the gas phase.39,42,43 Different solution structures can generate dissimilar gas-phase geometries.
Our group has carried out a number of studies involving ubiquitin ions.38–40,43–51 One category of those studies investigates gas-phase structural transitions of ubiquitin from compact to partially folded and unfolded structures as obtained by a variety of methods including heating ions as they are injected into the drift tube,40 storing ions inside an ion trap,46 as well as collisionally activating ions in a high electric field region in the middle of a drift tube.48,49 Another series of studies involves changing the solution conditions to favor different solution states of ubiquitin to explore the correlations between the gas-phase IMS distributions and populations of different solution structures.39,43,44 The present work is discussed in light of these prior results.
Below, we first present an analysis that compares the gas-phase unfolding of ubiquitin [M+7H]7+ ions generated from different water:methanol solutions. In all cases, the collision cross section distributions of [M+7H]7+ ions are dominated by compact conformations under gentle electrospray ionization (ESI) conditions, which makes the analysis less complicated without interference from initial ESI-formed partially folded or elongated states. Distributions for partially folded and elongated structures are reported, which are formed by collisional activation of the compact conformations of ubiquitin [M+7H]7+ ions with helium. This work indicates that the collisional activation process is influenced by antecedent structures emerging from solution. That is, the distribution of elongated conformers formed after ESI, ion mobility separation, selection, and activation still retains information about the solution populations.
Experimental
Instrumentation
Details of IMS theory,28–30, 52 – 54 experiments,15,16,27 and instrumentation 55, 56 have been described elsewhere. Here, only a brief summary of the experimental approach utilized in this work is presented. The experiments were carried out on two similar home-built ion mobility spectrometers coupled to time-of-flight (TOF) mass spectrometers. The main difference between the two instruments is the total length of the drift tube. Detailed descriptions of the instruments utilized here have been published previously.55,56 Figure 2 shows a schematic diagram of the IMS instrument which employs a drift tube of ~289 cm in length. A continuous beam of ubiquitin ions is generated by ESI and accumulated in the source ion funnel (F1). Ions are periodically pulsed into the drift tube via an electrostatic gate (G1). The drift tube is filled with ~3 Torr He buffer gas at 300 K and operated with a uniform electric field of ~10 V·cm−1. As ions move through the buffer gas, different structures are separated based on variances in their mobilities. Three ion funnels (F2-F4) that are placed along the drift tube radially focus the diffuse ion packet to improve the ion transmission. Upon exiting the drift tube, ions are extracted into a differentially pumped region and pulsed into a TOF mass spectrometer for mass-to-charge (m/z) measurements. The data were recorded in a nested fashion as described previously.57
Figure 2.
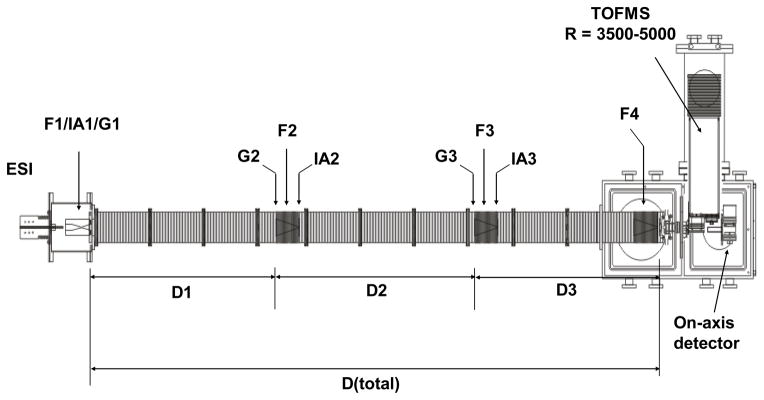
Schematic diagram of the IMS-IMS-MS instrument used in this work. The drift tube region incorporates ion funnels (F1-F4), ion gates (G1-G3), ion drift regions (D1-D3) and ion activation regions (IA1-IA3). More details on ion selection and activation are in text.
The drift tube segments that are separated by ion funnels can be utilized as a single drift region for one dimensional IMS analyses (IMS-MS) or operated independently for multidimensional IMS measurements. For IMS-MS measurements, G2 and G3 transmit all ions into the subsequent drift regions, such that the entire structural distribution can be examined. This work also utilized two dimensional IMS measurements (IMS-IMS-MS), where ions of a specified mobility can be selected at G2, activated at IA2, and the newly formed structures are separated in D2 and D3. In IMS-IMS experiments, a repulsive potential (~16 V) is applied to G2 and pulsed down to the common drift potential for a brief period (80 μs) at specified delay times with respect to the source pulse at G1, so that only ions of specific mobilities can pass through G2. The pulse for G2 is triggered by the source pulse at G1 with a pulse-delay generator. Those mobility-selected ions traverse F2 and then enter the activation region IA2, where ions can be accelerated by increasing the electric field between the two lenses (spaced ~0.3 cm apart) comprising IA2. Ions are activated by energetic collisions with buffer gas to produce a distribution of product states. The extent of activation can be controlled by varying the voltage drop between the two lenses in the activation region. In this work, a voltage of 5 V was used for passive transmission of ions and an activation voltage of 100 V (electric field of ~333 V·cm−1) was applied to induce structural transitions of ubiquitin ions. Upon entering the second drift tube region, ions are rapidly equilibrated to the temperature of the buffer gas and subsequently separated in D2 and D3 regions (~195 cm in length). The IMS-IMS-MS experiments were conducted on the instrument depicted in Figure 2.
Ion activation can also be achieved by applying a voltage (10 to 140 V) between the gate lens at G1 and the first lens of D1 which are about ~0.19 cm apart. The gate lens is constructed with a Ni mesh grid. These experiments were conducted on a similar instrument as that shown in Figure 2, but employed a shorter drift tube of ~182 cm in total length. The collisionally activated ions were separated in the entire drift region.
Generation of collision cross section distributions
It is useful to plot IMS distributions on a collision cross section scale. Because the charge state is known, this is straight forward. Experimental drift times (tD) are converted to collision cross sections (Ω) by using the following equation:54
| (1) |
where kb is Boltzmann’s constant, T is the temperature, and ze corresponds to the charge of the ion. mI and mB refer to the masses of the ion and buffer gas (He in this work), respectively. L and E are the drift length and electric field strength, respectively. P is the buffer gas pressure and N is the number density of the buffer gas at STP conditions. The current instrument contains ion funnels, which are operated at a disparate electric field from the drift regions. Thus, cross sections can be calculated using the selection time that ions require to travel from G1 to G2, where a highly uniform electric field is employed. This selection time is equivalent to the delay time of the pulse at G2 with respect to the G1 pulse in mobility selection experiments. A correlation curve of the selection time and the total drift time can be obtained by recording total drift times for various mobility-selected structures. The cross sections of new structures formed upon activation at IA2 are obtained by calibrating the drift time associated with D2+D3 to the selection time.
Sample preparation
Lyophilized ubiquitin from bovine erythrocytes (≥98%, Sigma-Aldrich Co., St. Louis, MO) was dissolved in water:methanol:formic acid solutions with a final concentration of ~1mg·mL−1. Water and methanol were mixed to the desired volume ratio (i.e., 100:0, 70:30, 40:60 and 10:90) and then formic acid was added to adjust the pH to 2.0 (direct measurement). The protein solution was electrosprayed directly into the IMS-MS instrument.
Results and Discussion
Selection and activation of ions
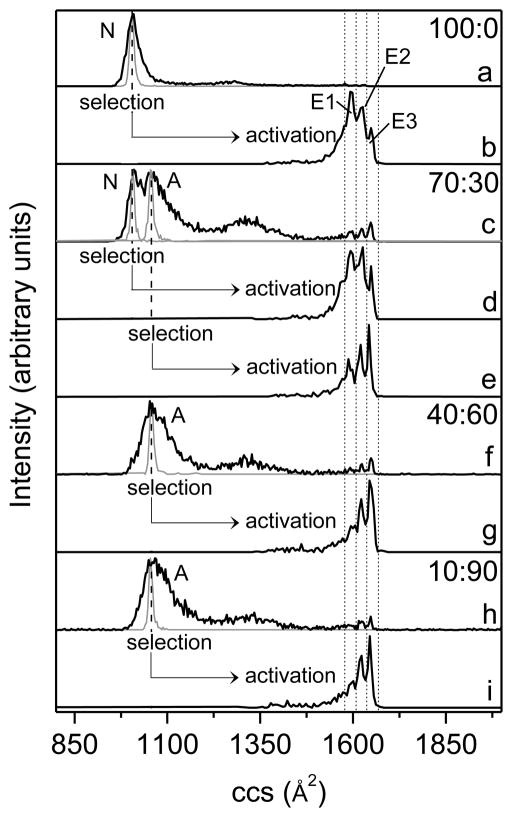
Figure 3 shows cross section distributions that were recorded under ion selection and activation for ubiquitin [M+7H]7+ ions electrosprayed from four water:methanol solutions (i.e., 100:0, 70:30, 40:60 and 90:10, pH = 2). Compared to previous studies,38,43 the present work uses gentler source conditions (lower source capillary voltage as well as RF frequency and voltage in F1) and the resulting distributions show a relatively low population of partially folded conformations. As shown in Figure 3, all solution conditions result in the formation of mainly compact structures. Consistent with the previous work,38,43 ubiquitin [M+7H]7+ ions generate a distribution centered at ~1010 Å2 for the 100:0 water:methanol solution which favors the N-state ubiquitin; the distributions shift to ~1060 Å2 for solutions with high methanol content (40:60 and 10:90 water:methanol) in which the A- and U-state ubiquitin are populated.18,21,58 The distribution generated from the 70:30 water:methanol solution shows two peaks that are partially resolved with cross sections of ~1010 Å2 and ~1060 Å2, indicating that ubiquitin exists as a mixture of N and A states in the solution.43 The theoretical cross sections calculated from coordinates for the crystal structure of N-state ubiquitin and a model Astate are 1090 Å2 and 1640 Å2, respectively.39 It appears that the N-state ubiquitin evolves into a slightly more compact structure (1010 Å2) during the ESI process. On the other hand, some of the extended A state appears to form a population of compact structures for [M+7H]7+ ions upon desolvation, which have cross sections around 1060 Å2 and are approximately 5% larger than that for ions formed from the N state (1010 Å2). In addition to compact conformers, a small fraction of ions are also observed as more extended structures (a broad feature from ~1210 to ~1470 Å2 and three peaks at 1600 Å2, 1625 Å2, and 1650 Å2, corresponding to the E1, E2, and E3 elongated states, respectively).
Figure 3.
Collision cross section (ccs) profiles for ubiquitin [M+7H]7+ ions obtained at different experimental conditions. (a), (c), (f), and (h) show the initial distributions of ions formed by electrospraying different water:methanol solutions (black lines) and the distributions for mobility-selected ions that are isolated at G2 (grey lines). Solution compositions (water:methanol) are provided as labels. Distributions (b) and (d) are obtained by collisionally activating (100 V, at IA2) the selected ions having cross sections around 1010 Å2, and distributions (e), (g), and (i) correspond to the activated ions with initial cross sections around 1060 Å2. The activated distributions are plotted underneath the corresponding selected distributions indicated by arrow-headed lines. Short dashed lines designate the integration regions for determining populations of elongated states E1-E3. The solution states of ubiquitin (N and A) from which ions are generated and the elongated states (E1-E3) obtained upon collisional heating have been labeled.
To investigate structural transitions for gas-phase ubiquitin ions produced from solutions that favor the N and A states, ions with a specified mobility are selected, collisionally heated, and cooled to render a new distribution of product states. A narrow population of structures at the center of the total distribution are selected for solutions of 100:0, 40:60 and 10:90 water:methanol. For the 70:30 water:methanol solution which generates a distribution of two partially resolved peaks, structures at each peak center have been selected. As shown in Figure 3, the distributions for selected ions remain narrow during separation in the subsequent drift region (D2+D3, Figure 2) for all solution conditions. The sharp feature for selected ubiquitin [M+7H]7+ ions has been reported previously.47 The data presented here provide evidence that the selected ions arising from solutions that favor either the N or A states have stable gas-phase conformations and do not undergo structural transitions during the time scale of the experiment.
It is possible to induce structural transitions with higher activation voltages. To probe the influence of solution compositions on produced extended states, the selected ubiquitin [M+7H]7+ ions from different solutions have been activated at IA2 (Figure 2). At an activation voltage of 100 V, ions are heated and the initial compact structures are primarily converted to elongated conformations. Structural distributions for the obtained unfolded ions are measured in the subsequent segments of the drift tube, as displayed in Figure 3. For all solution conditions, the compact ions produce three resolvable elongated states E1, E2, and E3 with cross sections of 1600 Å2, 1625 Å2, and 1650 Å2, respectively. Those three elongated states had been observed previously.48 It is notable that the relative abundances for the three extended structures vary between solutions. Conformer E1 with the cross section of 1600 Å2 is the most abundant species for ions formed from the 100:0 water:methanol solution, while conformer E3 is of the lowest intensity. Conversely, ions produced from the denaturing solutions (i.e., 40:60 and 10:90 water:methanol) unfold and form a distribution that is populated by E2 and E3 with E1 being the least abundant conformer. For the 70:30 water:methanol solution, activation of the selected ions with cross sections around 1010 Å2 forms E1 and E2 with similar peak intensities as well as a smaller amount of E3; ions that are selected at 1060 Å2 produce a distribution showing an increase in peak height from E1 to E3 upon activation.
Populations of elongated states produced upon activating compact ions from different solutions
The relative abundances for conformers E1-E3 under different experimental conditions have been estimated by integrating corresponding peak areas and listed in Table 1. Dashed lines drawn in Figure 3 designate the integration regions. Table 1 reveals that the relative abundances for E1-E3 from the 40:60 water:methanol solution are similar to those from the 10:90 solution, suggesting that mobility-selected ions at 1060 Å2 formed by ESI out of both denaturing solutions have similar conformational motifs. In contrast, ions that are selected at 1010 Å2 from the native solution of 100:0 water:methanol unfold to E1-E3 with different intensity profiles compared to ions at 1060 Å2 from denaturing solutions. The results indicate that the product distribution of activated conformers depends strongly on the solution condition.
Table 1.
Relative intensities for E1-E3 under different water:methanol solution conditions.
| 100:0 (N) a | 40:60 (A) a | 10:90 (A) | 70:30 (N) | 70:30 (A) | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Meas.b | Meas. | Meas. | Meas. | Cal.c | Meas. | Cal. | |
| E1 | 0.46 | 0.23 | 0.22 | 0.42 | 0.42 | 0.29 | 0.28 |
| E2 | 0.37 | 0.37 | 0.39 | 0.39 | 0.37 | 0.37 | 0.38 |
| E3 | 0.17 | 0.41 | 0.39 | 0.19 | 0.21 | 0.35 | 0.34 |
N and A correspond to ions selected at 1010 Å2 and 1060 Å2, respectively.
Measured relative intensities from the obtained cross section distributions upon activation.
Calculated relative intensities from fractions of native- and A-state gas conformers for the selected ions.
For the 70:30 water:methanol solution, it is interesting to note that the selected ions at 1010 Å2 produce E1-E3 with different intensities compared to those selected at the same mobility from the 100:0 water:methanol solution; in addition, the relative abundances for E1-E3 that are generated from precursor ions at 1060 Å2 are also dissimilar to those from the 40:60 and 10:90 water:methanol solutions.
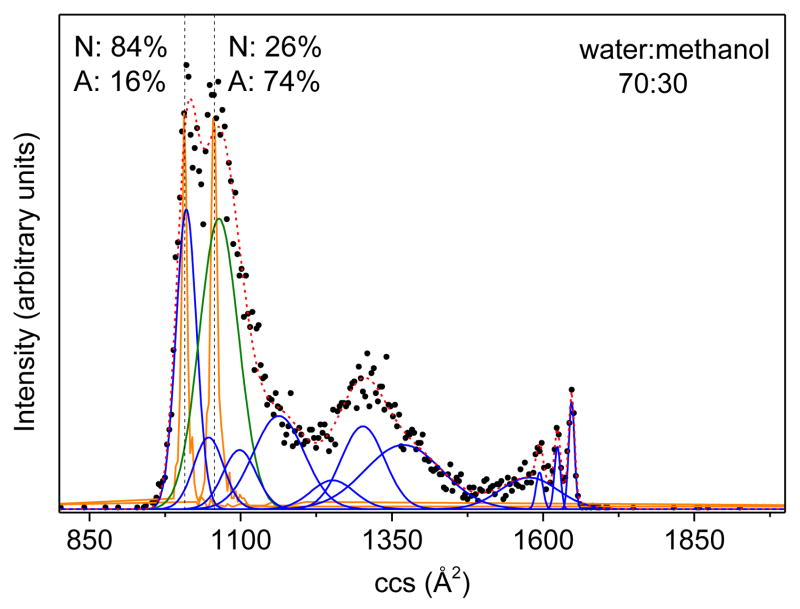
It is worthwhile to consider the origin of these variations. The initial distribution (Figure 3c) for ubiquitin [M+7H]7+ ions arising from the 70:30 water:methanol solution indicates that the N-state conformers with cross sections centered around 1010 Å2 partially overlap with the A-state ions at 1060 Å2. Consequently, ions isolated at each mobility are comprised of both N- and Astate conformers for the solution composition of 70:30 water:methanol. To measure the fractions of N- and A-state conformers over total selected ions, we employed a set of previously determined Gaussian distributions38 to model the source distribution, as displayed in Figure 4. At the cross section of 1010 Å2, N-state conformers dominate the distribution with a relative abundance of ~84% and the fraction for A-state ions is ~16%. Ions selected at 1060 Å2 consist of ~26% of N state and ~74% of A state. The relative intensities (IE) of E1-E3 for the 70:30 water:methanol solution can be calculated with the following equation.
Figure 4.
The collision cross section (ccs) distribution for ubiquitin [M+7H]7+ ions from the 70:30 water:methanol solution with Gaussian models. The experimental data are plotted as black circles. The Gaussian distributions employed in the modeling are drawn as blue and green lines, representing the N and A states, respectively, and the sum of the Gaussian functions is depicted as a red dashed line. The selected distributions of ions are also shown as orange lines. The dashed lines indicate the center of the selected distributions, at which the relative intensities for N and A states have been measured.
| (2) |
Here, FN and FA correspond to fractions of N- and A-state ions for the selected distribution, respectively; IE(N) and IE(A) is the relative intensity of the respective elongated state when generated from N- and A-state conformers, respectively. As shown in Table 1, the calculated relative abundances for E1-E3 are close to values measured from the experimental distribution for both selections. This provides further evidence that N- and A-state ubiquitin coexist in the 70:30 water:methanol solution and can be distinguished in gas-phase separation. Activation of those two conformers leads to different populations of elongated states.
Activation of source distributions
It is valuable to provide data regarding the activation of source distributions of [M+7H]7+ ions. Such information should make it possible to compare other experimental techniques and approaches such as ECD,12,59–61 photofragmentation,62,63 and infrared (IR)/ultraviolet spectroscopy.64,65 In this analysis, total distributions are activated with voltages ranging from 10 to 140 V. Collisional activations have been performed before the mobility separation. The drift time distributions for activated ions are collected with a drift tube of 183 cm in length, which has the resolution to separate the three elongated states. Instrument conditions have been tuned to be as gentle as possible in order to have [M+7H]7+ ions dominated by compact structures under nonactivating conditions (10V).
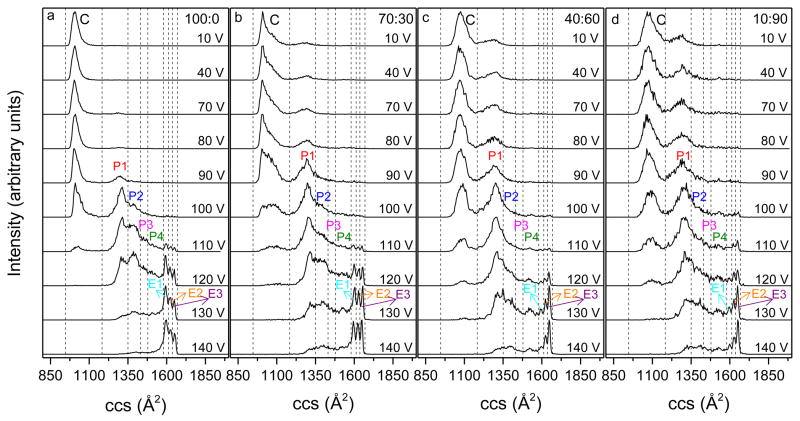
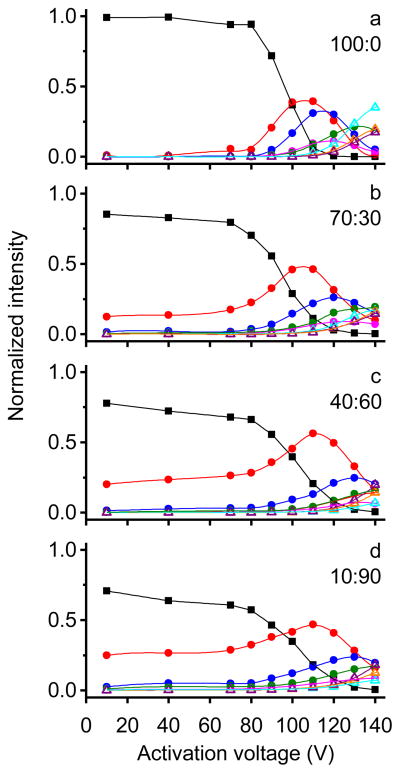
Figure 5 shows the obtained cross section distributions at different activation voltages and Figure 6 shows the normalized intensity of different conformations as a function of activation voltage for different solutions. For the 100:0 water:methanol solution (Figures 5a and 6a), distributions remain unchanged with activation voltages below 80 V and ubiquitin [M+7H]7+ ions exist primarily as the compact conformer (C). With higher activation voltages, the fraction of conformer C decreases. Upon application of 90 V a partially folded state (P1) with cross sections around 1300 Å2 emerges. The intensity of conformer P1 increases significantly (~40%) when the activation voltage is increased to 100 V. At this voltage, an unresolved feature corresponding to partially folded structures (P2) is also observed which has cross sections ranging from ~1350 to ~1430 Å2. As the activation voltage is increased to 110 V, the relative abundance of conformer P2 increases to ~30% and a small unresolved tail from ~1430 to 1570 Å2 (P3 and P4 conformers) is clearly present; additionally, small peaks corresponding to elongated states E1-E3 are produced. At 120 V activation, conformer C disappears and elongated states E1-E3 become more intense. With higher voltages, the fractions of conformers P1-P4 decrease and E1-E3 dominate the distribution. By 140 V, E1-E3 comprise ~70% of the distribution. Conformer E1 remains the most abundant species among the elongated states from 110 to 140 V with E3 being the least populated, which is consistent with results obtained from the selection and activation experiment presented above. These data indicate that the compact conformer of ubiquitin converts to partially folded conformers P1-P4 at gentle activation conditions. At even higher voltages, ions are further extended and form elongated states E1-E3.
Figure 5.
Collision cross section (ccs) distributions for ubiquitin [M+7H]7+ ions that are activated at IA1 with voltages from 10 to 140 V. Panels a-d show profiles for ions that are generated from 100:0, 70:30, 40:60 and 10:90 water:methanol solutions, respectively. Dashed lines are drawn to designate the integration regions for determining populations of different conformers that are compact (C), partially folded (P1-P4) and elongated (E1-E3). Different conformers have been labeled.
Figure 6.
Normalized intensity as a function of activation voltage for different conformers. Plots a–d show profiles for ions that are generated from 100:0, 70:30, 40:60 and 10:90 water:methanol solutions, respectively. Here closed square, closed circle and open triangle represent the normalized intensity of compact, partially folded and elongated conformations, respectively. The colors for each conformation are the same as its label given in Figure 5: C (black), P1 (red), P2 (blue), P3 (pink), P4 (green), E1 (cyan), E2 (orange), and E3 (purple).
When ions are generated from solutions with higher methanol content (70:30, 40:60 and 10:90 water:methanol), activated distributions display similar trends, as shown in Figures 5b–5d and Figures 6b–6d. Similar to ions formed by ESI a native solution, the compact structures of Astate ions also convert to partially folded conformers P1-P4 and elongated states E1-E3 via collisional activation. Conformers with larger cross sections become more abundant as activation voltages increase. However, differences in unfolding behaviors between N- and Astate ions are also observed. Conformer P2 from 40:60 and 10:90 water:methanol solutions has a significantly lower intensity at 110 and 120 V activation than that formed from the aqueous solution. For example, by 110 V, a considerable amount (~30%) of conformer P2 is observed for the aqueous solution, but only ~15% is present for denaturing solutions. In addition, at 120–140 V activation the fraction of partially folded structures P1-P4 is greater for A-state ions, whereas, N-state ions have a higher relative intensity for elongated states E1-E3. At 140 V, ~60% of Astate ions still remain as partially folded structures P1-P4, while 70% of N-state ions exist as elongated states E1-E3. This indicates that N-state ions are more likely to unfold to elongated states when collisionally heated. For the elongated states, conformer E3 is the most intense feature while E1 has the lowest intensity when 40:60 and 10:90 water:methanol solutions are electrosprayed and collisionally activated. In contrast, the 100:0 solution shows the reversed order with E1 being most intense and E3 being least intense, which is similar to the selection and activation distributions as described above. It is interesting to note that although A-state ions require higher energy to get the same amount of elongated states as N-state ions, the elongated states generated from the A-state ubiquitin display a higher portion of conformer E3, which is the most extended conformer. In other words, the A state generates a smaller amount of elongated states, but the elongated states that it produces are more extended compared to the N state when the same activation voltage is applied. For the 70:30 water:methanol solution, the behavior of the unfolding process is between that of the aqueous and denaturing solutions because both N- and A-state gas-phase conformers are formed for [M+7H]7+ ions.
The similarities in product conformers P1-P4 and E1-E3 between aqueous and denaturing solutions suggest that those are favored gas-phase structures for ubiquitin [M+7H]7+ ions. However, differences in the initial ESI-formed structures lead to different energy barriers associated with the formation of more extended structures, thus, partially folded and elongated states are observed at different intensities for different solutions.
Summary and Conclusions
The unfolding of ubiquitin [M+7H]7+ ions produced by electrospraying different water:methanol solutions has been studied in the gas phase by collisional heating. In the first set of experiments, a selected distribution of compact structures is activated at a voltage of 100 V. Three resolved elongated states are formed, the relative intensity of which is observed to be dependent on the solution state of ubiquitin. In the second set of experiments, the total distribution of ions formed by ESI is activated at variable voltages ranging from 10 to 140 V to probe the unfolding behavior under different activation conditions. This work provides a detailed description about how compact structures of ubiquitin evolve into elongated states by energetic collisions in the gas phase. Ions from both the aqueous solution that is dominated by the N state and denaturing solutions that favor the A state are converted to partially folded structures under gentle activation conditions and to elongated states at higher voltages. Product states formed by activation are similar in terms of cross sections across all solutions, however, differ in the population. Compact structures for A-state ions are more prone to unfolding from 10 to 80 V activation, however, their intensities decrease slower with a further increase in activation voltage. In addition, a greater fraction of A-state ions remain as partially folded conformers at high activation voltages (130 and 140 V), while the majority of N-state ions exist as elongated states at these activation voltages. This indicates that partially folded conformers generated from the denaturing solutions are more stable than those from the native solution during the activation process. For elongated states, E1 is most abundant and E3 is least abundant when the aqueous solution is electrosprayed, however, E1 is least abundant and E3 is most abundant for denaturing solutions.
These results provide strong evidence that different solution states of ubiquitin not only generate dissimilar compact structures during the ESI process, but also lead to different distributions of partially folded and elongated states upon collisional heating. That is, the final states of ubiquitin [M+7H]7+ ions can reflect the solution condition from which the ions were electrosprayed. Studies by ECD suggest that salt bridges and hydrogen bonds play an important role in stabilizing the gas-phase structures of proteins.12,66 Thus, we speculate that some of the salt bridges and hydrogen bonds that are present in solution can be retained in the gas phase and are disrupted upon collisional activation that influences the unfolding of ubiquitin [M+7H]7+ ions. This work provides further evidence that ions retain some memory of their solution conformations. Such insight suggests that other techniques such as ECD, photofragmentation, and IR spectroscopy can provide insights about states from solutions.
Acknowledgments
We gratefully acknowledge partial funding of this work from grants that support instrumentation development. These include grants from the NIH (1RC1GM090797-02) and funds from the Indiana University METACyt initiative that is funded by a grant from the Lilly Endowment.
References
- 1.Dobson CM. Protein Folding and Misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Brockwell DJ, Radford SE. Intermediates: Ubiquitous Species on Folding Energy Landscapes? Curr Opin Struct Biol. 2007;17:30–37. doi: 10.1016/j.sbi.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dill KA, Ozkan SB, Shell MS, Weikl TR. The Protein Folding Problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freddolino PL, Harrison CB, Liu Y, Schulten K. Challenges in Protein-Folding Simulations. Nat Phys. 2010;6:751–758. doi: 10.1038/nphys1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roder H, Maki K, Cheng H. Early Events in Protein Folding Explored by Rapid Mixing Methods. Chem Rev. 2006;106:1836–1861. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett AI, Radford SE. An Expanding Arsenal of Experimental Methods Yields and Explosion of Insights into Protein Folding Mechanisms. Nat Struct Mol Biol. 2009;16:582–588. doi: 10.1038/nsmb.1592. [DOI] [PubMed] [Google Scholar]
- 7.Schuler B, Eaton WA. Protein Folding Studied by Single-Molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A Transient and Low-Populated Protein-Folding Intermediate at Atomic Resolution. Science. 2010;329:1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 9.Gianni S, Ivarsson Y, De Simone A, Travaglini-Allocatelli C, Brunori M, Vendruscolo M. Structural Characterization of a Misfolded Intermediate Populated during the Folding Process of a PDZ domain. Nat Struct Mol Biol. 2010;17:1431–1438. doi: 10.1038/nsmb.1956. [DOI] [PubMed] [Google Scholar]
- 10.Rennella E, Cutuil T, Schanda P, Ayala I, Forge V, Brutscher B. Real-Time NMR Characterization of Structure and Dynamics in a Transiently Populated Protein Folding Intermediate. J Am Chem Soc. 2012;134:8066–8069. doi: 10.1021/ja302598j. [DOI] [PubMed] [Google Scholar]
- 11.Konermann L, Pan J, Liu Y-H. Hydrogen Exchange Mass Spectrometry for Studying Protein Structure Dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 12.Skinner OS, McLafferty FW, Breuker K. How Ubiquitin Unfolds after Transfer into the Gas Phase. J Am Soc Mass Spectrom. 2012;23:1011–1014. doi: 10.1007/s13361-012-0370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Cui W, Gross ML. Native Electrospray Ionization and Electron-Capture Dissociation for Comparison of Protein Structure in Solution and the Gas Phase. Int J Mass Spectrom. 2013;354–355:288–291. doi: 10.1016/j.ijms.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Rempel DL, Gross ML. Temperature Jump and Fast Photochemical Oxidation Probe Submillisecond Protein folding. J Am Chem Soc. 2010;132:15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoaglund-Hyzer CS, Counterman AE, Clemmer DE. Anhydrous Protein Ions. Chem Rev. 1999;99:3037–3079. doi: 10.1021/cr980139g. [DOI] [PubMed] [Google Scholar]
- 16.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Biomolecule Analysis by Ion Mobility Spectrometry. Annu Rev Anal Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenkinski RE, Chen DM, Glickson JD, Goldstein G. Nuclear Magnetic Resonance Studies of the Denaturation of Ubiquitin. Biochim Biophys Acta. 1977;494:126–130. doi: 10.1016/0005-2795(77)90140-4. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson KD, Mayer AN. Alcohol-Induced Conformational Changes of Ubiquitin. Arch Biochem Biophys. 1986;250:390–399. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]
- 19.Harding MM, Williams DH, Woolfson DN. Characterization of a Partially Denatured State of a Protein by Two-Dimensional NMR: Reduction of the Hydrophobic Interactions in Ubiquitin. Biochemistry. 1991;30:3120–3128. doi: 10.1021/bi00226a020. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of Ubiquitin Refined at 1.8 Å Resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 21.Brutscher B, Brüschweiler R, Ernst RR. Backbone Dynamics and Structural Characterization of the Partially Folded A State of Ubiquitin by 1H, 13C, and 15N Nuclear Magnetic Resonance Spectroscopy. Biochemistry. 1997;36:13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 22.Pan Y, Briggs MS. Hydrogen Exchange in Native and Alcohol Forms of Ubiquitin. Biochemistry. 1992;31:11405–11412. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 23.Stockman BJ, Euvrard A, Scahill TA. Heteronuclear Three-Dimensional NMR Spectroscopy of a Partially Denatured Protein: the A-state of Human Ubiquitin. J Biomol NMR. 1993;3:285–296. doi: 10.1007/BF00212515. [DOI] [PubMed] [Google Scholar]
- 24.Cox JPL, Evans PA, Packman LC, Williams DH, Woolfson DN. Dissecting the Structure of a Partially Folded Protein: Circular Dichroism and Nuclear Magnetic Resonance Studies of Peptides from Ubiquitin. J Mol Biol. 1993;234:483–492. doi: 10.1006/jmbi.1993.1600. [DOI] [PubMed] [Google Scholar]
- 25.Cordier F, Grzesiek S. Quantitative Comparison of the Hydrogen Bond Network of AState and Native Ubiquitin by Hydrogen Bond Scalar Couplings. Biochemistry. 2004;43:11295–11301. doi: 10.1021/bi049314f. [DOI] [PubMed] [Google Scholar]
- 26.Kony DB, Hünenberger PH, van Gunsteren WF. Molecular Dynamics Simulations of the Native and Partially Folded States of Ubiquitin: Influence of Methanol Cosolvent, pH and Temperature on the protein Structure and Dynamics. Protein Sci. 2007;16:1101–1118. doi: 10.1110/ps.062323407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemmer DE, Jarrold MF. Ion Mobility Measurements and their Applications to Clusters and Biomolecules. J Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 28.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 29.Wyttenbach T, von Helden G, Batka JJ, Carlat D, Bowers MT. Effect of the Long-Range Potential on Ion Mobility Measurements. J Am Soc Mass Spectrom. 1997;8:275–282. [Google Scholar]
- 30.Shvartsburg AA, Jarrold MF. An Exact Hard-Spheres Scattering Model for the Mobilities of Polyatomic Ions. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 31.Wyttenbach T, Bowers MT. Gas-phase Conformations: The Ion Mobility/Ion Chromatography Method. Top Curr Chem. 2003;225:207–232. [Google Scholar]
- 32.Loo JA. Studying Noncovalent Protein Complexes by Electrospray Ionization Mass Spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 33.Loo JA, He JX, Cody WL. Higher Order Structure in the Gas Phase Reflects Solution Structure. J Am Chem Soc. 1998;120:4542–4543. [Google Scholar]
- 34.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 35.Ruotolo BT, Robinson CV. Aspects of Native Proteins are Retained in Vacuum. Curr Opin Chem Biol. 2006;10:402–408. doi: 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-β Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer’s Disease. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyttenbach T, Bowers MT. Structural Stability from Solution to the Gas Phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility-Mass Spectrometry Environment. J Phys Chem B. 2011;115:12266–12275. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- 38.Shi H, Gu L, Clemmer DE, Robinson RAS. Effects of Fe(II)/H2O2 Oxidation on Ubiquitin Conformers Measured by Ion Mobility-Mass Spectrometry. J Phys Chem B. 2013;117:164–173. doi: 10.1021/jp3099544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Pierson NA, Valentine SJ, Clemmer DE. Conformation Types of Ubiquitin [M+8H]8+ Ions from Water:Methanol Solutions: Evidence for the N and A States in Aqueous Solution. J Phys Chem B. 2012;116:3344–3352. doi: 10.1021/jp210797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valentine SJ, Counterman AE, Clemmer DE. Conformer-Dependent Proton-Transfer Reactions of Ubiquitin Ions. J Am Soc Mass Spectrom. 1997;8:954–961. [Google Scholar]
- 41.Shelimov KB, Clemmer DE, Hudgins RR, Jarrold MF. Protein Structure in Vacuo: Gas-Phase Conformations of BPTI and Cytochromec. J Am Chem Soc. 1997;119:2240–2248. [Google Scholar]
- 42.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. Number of Solution States of Bradykinin from Ion Mobility and Mass Spectrometry Measurements. J Am Chem Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi H, Clemmer DE. Evidence for Two New Solution States of Ubiquitin by IMS-MS Analysis. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Taraszka JA, Counterman AE, Clemmer DE. Influence of Solvent Composition and Capillary Temperature on the Conformations of Electrosprayed Ions: Unfolding of Compact Ubiquitin Conformers from Pseudonative and Denatured Solutions. Int J Mass Spectrom. 1999;185/186/187:37–47. [Google Scholar]
- 45.Badman ER, Hoaglund-Hyzer CS, Clemmer DE. Dissociation of Different Conformations of Ubiquitin Ions. J Am Soc Mass Spectrom. 2002;13:719–723. doi: 10.1016/S1044-0305(02)00374-4. [DOI] [PubMed] [Google Scholar]
- 46.Myung S, Badman ER, Lee YJ, Clemmer DE. Structural Transitions of Electrosprayed Ubiquitin Ions Stored in an Ion Trap over ~ 10 ms to 30 s. J Phys Chem A. 2002;106:9976–9982. [Google Scholar]
- 47.Koeniger SL, Merenbloom SI, Clemmer DE. Evidence for Many Resolvable Structures within Conformation Types of Electrosprayed Ubiquitin Ions. J Phys Chem B. 2006;110:7017–7021. doi: 10.1021/jp056165h. [DOI] [PubMed] [Google Scholar]
- 48.Koeniger SL, Merenbloom SI, Sevugarajan S, Clemmer DE. Transfer of Structural Elements from Compact to Extended States in Unsolvated Ubiquitin. J Am Chem Soc. 2006;128:11713–11719. doi: 10.1021/ja062137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koeniger SL, Clemmer DE. Resolution and Structural Transitions of Elongated States of Ubiquitin. J Am Soc Mass Spectrom. 2007;18:322–331. doi: 10.1016/j.jasms.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Lee S, Ewing MA, Nachtigall FM, Kurulugama RT, Valentine SJ, Clemmer DE. Determination of Cross Sections by Overtone Mobility Spectrometry: Evidence for Loss of Unstable Structures at Higher Overtones. J Phys Chem B. 2010;114:12406–12415. doi: 10.1021/jp1060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bohrer BC, Atlasevich N, Clemmer DE. Transitions between Elongated Conformations of Ubiquitin [M+11H]11+ Enhance Hydrogen/Deuterium Exchange. J Phys Chem B. 2011;115:4509–4515. doi: 10.1021/jp2008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mack E. Average Cross-Sectional Areas of Molecules by Gaseous Diffusion Methods. J Am Chem Soc. 1925;47:2468–2482. [Google Scholar]
- 53.Revercomb HE, Mason EA. Theory of Plasma Chromatography/Gaseous Electrophoresis – A Review. Anal Chem. 1975;47:970–983. [Google Scholar]
- 54.Mason EA, McDaniel EW. Transport Properties of Ions in Gases. Wiley; New York: 1988. [Google Scholar]
- 55.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. IMS-IMS and IMS-IMS-IMS/MS for Separating Peptide and Protein Fragment Ions. Anal Chem. 2006;78:2802–2809. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]
- 56.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth H, Smith RD, Clemmer DE. An IMS-IMS Analogue of MS-MS. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 57.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-Dimensional Ion Mobility/TOFMS Analysis of Electrosprayed Biomolecules. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 58.Mohimen A, Dobo A, Hoerner JK, Kaltashov IA. A Chemometric Approach to Detection and Characterization of Multiple Protein Conformers in Solution Using Electrospray Ionization Mass Spectrometry. Anal Chem. 2003;75:4139–4147. doi: 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- 59.Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 60.Breuker K, Oh HB, Horn DM, Cerda BA, McLafferty FW. Detailed Unfolding and Folding of Gaseous Ubiquitin Ions Characterized by Electron Capture Dissociation. J Am Chem Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 61.Robinson EW, Leib RD, Williams ER. The Role of Conformation on Electron Capture Dissociation of Ubiquitin. J Am Soc Mass Spectrom. 2006;17:1469–1479. doi: 10.1016/j.jasms.2006.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reilly JP. Ultraviolet Photofragmentation of Biomolecular Ions. Mass Spectrom Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee S, Li Z, Valentine SJ, Zucker SM, Webber N, Reilly JP, Clemmer DE. Extracted Fragment Ion Mobility Distributions: A New Method for Complex Mixture Analysis. Int J Mass Spectrom. 2012;309:154–160. doi: 10.1016/j.ijms.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papadopoulos G, Svendsen A, Boyarkin OV, Rizzo TR. Conformational Distribution of Bradykinin [bk + 2H]2+ Revealed by Cold Ion Spectroscopy Coupled with FAIMS. J Am Soc Mass Spectrom. 2012;23:1173–1181. doi: 10.1007/s13361-012-0384-0. [DOI] [PubMed] [Google Scholar]
- 65.Nagornova NS, Rizzo TR, Boyarkin OV. Exploring the Mechanism of IR-UV Double-Resonance for Quantitative Spectroscopy of Protonated Polypeptides and Proteins. Angew Chem Int Ed. 2013;52:6002–6005. doi: 10.1002/anie.201301656. [DOI] [PubMed] [Google Scholar]
- 66.Breuker K, Brüschweiler S, Tollinger M. Electrostatic Stabilization of a Native Protein Structure in the Gas Phase. Angew Chem Int Ed. 2011;50:873–877. doi: 10.1002/anie.201005112. [DOI] [PMC free article] [PubMed] [Google Scholar]