Abstract
RNA interference (RNAi) is an evolutionally conserved gene silencing mechanism present in a variety of eukaryotic species. RNAi uses short double-stranded RNA (dsRNA) to trigger degradation or translation repression of homologous RNA targets in a sequence-specific manner. This system can be induced effectively in vitro and in vivo by direct application of small interfering RNAs (siRNAs), or by expression of short hairpin RNA (shRNA) with non-viral and viral vectors. To date, RNAi has been extensively used as a novel and effective tool for functional genomic studies, and has displayed great potential in treating human diseases, including human genetic and acquired disorders such as cancer and viral infections. In the present review, we focus on the recent development in the use of RNAi in the prevention and treatment of viral infections. The mechanisms, strategies, hurdles and prospects of employing RNAi in the pharmaceutical industry are also discussed.
Keywords: RNA interference, Short hairpin RNA, Micro RNA, Antiviral therapy, Viral infection, Human immunodeficiency virus, Hepatitis C virus, Hepatitis B virus, SARS-coronavirus
INTRODUCTION
RNA interference (RNAi), a highly conserved gene silencing mechanism plays an important role in the regulation of gene expression. This system was examined in a broad variety of species including plants, fungi, yeasts, nematodes, flies and mammals. In fact, RNAi serves as a safeguard for the preservation of genomic integrity. It protects the host from viral infections and invasion by mobile genetic elements by degrading the exogenous genomic material (e.g., viral RNAs).
RNAi is triggered by small double-stranded RNA (dsRNA) and functions at all levels, including transcription[1], post-transcription[2] and translation[3]. The first reports on RNA-induced post-transcriptional gene silencing (PTGS) phenomena were published in the early 90s, when Napoli[4] and Van der Krol[5] described the co-suppression of both viral transgenes and their homologous endogenous genes in transgenic plants. Similar inactivation of gene expression called “Quelling” was observed in Neurospora crassa by transformation with homologous sequences[6]. In 1995, sense RNA was demonstrated to be as effective as antisense RNA in disrupting the expression of par-1 in Caenorhabditis elegans[7]. The mechanism of action remained enigmatic until 1998, when Fire and Mello discovered that dsRNA, instead of the single-stranded sense or antisense RNA, mediated gene silencing by degrading endogenous mRNAs in a sequence-specific manner[8]. They also challenged a previous report published in 1995 claiming it to be an artificial effect of dsRNA contamination. Further studies have revealed that RNAi can occur at both the transcription and post-transcription levels. Transcriptional gene silencing involves histone H3 methylation and the formation of heterochromatin[9-11]. Post-transcriptional gene silencing includes small interfering RNA (siRNA) that mediates sequence-specific target RNA degradation, and micro RNA (miRNA) which promotes blockage of protein translation at the 3'-untranslated region (3'UTR)[12].
In recent years, RNAi has become a powerful tool to probe gene functions and to rationalize drug design. It has been employed as a prophylactic and therapeutic agent for combating a wide range of disorders, including infectious diseases, tumors and metabolic disorders. Several lethal viruses, including human immunodeficiency virus (HIV), the hepatitis C and B viruses (HCV & HBV), coronavirus, influenza A virus (IAV), human papillomavirus (HPV), have been shown to be inhibited or eliminated by RNAi. These findings have emphasized the potential of RNAi in clinical applications. In the present review, we discuss the mechanism of RNAi, and its role in the prevention and the treatment of viral infections.
Mechanisms of RNAi
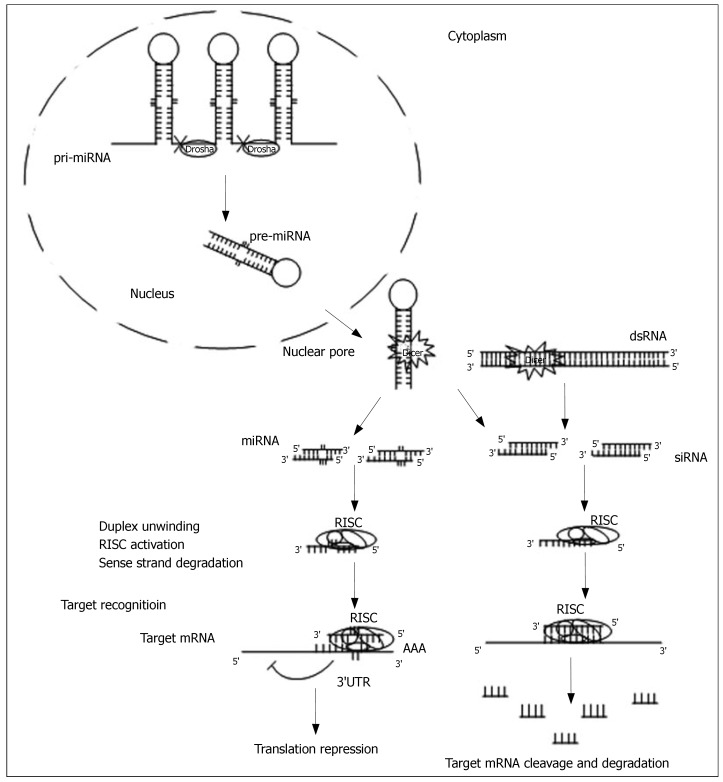
Biochemical and genetic studies have revealed the detailed mechanism by which dsRNA-mediated gene silencing takes place. In general, the mechanism includes two major steps: the initiator step and the effector step (Figure 1).
Figure 1.
The RNA interference pathways.
In the initiator step, long dsRNAs, which are produced by endogenous genes, invading viruses, transposons or experimental transgenes, are initially recognized by a dsRNA-binding protein, RDE-4/R2D2[13,14]. They are then submitted to and cleaved by the RNase III-like nuclease Dicer[15], which generates 21-23 nucleotide duplex RNAs with overhanging 3’ ends[16], called small interfering RNAs (siRNAs). The presence of highly conserved Dicer in yeast[11], plants[17,18], C. elegans[19], Drosophila[15], mice[20] and humans[21,22], suggests that the RNAi pathways share similar basic mechanisms in these organisms.
In the effector step, siRNAs are incorporated into a multicomponent nuclease complex, the RNA-induced silencing complex (RISC)[23]. The antisense strand of the duplex directs RISC to recognize and to cleave cognated target RNAs, which undergoes specific base pairing and endonucleolytic cleavage. This leads to the degradation of the unprotected and single-stranded target RNA. To date, several components of the RISC have been identified, including some conserved argonaute proteins that share the PAZ domain with Dicer family proteins[24].
The Dicer also cleaves the 60-70nt long precursor miRNA (pre-miRNAs) into miRNAs, which are of similar size as siRNAs. This pathway is referred to as miRNA-dependent gene silencing. The pre-miRNAs, whose structures are imperfectly complementary to each strand, are generated from endogenous stem loop precursors or hairpins, named primary-miRNA (pri-miRNA). The pri-miRNAs are first cleaved by Drosha RNase III in the nucleus[25]. The resulting pre-miRNAs are then exported into the cytoplasm for further processing by Dicer. The complex of the activated RISC and miRNA binds the 3’UTR of specific mRNAs, which triggers cleavage by perfect base-pairing, or translational repression by partial base-pairing recognition[26-29].
Strategies for RNA interference
In order to study the functional genomics and biology of RNA interference, much effort has gone into the study of artificial RNAi-inducing gene silencing. Strategies for delivery of RNAi reagents into mammalian cells can be divided into two types, the transient RNAi and the stable/inducible RNAi.
The methods commonly used in producing siRNA extraneously include chemical synthesis, in vitro transcription, and recombinant human Dicer/E. coli RNase III digestion of long dsRNAs. These siRNAs can be transiently transfected into target cells. Alternatively, the short hairpin RNAs (shRNAs) are expressed endogenously from plasmids and viral vectors. The shRNA expression cassettes can be stably integrated into the genome of target cells, transcribed intranuclearly and processed into siRNAs by Dicer in the cytosol. In general, RNA Pol III promoters (i.e., U6, H1 and tRNA promoters) are commonly used to drive shRNA expression in the RNAi studies. The viral vectors including retro-[30-33], lenti-[34-36], adeno- and adeno-associated viral vectors[37-39] have been demonstrated to feature high-efficiency gene delivery and can overcome the obstacles of cell-type-dependent transfection. The development of an inducible RNAi system has certainly enhanced our understanding of candidate genes’ functions, as it provides an invaluable genetic switcher that allows the inducible and reversible control of specific gene’s expression in vitro[40-42] and in vivo[43-45].
RNAi applications to combat viral infection
Viral infection is a serious public health, social and economic problem. More effective approaches are urgently needed to prevent viral propagation. Several studies have shown that RNAi technology has potential advantages over traditional measures such as the use of anti-viral drugs and vaccines, because of its ease of use, rapidity of action, high efficiency and specificity of activity when applied to the different stages of virus-host interactions[46]. In this section we will focus on the prospective use of RNAi in several common human pathogens such as HIV, HCV, HBV, SARS-coronavirus and influenza virus.
Human immunodeficiency virus
Human immunodeficiency virus type 1 (HIV-1) is the first primate virus shown to be inhibited by RNAi. HIV is a retrovirus that has been categorized into the subgroup of lentiviruses. Upon infection, the positive strand of the HIV’s RNA genome is reversely transcripted into a linear dsDNA soon after the virus enters the host cells by receptor recognition and cell adhesion. The linear dsDNA becomes circularized, is then transported into the nucleus and integrated into the host chromosome as a provirus. By utilizing the host enzymes, HIV provirus converts viral genes into mRNAs, which are used as blueprints for the subsequent expression of viral structure proteins and enzymes. It has been suggested that the genomic RNA or the newly transcribed viral mRNAs are good targets for siRNAs intervention.
It is unclear whether RNAi can target RNA genome of HIV-1 infectious particles. Jacque reported siRNA-mediated inhibition of the early and late steps of HIV-1 replication, by targeting various regions of the HIV-1 genome and by preventing the formation of viral complementary-DNA intermediates[47]. Other workers have suggested that the incoming HIV-1 RNA genome may not be accessible to siRNAs[48,49]. To date, several viral target sequences have been identified. These include the structure proteins, Gag[48,50-52] and Env[52,53]; the reverse transcriptase Pol[48]; the regulatory proteins, Tat[54,55] and Rev[54,56], and the two accessory proteins Nef[47,57,58] and Vif[47] (Table 1). The long terminal repeats that the integrase employs to insert HIV’s DNA genome into host DNA, have also been targeted[47,51].
Table 1.
Strategies designed to inhibit HIV replication via RNA interference
| Target gene | RNAi inducer (length) | Promoter | Vector | Cell type | Delivery methods | Inhib. of virus prod. (fold) | Reference |
| Viral Gene | |||||||
| LTR, Vif, Nef | siRNA (21 bp)/shRNA (19 bp)1 | T7 | Plasmid | Magi, PBLs | Transfection | > 20 | [47] |
| Gag, Pol | siRNA (21 bp) | - | - | HOS.T4.CXCR4 | Transfection | > 10 | [48] |
| Gag, LTR | siRNA (23 bp)/dsRNA (21nt)2 | - | - | U87-CD4+-CCR5+/CXCR4+, PBMC | Transfection | 4 | [51] |
| Gag, Env | dsRNA (441-531nt)3 | - | - | COS, Hela-CD4+, PBMC, ACH2 | Transfection | 70 | [52] |
| Tat + Rev | siRNA (21 bp) | - | - | 293T, Jurkat, PBMC | Transfection | > 15 | [54] |
| Rev (Tat) | siRNA (21 bp)5 | Dual U6 | Plasmid | 293/EcR | Transfection | 10000 | [56] |
| Nef | dsRNA (556nt)3 | - | - | MT4-T, U937 | Transfection | 2.5 | [57] |
| Env | siRNA (20 bp)/shRNA (20 bp)4 | U6 | Plasmid, Lentivirus | COS, MT-4 | Transfection /Transduction | > 10 | [53] |
| Nef | shRNA (21 bp) | H1 | Retrovirus | SupT1 | Transduction | > 10 | [58] |
| Gag, Pol, Int, Vpu | shRNA (21 bp) | H1 | Lentivirus | 293T, Magi, GHOST hi5, CEM-A, Molt-4, PBMC | Transduction | > 20 | [61] |
| Cellular gene | |||||||
| Tsg101 | siRNA (21 bp) | - | - | 293T | Transfection | 10-20 | [63] |
| LEDGF/p75 | siRNA (21 bp) | - | - | Hela | Transfection | NR | [64] |
| P-TEFb (CDK9/CyclinT1) | siRNA (21 bp) | - | - | Hela, Magi | Transfection | 3-5 | [65] |
| hRIP | siRNA (21 bp) | - | - | Hela, Jurkat, Macrophages | Transfection | -100 | [66] |
| Emerin | siRNA (21 bp) | - | - | Hela, Macrophages | Transfection | > 10 | [67] |
| LEDGF/p75, HRP2 | siRNA (21 bp) | - | - | Hela-P4 | Transfection | 2-3 | [68] |
| CXCR4 | siRNA (21 bp) | - | - | HOS-CD4+, HOS-CD4+-CXCR4+/CCR5+ | Transfection | 3-5 | [70] |
| Importin 7 | siRNA (21 bp) | - | - | Hela, Macrophages | Transfection | -10 | [69] |
| CXCR4+ CD4, | |||||||
| CXCR4+ CCR5 | shRNA (19/21 bp)6 | - | - | Magi-CXCR4/CCR5, PBMC | Transfection | > 15 | [71] |
| CCR5 | shRNA (19 bp) | U6 | Lentivirus | Magi-CCR5, PBLs | Transduction | 3-7 | [34] |
| Combination of viral and cellular genes | |||||||
| Gag, CD4 | siRNA (21 bp) | - | - | Magi-CCR5, Hela-CD4 | Transfection | 4-25 | [50] |
| Tat, RT, NF-κB (p65) | siRNA (21 bp) | - | - | Magi, Jurkat | Transfection | 5-500 | [74] |
The fold inhibition of virus production refers to the results obtained with the most potent siRNA/shRNA tested in a specific cell model. All siRNAs were prepared by chemical synthesis unless indicated otherwise. LTR: Long terminal repeat; PBLs: Peripheral blood lymphocytes; PBMC: Peripheral blood mononuclear cell; Pol: Polymerase; Env: Envelope; Tsg101: Tumor susceptibility gene 101; LEDGF/P75: Lens epithelium-derived growth factor/transcription co-activator p75; NR: Not reported; P-TEF: Positive transcription elongation factors; hRIP: Human Rev-interacting protein; HRP2: Hepatoma-derived growth factor related protein 2; RT: Reverse transcriptase; NF-κB: Nuclear factor-NF-κB.
shRNA expressed from a transfected plasmid under the control of a T7 promoter.
dsRNA produced by in vitro T7 promoter-mediated transcription.
dsRNA produced by in vitro SP6/T7 promoter-mediated transcription.
shRNA and siRNA expressed from transfected plasmids under the control of one and two U6 promoters respectively, shRNA further stably expressed from a recombinant lentiviral vector driven by a U6 promoter.
siRNA expressed from a transfected plasmid under the control of two U6 tandem promoters that drive the synthesis of each of the siRNA strand.
shRNA produced by in vitro T7 promoter-mediated transcription.
Several studies have demonstrated that HIV may be able to escape RNAi target by mutations[58-60]. To overcome this problem, lentiviral vectors incorporated with different shRNA-expressing-cassettes, which can simultaneously target multiple sequences including conserved sequences of the HIV genome, have been constructed[61,62]. Another proposed strategy using RNAi application is the targeting of host genes. Some host genes are essential for viral replication but have a much slower mutation rate than the viral genes. These genes have been targeted by RNAi, and the results are very encouraging[63-69] (Table 1). Down-regulation of the cell surface CD4 receptor and/or one of the co-receptors CCR5 and CXCR4 by RNAi has led to dramatic reduction of viral entry into cells[34,70,71]. Compared with CD4 and CXCR4, CCR5 has been found to be a preferential target, since no immune defects or host mortality was observed on its deletion[72,73]. Therefore, careful selection of host immutable co-factors that are important for viral replication, but not for host survival, is of prime importance in the development of anti-HIV strategies. Furthermore, simultaneous targeting by RNAi of both the virus and host factors[50,74] has been shown to be more effective in inhibiting HIV-1 replication than the targeting of either virus or host factors alone.
Hepatitis C virus
Hepatitis C virus infection is a major cause of chronic liver diseases, including liver cirrhosis and hepatocellular carcinoma (HCC). The estimated number of infected individuals are about 170 million worldwide[75], which accounts for nearly 3% of the world’s population. The World Health Organization (WHO) has recognized HCV infection as a global health problem.
HCV is a small, enveloped RNA virus that belongs to the Flaviviridae family. The cytoplasmic replicating virus contains a 9.6 kb RNA genome that functions as the messenger RNA and replication template. The development of anti-HCV drugs has accelerated since the replicon-based culture system was established a few years ago[76,77]. Several regions of the HCV’s RNA genome, including 5'UTR and the coding sequences of Core, NS3, NS4B and NS5B, are sensitive to the action of siRNA[78-83] (Table 2). The therapeutic potential of RNAi was further emphasized by in vivo studies[84,85]. The administration of siRNA and shRNA to target cell surface receptor FAS[86], caspase 8[87] and NS5B[84], has resulted in the destruction of cognate mRNAs and protection of mice from liver failure. The use of multiple siRNAs against highly conserved HCV sequences with and without host cell cofactors may limit the emergence of resistant viruses as has been demonstrated in several studies[88-92] (Table 2).
Table 2.
Strategies designed to inhibit HCV replication via RNA interference
| Target gene | RNAi inducer (length) | Promoter | Vector | Model | Delivery methods | Inhib. of virus prod. (fold) | Reference |
| In vitro studies | |||||||
| Viral gene | |||||||
| 5’-UTR | siRNA (21 bp) | - | - | 5-2 cells (Huh-7) | Transfection | -6 | [79] |
| siRNA (21 bp)/shRNA(19 bp)1 | U6 | Plasmid | 293T, Huh 7 | Transfection | > 10 | [78] | |
| NS4B | siRNA (23 bp) | - | - | Huh-7.5 | Transfection | -80 | [80] |
| NS3, NS4B, NS5A, NS5B | siRNA (21 bp) | - | - | S1179I (Huh-7) | Transfection | -23 | [81] |
| IRES, NS3, NS5B | siRNA (23bp)/shRNA (21 bp)2 | Dual H1 | Plasmid | Huh-7 | Transfection | > 9 | [82] |
| 5’-UTR, C, NS4B, NS5A, NS5B | esiRNA (15-40 bp)3/shRNA (19bp) | H1 | Mo-MuLV | Huh-7 | Transfection / Transduction | -100 | [88] |
| 5’-UTR, C, NS3, NS5B | siRNA (21 bp)/shRNA (19 bp)4 | U6 | Plasmid/Lentivirus | Huh-7 | Transfection / Transduction | -7 | [83] |
| Cellular gene | |||||||
| Lα, PTB, eIF2Bγ, hVAP33 | shRNA (19 bp) | U6 | Plasmid/Adenovirus | Huh-7 | Transfection / Transduction | -13 | [91] |
| Cyp-A,B,C | shRNA (NR) | U6 | Plasmid/Retrovirus | Huh-7 | Transfection / Transduction | -10 | [92] |
| Combination of viral and cellular genes | |||||||
| 5’-UTR, 3’-UTR, PSMA7, HuR | shRNA (19-21 bp) | U6 | Plasmid/Retrovirus | Huh-7 | Transfection / Transduction | > 2 | [89] |
| CD81, IRES, NS5B | shRNA (19-21 bp) | H1 | Lentivirus | Huh-7 | Transduction | > 32 | [90] |
| In vivo studies | |||||||
| NS5B | siRNA (23 bp) | - | - | Mice | Hydrodynamic transfection | 3 | [84] |
| IRES | shRNA (19-25 bp)5 | - | - | Mice | Hydrodynamic transfection | -50 | [85] |
The fold inhibition of virus production represents the most potent effect caused by a specific siRNA or combinatorial siRNAs. All siRNAs were prepared by chemical synthesis unless indicated otherwise. UTR: Untranslated region; NS: Non-structural; IRES: Internal ribosomal entry site; C: Core protein; esiRNA: Endoribonuclease-prepared siRNA; Mo-MuLV: Moloney murine leukemia virus; PTB: Polypyrimidine tract-binding protein; eIF2Bγ: Subunit gamma of human eukaryotic initiation factors 2B; hVAP-33: Human VAMP-associated protein of 33 kDa; Cyp: Cyclophilin; PSMA7: Proteasome a-subunit 7; HuR: Hu antigen R; N.R: not reported.
stem-loop- and tandem-type siRNA expressed from DNA-based vectors driven by one and two U6 promoters respectively.
shRNA expressed from a transfected plasmid under the control of two H1 tandem promoters that drive the synthesis of each of the siRNA strand.
esiRNA generated by in vitro T3/T7 promoter-mediated transcription.
shRNA expressed from a transfected plasmid or a lentivirus vector respectively under the control of a U6 promoter. 5 shRNA generated by in vitro T7 promoter-mediated transcription.
Hepatitis B virus
Hepatitis B virus infection is a major public health problem. It is estimated that, approximately 2 billion people are infected with HBV worldwide, and about 400 million are HBV chronic carriers[93]. HBV infection is highly prevalent in Asia and South Africa and results in over one million deaths worldwide annually.
Although the clinical symptoms caused by HBV and HCV infection are very similar, the viruses are completely unrelated[94]. HBV, the prototypical member of the Hepadnaviridae family, is one of the smallest DNA viruses (-3.2 kb), which can undergo reverse-transcription for viral replication. The HBV genome contains four overlapping open reading frames: P (polymerase-reverse transcriptase), C (core structure protein), S (surface glycoprotein) and X (HBx protein). After the uncoated nucleocapsids enter the nucleus, the HBV genome is repaired to form a covalently closed circular DNA (cccDNA), which is a template for messenger RNA transcription. The RNA intermediates-pregenomic and subgenomic RNAs, coding for viral multifunctional proteins, are transported into the cytoplasm where translation is initiated. After the pregenomic transcript is packaged into virion core particle, it is reversely transcribed by viral reverse transcriptase, thus producing a single stranded (-) DNA. Based on the structure of the (-) stranded DNA, a complementary (+) DNA strand is synthesized. Due to the lack of proofreading function of its polymerase, HBV undergoes rapid mutagenesis, with the creation of a large number of drug-resistant variants. These drug-resistant variants are further amplified under selective pressure during antiviral treatment, resulting in the elimination of the anti-viral effect and virus rebound during treatment. In severe cases, this can lead to death, even after cessation of treatment. Because of this challenge, new drugs with different targets or drug metabolism mechanisms are urgently required for better treatment outcome.
Several sites of the HBV genome including the P, Pre C/C, PreS/S, X gene, have been employed as targets to examine the in vitro efficacy of RNAi[95-99] (Table 3). Some sites have also been tested in hydrodynamic HBV model and transgenic HBV model[100-104] (Table 3). Our group has successfully designed multiple shRNAs that target DR elements and regions that code for core, polymerase, PreS, S, and X proteins. These shRNA were found to potently inhibit HBV replication and showed synergistic antiviral effects with the commonly used antiviral drug, lamivudine[105]. In a recent study, we showed that simultaneous delivery of two shRNAs that target different regions, exhibited strong synergistic antiviral effects in a hydrodynamic transgenic mice model. In this study, both S and e antigens were reduced to undetectable levels, and the viral load was reduced by greater than one hundred-fold (He et al unpublished observations). These results clearly demonstrate the potential of RNAi application in anti-HBV therapy.
Table 3.
Strategies designed to inhibit HBV replication via RNA interference
| Target gene | RNAi inducer (length) | Promoter | Vector | Model | Delivery methods | Inhib. of virus prod. (fold) | Reference |
| In vitro studies | |||||||
| C | siRNA (21 bp) | - | - | Huh-7, HepG2 | Transfection | -4-5 | [95] |
| siRNA (19 bp) | - | - | HepAD38, HepAD79 | Transfection | -50 | [96] | |
| C, X | shRNA (19 bp) | hH1 | Plasmid | Huh-7, HepG2.2.15 | Transfection | 2-20 | [97] |
| C, S, P, X, DR | shRNA (21-24 bp) | mU6 | Plasmid | HepG2.2.15 | Transfection | -2 | [98] |
| shRNA (21 bp) | hU6 | Plasmid | HepG2 | Transfection | > 30 | [105] | |
| S | shRNA (19 bp) | hH1 | PFV, AAV | 293T.HBs, HepG2.2.15 | Transduction | 4-9 | [99] |
| In vivo studies | |||||||
| C, S, P, X | shRNA (25 bp) | hU6 | Plasmid | Immunocompetent C57BL/6J mice, Immunocompromised NOD/SCID mice | Hydrodynamic transfection1 | 3-12 | [100] |
| C, S | siRNA (21 bp) | - | - | Male NMRI mice | High-volume injection via tail vein1 | -4 | [101] |
| S | shRNA (19 bp) | hH1, hU6 | Plasmid | BALB/c mice, HBsAg-transgenic FVB/N mice | Hydrodynamic transfection2 | -9 | [102] |
| P, S, X | shRNA (20 bp) | hH1 | Plasmid | C57BL/6 HBV-transgenic mice | Hydrodynamic transfection1 | 19-99 | [103] |
| P, S, X | shRNA (NR) | mU6 | Adenovirus | HBV-transgenic mice | Hydrodynamic transfection | > 9 | [104] |
The fold inhibition of virus production refers to the results obtained with the most potent siRNA/shRNA. All siRNAs were prepared by chemical synthesis unless indicated otherwise. C: Core antigen; S: Surface antigen; P: Polymerase; X: X protein; DR: Direct repeat element; PFV: Prototype foamy virus; AAV: Adeno-associated virus; mU6: Mouse U6; hU6: Human U6; hH1: Human H1. 1shRNA expression plasmid/naked siRNA coinjected with the pHBV construct. 2shRNA expression plasmid simultaneously or subsequently injected with the pHBV/pSAg construct in BALB/c mice.
SARS-coronavirus
Severe acute respiratory syndrome (SARS) outbreak affected nearly 30 countries during the years 2002-2003. This epidemic was caused by a novel SARS-associated coronavirus (SARS-CoV)[106-108]. SARS-CoV is a large (-30 kb), enveloped, positive-stranded RNA virus and its genome is composed of replicase (rep), spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes. The prophylactic and therapeutic efficacies of siRNAs were tested because of the absence of any effective drugs or vaccines against SARS-CoV infection. Both in vitro and in vivo applications proved satisfactory, using synthetic siRNAs as well as vector-based shRNAs against leader sequence[109,110], 3'-UTR[110], non-structural[111] and structural genes[110,112-115] of SARS-CoV (Table 4). Another recent report revealed that the siRNA-mediated depletion of the host cellular clathrin heavy chain gene, reduced the SARS-CoV infectivity[116]. Locked nucleic acid (LNA)-modified siRNAs, an RNA-like high affinity nucleotide analogue, has been found to improve the performance of gene silencing via enhancement of siRNA biostability and specialty. The improvement was clearly apparent when siRNA was transfected into Vero cells prior to a lethal SARS-CoV attack[117].
Table 4.
Strategies designed to inhibit SARS-CoV replication via RNA interference
| Target gene | RNAi inducer (length) | Promoter | Vector | Model | Delivery methods | Inhib. of virus prod. (fold) | Reference |
| In vitro studies Viral gene | |||||||
| Leader, TRS, 3’-UTR, S | siRNA (21 bp) | - | - | Vero E6 | Transfection | 9 | [110] |
| N | shRNA (20 bp) | U6 | Plasmid | 293 | Transfection | NR | [112] |
| E, M, N | siRNA (21 bp) | - | - | Vero E6 | Transfection | > 4 | [113] |
| P | shRNA (19 bp) | H1 | Plasmid | Vero | Transfection | > 100 | [114] |
| S | shRNA (22 bp) | U6 | Plasmid | Vero E6, 293T | Transfection | -6 | [115] |
| Rep | siRNA (21 bp) | - | - | FRhk-4 | Transfection | > 12 | [118] |
| Cellular gene | |||||||
| CHC | siRNA (25 bp) | - | - | HepG2, COS7 | Transfection | -1 | [116] |
| In vivo studies | |||||||
| S, NSP12 | siRNA (21 bp) | - | - | BALB/C mouse, Rhesus macaque (Macaca mulatta) | i.t.1 and i.n.2 administration | 3 | [119] |
The fold inhibition of virus production refers to the results obtained with the most potent siRNA/shRNA. All siRNAs were prepared by chemical synthesis unless indicated otherwise. TRS: Transcription-regulating sequence; UTR: Untranslated region; S: Spike protein; N: Nucleocapsid protein; NR: Not reported; E: Envelope protein; M: Membrane protein; P: RNA polymerase; Rep: Replicase; FRhk-4: Fatal Rhesus monkey kidney cells; CHC: Clathrin heavy chain; NSP: Non-structural protein; i.t.: Intratracheal; i.n.: Intranasal.
siRNA and target-sequence containing reporter plasmid co-administered intratracheally into mouse lungs in D5W or Infasurf solution;
siRNA instilled intranasally to monkey in D5W solution with different dosing regimens.
It is worth mentioning that our group was the first to demonstrate in 2003 the remarkable inhibition and replication of SARS-CoV infection by siRNAs against rep gene[118]. Subsequently, we designed siRNAs that could target both rep and structural genes. We also evaluated the antiviral effect, dose response, duration and viral kinetics of siRNAs in foetal rhesus kidney (FRhK-4) cells[119,120]. Two of the siRNAs were further evaluated for safety and antiviral efficacy in a rhesus macaque SARS model[119]. These siRNAs relieved SARS-like symptoms, and were safe for prophylaxis and therapeutic treatment. These findings greatly encouraged the clinical testing of siRNAs as an anti-SARS therapy.
Influenza virus
Influenza virus is one of the public health scourges worldwide. Three influenza epidemics have occurred in the last century and have caused tens of millions of deaths globally. Recent outbreaks of highly pathogenic avian influenza in Asia and Europe have greatly increased public awareness, and accelerated the development of measures for the prophylaxis and therapy of this infection.
Influenza viruses are enveloped, single-stranded, segmented (7-8) RNA viruses which belong to the Orthomyxoviridae family[121]. They are classified into influenza virus types A, B, and C, based on their nucleoproteins and matrix proteins. Influenza A virus (IAV) is the most prevalent respiratory pathogen worldwide.
Since it is an RNA virus, IAV has the ability for rapid genetic changes through antigen drift[122] or antigen shift[123]. This involves the accumulation of minor mutations within the viral genome, or reassortment of RNA segments between different viruses, which results in the emergence of new viral strains. Ge et al[124,125] and Tompkins et al[126] verified the efficacy of siRNAs which specifically target the conserved regions of the influenza virus genome (nucleocapsid and acid polymerase). They confirmed that siRNAs were potent inhibitors of the influenza virus both in vitro and in vivo, and could be administered both prior to and subsequent to a lethal IAV challenge. Moreover, Ge developed an unconventional delivery system, administering small volumes of siRNAs or DNA vectors encoding shRNA in complex with polyethyleneimine (PEI) by slow intravenous infusion[127]. This system was effective in reducing virus production in infected mice and provided helpful suggestions for future clinical application of siRNAs.
Progress of RNAi for clinical application
Since RNAi was found to have antiviral activity in transgenic plants, much evidence has emerged with regard to its pivotal role in antiviral therapeutic applications. Numerous investigations have reported successful inhibition of viral replication in cultured cells and in murine/nonhuman primate models using both transient transfection of synthetic siRNA and stable expression of shRNA. To harness the full potential of RNAi for therapeutic applications, pharmaceutical companies are actively engaged in clinical trials. In 2004, Acuity Pharmaceuticals initiated a clinical trial using RNAi in the treatment of macular degeneration; encouraging results have been obtained in the Phase I/II studies[128]. In 2006, Alnylam Pharmaceuticals launched a Phase I clinical trial in the U.S. of an inhaled formulation of ALN-RSVO1 (an RNAi-based drug) to combat respiratory syncytial virus (RSV) infection[129]. Other potential indications for RNAi use include asthma, Huntington’s disease, spinocerebellar ataxia, and HIV, HAV, HBV and influenza virus infections, and clinical trials are under consideration in many of these conditions[130].
Challenges and perspectives
Despite the rapid progress in RNAi use, its clinical application still poses several challenges. These include target specificity, biostability, biosafety, and delivery efficacy of the RNAi system in various diseases. Recent studies have indicated off-target effects associated with the use of siRNA[131-133]. In order to improve the power of gene silencing and to avoid undesirable adverse effects induced by siRNAs, such as nonspecific gene silencing and immunoactivation[134,135], great effort has been made to improve siRNA design, including its sequence[136], size[137] and structure[138]. However, the poor pharmacokinetic properties of siRNAs have added another hurdle in the development of RNAi-based therapies. Multiple chemical modifications at different positions of the siRNA duplexes, including sugars[117,139-141], backbones[142,143], and bases of oligonucleotides[144,145] have been found to prolong siRNA half-life in serum. Conjugation of one or both strands of siRNAs with lipids[146,147] and peptides[148,149], has been shown to enhance nuclease stability and improve cellular uptake.
The systematic and site-specific deliveries of siRNA also need to be addressed. Non-viral vectors, such as cationic lipids[150-152] and polymers[153-156], have been widely used for in vitro and in vivo siRNA delivery. It has been reported that siRNAs encapsulated into stable nucleic acid lipid particles (SNALPs) improve the potency, lengthen the half-life, lower the effective dose and reduce the dosing frequency. This was observed in a study comparing unformulated siRNAs in rodents challenged with replicating virus[157,158] and non-human primates[159]. Besides, Song et al designed a protamine-antibody fusion protein to deliver siRNA to HIV-infected or envelope-transfected cells. This study established a systemic, cell-type specific, antibody-mediated in vivo delivery system of siRNAs via cell surface receptors[160]. The current advances have brought siRNA close to the era of clinical trials and real-life therapeutic applications in infected human subjects.
However, before RNAi-based clinical trials can be carried out, the toxicity and side-effects of RNAi, and the harmful potential of viral vectors need careful attention. It has been shown that over-expression of shRNA by double-stranded AAV8 viral vectors resulted in severe hepatic toxicity and even death. Moreover, it has been observed that over-expressed shRNA can saturate the miRNA pathway[161]. Our studies have shown that simultaneous delivery of two shRNAs using a weaker expressing viral vector (AAV2) did not produce any obvious liver toxicity or side-effects (He et al unpublished). Therefore, it is essential to use safer vectors and in this respect we believe that inducible viral vectors may be good candidates for future clinical studies.
Scientists in different fields, including geneticists, biochemists, pharmacologists, chemists and materials scientists, have supported the use of RNAi in clinical applications. As a part of the research force, our team while being cautious, is optimistic regarding the use of RNAi in human diseases.
Footnotes
Supported by RFCID, No 01030152, RGC, CUHK4428/06M, ITF ITS091/03 of Hong Kong Government, and Faculty Direct Fund of the Chinese University of Hong Kong
S- Editor Ma N L- Editor Anand BS E- Editor Yin DH
References
- 1.Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 3.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 4.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 7.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson DS, Jarvis P. Chromatin silencing: RNA in the driving seat. Curr Biol. 2003;13:R13–R15. doi: 10.1016/s0960-9822(02)01380-5. [DOI] [PubMed] [Google Scholar]
- 11.Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 16.Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 17.Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A. Short integuments1/suspensor1/carpel factory, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park W, Li J, Song R, Messing J, Chen X. Carpel factory, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 21.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 24.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 27.Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 28.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 29.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devroe E, Silver PA. Retrovirus-delivered siRNA. BMC Biotechnol. 2002;2:15. doi: 10.1186/1472-6750-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton GM, Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc Natl Acad Sci USA. 2002;99:14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 33.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, et al. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46:200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 37.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 38.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Liu X, Li CY, Ding Y, Chau D, Li G, Kung HF, Lin MC, Peng Y. Recombinant adeno-associated virus mediated RNA interference inhibits metastasis of nasopharyngeal cancer cells in vivo and in vitro by suppression of Epstein-Barr virus encoded LMP-1. Int J Oncol. 2006;29:595–603. [PubMed] [Google Scholar]
- 40.Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu RH, Cheng TL, Lo SR, Hsu HC, Hung CF, Teng CF, Wu MP, Tsai WH, Chang WT. A tightly regulated and reversibly inducible siRNA expression system for conditional RNAi-mediated gene silencing in mammalian cells. J Gene Med. 2007;9:620–634. doi: 10.1002/jgm.1048. [DOI] [PubMed] [Google Scholar]
- 42.Henriksen JR, Løkke C, Hammerø M, Geerts D, Versteeg R, Flaegstad T, Einvik C. Comparison of RNAi efficiency mediated by tetracycline-responsive H1 and U6 promoter variants in mammalian cell lines. Nucleic Acids Res. 2007;35:e67. doi: 10.1093/nar/gkm193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L, Lin X, Staver M, Shoemaker A, Semizarov D, Fesik SW, Shen Y. Evaluating hypoxia-inducible factor-1alpha as a cancer therapeutic target via inducible RNA interference in vivo. Cancer Res. 2005;65:7249–7258. doi: 10.1158/0008-5472.CAN-04-4426. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, McMahon AP. Reproducible and inducible knockdown of gene expression in mice. Genesis. 2006;44:252–261. doi: 10.1002/dvg.20213. [DOI] [PubMed] [Google Scholar]
- 45.Seibler J, Kleinridders A, Küter-Luks B, Niehaves S, Brüning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan FL, Yin JQ. RNAi, a new therapeutic strategy against viral infection. Cell Res. 2004;14:460–466. doi: 10.1038/sj.cr.7290248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu WY, Myers CP, Kilzer JM, Pfaff SL, Bushman FD. Inhibition of retroviral pathogenesis by RNA interference. Curr Biol. 2002;12:1301–1311. doi: 10.1016/s0960-9822(02)00975-2. [DOI] [PubMed] [Google Scholar]
- 49.Westerhout EM, ter Brake O, Berkhout B. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006;3:57. doi: 10.1186/1742-4690-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 51.Capodici J, Karikó K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 52.Park WS, Miyano-Kurosaki N, Hayafune M, Nakajima E, Matsuzaki T, Shimada F, Takaku H. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 2002;30:4830–4835. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayafune M, Miyano-Kurosaki N, Takaku H, Park WS. Silencing of HIV-1 gene expression by siRNAs in transduced cells. Nucleosides Nucleotides Nucleic Acids. 2006;25:795–799. doi: 10.1080/15257770600726083. [DOI] [PubMed] [Google Scholar]
- 54.Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Efficient gene transfer of HIV-1-specific short hairpin RNA into human lymphocytic cells using recombinant adeno-associated virus vectors. Mol Ther. 2004;9:396–402. doi: 10.1016/j.ymthe.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 56.Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto T, Omoto S, Mizuguchi M, Mizukami H, Okuyama H, Okada N, Saksena NK, Brisibe EA, Otake K, Fuji YR. Double-stranded nef RNA interferes with human immunodeficiency virus type 1 replication. Microbiol Immunol. 2002;46:809–817. doi: 10.1111/j.1348-0421.2002.tb02768.x. [DOI] [PubMed] [Google Scholar]
- 58.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabariegos R, Giménez-Barcons M, Tàpia N, Clotet B, Martínez MA. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J Virol. 2006;80:571–577. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang LJ, Liu X, He J. Lentiviral siRNAs targeting multiple highly conserved RNA sequences of human immunodeficiency virus type 1. Gene Ther. 2005;12:1133–1144. doi: 10.1038/sj.gt.3302509. [DOI] [PubMed] [Google Scholar]
- 62.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 64.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 65.Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1) J Virol. 2004;78:2517–2529. doi: 10.1128/JVI.78.5.2517-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu Z, Sánchez-Velar N, Catrina IE, Kittler EL, Udofia EB, Zapp ML. The cellular HIV-1 Rev cofactor hRIP is required for viral replication. Proc Natl Acad Sci USA. 2005;102:4027–4032. doi: 10.1073/pnas.0408889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacque JM, Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- 68.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analyses of integrase-interacting proteins lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 69.Fassati A, Görlich D, Harrison I, Zaytseva L, Mingot JM. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 2003;22:3675–3685. doi: 10.1093/emboj/cdg357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou N, Fang J, Mukhtar M, Acheampong E, Pomerantz RJ. Inhibition of HIV-1 fusion with small interfering RNAs targeting the chemokine coreceptor CXCR4. Gene Ther. 2004;11:1703–1712. doi: 10.1038/sj.gt.3302339. [DOI] [PubMed] [Google Scholar]
- 71.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 72.O'Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- 73.Nansen A, Christensen JP, Andreasen SØ, Bartholdy C, Christensen JE, Thomsen AR. The role of CC chemokine receptor 5 in antiviral immunity. Blood. 2002;99:1237–1245. doi: 10.1182/blood.v99.4.1237. [DOI] [PubMed] [Google Scholar]
- 74.Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency Virus Type 1 replication. J Virol. 2002;76:12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 76.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 77.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 78.Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seo MY, Abrignani S, Houghton M, Han JH. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J Virol. 2003;77:810–812. doi: 10.1128/JVI.77.1.810-812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takigawa Y, Nagano-Fujii M, Deng L, Hidajat R, Tanaka M, Mizuta H, Hotta H. Suppression of hepatitis C virus replicon by RNA interference directed against the NS3 and NS5B regions of the viral genome. Microbiol Immunol. 2004;48:591–598. doi: 10.1111/j.1348-0421.2004.tb03556.x. [DOI] [PubMed] [Google Scholar]
- 84.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 86.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 87.Zender L, Hutker S, Liedtke C, Tillmann HL, Zender S, Mundt B, Waltemathe M, Gosling T, Flemming P, Malek NP, et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci USA. 2003;100:7797–7802. doi: 10.1073/pnas.1330920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krönke J, Kittler R, Buchholz F, Windisch MP, Pietschmann T, Bartenschlager R, Frese M. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korf M, Jarczak D, Beger C, Manns MP, Krüger M. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol. 2005;43:225–234. doi: 10.1016/j.jhep.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 90.Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Yamada O, Sakamoto T, Yoshida H, Iwai T, Matsushita Y, Shimamura H, Araki H, Shimotohno K. Down-regulation of viral replication by adenoviral-mediated expression of siRNA against cellular cofactors for hepatitis C virus. Virology. 2004;320:135–143. doi: 10.1016/j.virol.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 92.Nakagawa M, Sakamoto N, Tanabe Y, Koyama T, Itsui Y, Takeda Y, Chen CH, Kakinuma S, Oooka S, Maekawa S, et al. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129:1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 93.Akbar SM, Horiike N, Onji M. Immune therapy including dendritic cell based therapy in chronic hepatitis B virus infection. World J Gastroenterol. 2006;12:2876–2883. doi: 10.3748/wjg.v12.i18.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 95.Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51–54. doi: 10.1016/s0014-5793(03)00400-9. [DOI] [PubMed] [Google Scholar]
- 96.Ying C, De Clercq E, Neyts J. Selective inhibition of hepatitis B virus replication by RNA interference. Biochem Biophys Res Commun. 2003;309:482–484. doi: 10.1016/j.bbrc.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 97.Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–770. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- 98.Ren XR, Zhou LJ, Luo GB, Lin B, Xu A. Inhibition of hepatitis B virus replication in 2.2.15 cells by expressed shRNA. J Viral Hepat. 2005;12:236–242. doi: 10.1111/j.1365-2893.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 99.Moore MD, McGarvey MJ, Russell RA, Cullen BR, McClure MO. Stable inhibition of hepatitis B virus proteins by small interfering RNA expressed from viral vectors. J Gene Med. 2005;7:918–925. doi: 10.1002/jgm.739. [DOI] [PubMed] [Google Scholar]
- 100.McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 101.Klein C, Bock CT, Wedemeyer H, Wüstefeld T, Locarnini S, Dienes HP, Kubicka S, Manns MP, Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9–18. doi: 10.1016/s0016-5085(03)00720-0. [DOI] [PubMed] [Google Scholar]
- 102.Cheng TL, Chang WW, Su IJ, Lai MD, Huang W, Lei HY, Chang WT. Therapeutic inhibition of hepatitis B virus surface antigen expression by RNA interference. Biochem Biophys Res Commun. 2005;336:820–830. doi: 10.1016/j.bbrc.2005.08.173. [DOI] [PubMed] [Google Scholar]
- 103.Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708–716. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773–778. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen Y, Du D, Wu J, Chan CP, Tan Y, Kung HF, He ML. Inhibition of hepatitis B virus replication by stably expressed shRNA. Biochem Biophys Res Commun. 2003;311:398–404. doi: 10.1016/j.bbrc.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 106.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhong NS, Zheng BJ, Li YM, Poon ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li T, Zhang Y, Fu L, Yu C, Li X, Li Y, Zhang X, Rong Z, Wang Y, Ning H, et al. siRNA targeting the leader sequence of SARS-CoV inhibits virus replication. Gene Ther. 2005;12:751–761. doi: 10.1038/sj.gt.3302479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ni B, Shi X, Li Y, Gao W, Wang X, Wu Y. Inhibition of replication and infection of severe acute respiratory syndrome-associated coronavirus with plasmid-mediated interference RNA. Antivir Ther. 2005;10:527–533. [PubMed] [Google Scholar]
- 112.Tao P, Zhang J, Tang N, Zhang BQ, He TC, Huang AL. Potent and specific inhibition of SARS-CoV antigen expression by RNA interference. Chin Med J (Engl) 2005;118:714–719. [PubMed] [Google Scholar]
- 113.Shi Y, Yang DH, Xiong J, Jia J, Huang B, Jin YX. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 2005;15:193–200. doi: 10.1038/sj.cr.7290286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Z, Ren L, Zhao X, Hung T, Meng A, Wang J, Chen YG. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, Li T, Fu L, Yu C, Li Y, Xu X, Wang Y, Ning H, Zhang S, Chen W, et al. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Inoue Y, Tanaka N, Tanaka Y, Inoue S, Morita K, Zhuang M, Hattori T, Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Ørum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He ML, Zheng B, Peng Y, Peiris JS, Poon LL, Yuen KY, Lin MC, Kung HF, Guan Y. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA. 2003;290:2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- 119.Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.He ML, Zheng BJ, Chen Y, Wong KL, Huang JD, Lin MC, Peng Y, Yuen KY, Sung JJ, Kung HF. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett. 2006;580:2414–2420. doi: 10.1016/j.febslet.2006.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, eds. Fundamental Virology. Philadelphia Lippincott Williams & Wilkins; 2001. pp. 725–770. [Google Scholar]
- 122.Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 123.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ge Q, Eisen HN, Chen J. Use of siRNAs to prevent and treat influenza virus infection. Virus Res. 2004;102:37–42. doi: 10.1016/j.virusres.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 125.Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA. 2003;100:2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA. 2004;101:8682–8686. doi: 10.1073/pnas.0402630101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wallach T. Acuity pharmaceuticals innovative therapies for ophthalmic diseases. Acuity Pharmaceuticals Corporate Fact Sheet online, 2006-08, cited 2007-05-01; 2 screens. Available from: http: //www.bioadvance.com/downloads/fact-sheets/ACU_FS_0807.pdf.
- 129.DeVincenzo JP. Respiratory syncytial virus.Alnylam RSV Fact Sheet online, 2007-07-09, cited 2007-07-11, 1 screen. Available from: http: //www.alnylam.com/therapeutic-programs/programs.asp.
- 130.Check E. A crucial test. Nat Med. 2005;11:243–244. doi: 10.1038/nm0305-243. [DOI] [PubMed] [Google Scholar]
- 131.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 132.Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Snøve O, Holen T. Many commonly used siRNAs risk off-target activity. Biochem Biophys Res Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 134.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 135.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 136.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 137.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 138.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 139.Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2'-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J Med Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 141.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 143.Hall AH, Wan J, Shaughnessy EE, Ramsay Shaw B, Alexander KA. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 145.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 147.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J Pharmacol Exp Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 148.Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 150.Mahato RI, Rolland A, Tomlinson E. Cationic lipid-based gene delivery systems: pharmaceutical perspectives. Pharm Res. 1997;14:853–859. doi: 10.1023/a:1012187414126. [DOI] [PubMed] [Google Scholar]
- 151.Zhang C, Tang N, Liu X, Liang W, Xu W, Torchilin VP. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J Control Release. 2006;112:229–239. doi: 10.1016/j.jconrel.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Löffler K, Fechtner M, Arnold W, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 153.Han S, Mahato RI, Sung YK, Kim SW. Development of biomaterials for gene therapy. Mol Ther. 2000;2:302–317. doi: 10.1006/mthe.2000.0142. [DOI] [PubMed] [Google Scholar]
- 154.Choi Y, Thomas T, Kotlyar A, Islam MT, Baker JR. Synthesis and functional evaluation of DNA-assembled polyamidoamine dendrimer clusters for cancer cell-specific targeting. Chem Biol. 2005;12:35–43. doi: 10.1016/j.chembiol.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 155.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 156.Grzelinski M, Urban-Klein B, Martens T, Lamszus K, Bakowsky U, Höbel S, Czubayko F, Aigner A. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- 157.Geisbert TW, Hensley LE, Kagan E, Yu EZ, Geisbert JB, Daddario-DiCaprio K, Fritz EA, Jahrling PB, McClintock K, Phelps JR, et al. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J Infect Dis. 2006;193:1650–1657. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 159.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 160.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 161.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]