Abstract
In aged mice we assessed how intensive exercise affects brain bioenergetics, inflammation, and neurogenesis-relevant parameters. After 8 weeks of a supra-lactate threshold treadmill exercise intervention, 21-month old C57BL/6 mice showed increased brain PGC-1α protein, mTOR and phospho-mTOR protein, citrate synthase mRNA, and mtDNA copy number. Hippocampal VEGF-A gene expression trended higher, and a positive correlation between VEGF-A and PRC mRNA levels was observed. Brain DCX, BDNF, TNF-α, and CCL11 gene expression, as well as plasma CCL11 protein levels, were unchanged. Despite these apparent negative findings, a negative correlation between plasma CCL11 protein levels and hippocampal DCX gene expression was observed; further analysis indicated exercise may mitigate this relationship. Overall, our data suggest supra-lactate threshold exercise activates a partial mitochondrial biogenesis in aged mice, and a gene (VEGF-A) known to support neurogenesis. Our data are consistent with another study that found systemic inflammation in general, and CCL11 protein specifically, suppresses hippocampal neurogenesis. Our study supports the view that intense exercise above the lactate threshold may benefit the aging brain; future studies to address the extent to which exercise-generated lactate mediates the observed effects are warranted.
Keywords: exercise, aging brain, mitochondrial biogenesis, neurogenesis, inflammation
1. Introduction
Aerobic exercise enhances muscle aerobic metabolism in young and old mice (Lira, et al., 2010). It also modifies brain bioenergetic infrastructures, although less robustly. Studies report long-term intensive exercise training induces brain mitochondrial biogenesis in young adult mice, an effect that associates with increased levels of peroxisome-proliferator activated receptor gamma co-activator 1 (PGC-1) family members (E, et al., 2013a,Steiner, et al., 2011). In aged mice, exercise mitigates an age-associated reduction in brain electron transport chain (ETC) enzyme activities (Boveris and Navarro, 2008), but beyond this little is known about how aerobic exercise modifies brain bioenergetic infrastructures in aged mice.
Aerobic exercise also induces dentate gyrus neurogenesis in both young and aged mice (Fabel, et al., 2003,van Praag, et al., 2005). Neurotrophic factors such as brain derived neurotrophic factor (BDNF) are said to mediate this phenomenon, although other proteins such as vascular endothelial growth factor A (VEGF-A) are also believed to contribute (Fabel, et al., 2003,Jin, et al., 2002,Rothman, et al., 2012).
Inflammation-associated molecules, on the other hand, reportedly retard mouse hippocampal neurogenesis. A specific member of the CC chemokine ligand (CCL) family, CCL11, suppresses neurogenesis; plasma levels of this chemokine increase with age in both rodents and humans (Villeda, et al., 2011). Microglial activation also suppresses neurogenesis (Ekdahl, et al., 2003). Inflammation similarly impairs mitochondrial function, and cytokines such as tumor necrosis factor α (TNF-α) reduce ETC enzyme activities (Samavati, et al., 2008,Stadler, et al., 1992). Since ETC enzyme inhibition can increase oxidative stress, and oxidative stress can also activate inflammatory responses (Mukhopadhyay, et al., 2012), a harmful feedback loop may result. Overall, a robust interplay between bioenergetics, inflammation, and neurogenesis is increasingly recognized.
In this study, we exercised aged mice and explored relationships between brain bioenergetic infrastructures, inflammation, and neurogenesis. We used a treadmill-based, supra-lactate threshold exercise protocol, as previous studies conducted in young mice by our group suggest supra-lactate treadmill exercise training impacts brain bioenergetic infrastructures to a greater extent than sub-lactate threshold treadmill exercise (E, et al., 2013a,E, et al., 2013b).
2. Methods
2.1. Animals
The animal work described in this study was approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center. Whenever possible, efforts were made to minimize animal discomfort. Twenty-four C57BL/6 male mice were included in these studies. Our mice were obtained from the National Institute of Aging, and were aged 18 months when they reached our vivarium. All mice were maintained on an ad libitum diet, and were housed 2 per cage on a 12: 12 hour light: dark schedule. After a one-month accommodation period, the mice were randomly placed into 2 groups, a control group (CT, n = 12) and an exercise group (EX, n = 12).
2.2. Exercise training
EX mice were exercised for 8 weeks, 5 days per week, 2 sessions per day, on a six-lane treadmill designed for mice (Columbus Instruments, Columbus, OH). The back of each treadmill lane contained an electrified grid, which delivered a shock stimulus to stationary mice (0.2 mA, 200 ms pulses, 1 Hz). The treadmill platform was set at a 5° incline.
The EX mice were subjected to an incremental exercise training protocol to ensure that exercise intensity exceeded the lactate threshold. For the first week each session consisted of a 10 minute warm-up at 15 m/min followed by 30 minutes at 18 m/min. This speed approximates the lactate threshold for untrained C57BL/6 mice (Billat, et al., 2005). During the following 7 weeks, treadmill speed was progressively increased every week, based on the blood lactate levels measured immediately after a running session. Specifically, for weeks 2, 3, 4, 5, 6, 7, and 8 the treadmill speed was set to 21 m/min, 22 m/min, 23 m/min, 24 m/min, 25 m/min, 25 m/min, and 26 m/min, respectively. Starting from the second week, warm-up was reduced to 5 min at 15 m/min, and the duration of each running session was adjusted every week in order to maintain a constant work load throughout the study. For example, the work load for each session during the first week was (10 min x 15 m/min) + (30 min x 18 m/min) = 690 m. During the sixth week, running time at 25 m/min was calculated as [690 m – (5 min × 15 m/min)] ÷ 25 m/min = 25 min.
The CT mice did not receive any exercise training. To minimize potential confounding factors such as differences in sound and light exposure, during the EX mice training sessions CT mice were placed in the same room as the EX mice.
At the conclusion of the 8-week training period, EX mice were sacrificed by decapitation 1 hour after the last session, and CT mice were also decapitated at approximately the same time on the same day. Brain tissue was immediately harvested and frozen in liquid nitrogen, and saved at −80°C for subsequent analysis.
2.3. Lactate, glucose, and insulin levels
Blood lactate levels were measured using a Lactate Scout Analyzer (Senslab, Leipzig, Germany). Blood glucose levels were measured using a One-Touch Ultra Blood Glucose Monitoring System (LifeScan, Milpitas, CA). Plasma samples were also prepared from tail vein blood that was collected in heparinized micro-hematocrit capillary tubes (Fisher Scientific, Pittsburgh, PA). Plasma insulin levels were measured using an insulin enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer’s instructions (American Laboratory Products Company, Salem, NH). Blood glucose was measured during a period of unrestricted food access (non-fasting) and after a 17 hour fast (fasting blood glucose), and plasma insulin was measured after a 17 hour fast. All blood glucose and plasma insulin levels were measured 4 hours before the running session on the measurement day. Values for the homeostasis model assessment of insulin resistance (HOMA-IR) were calculated as the product of fasting blood glucose (mM) and plasma insulin (microunits/ml) divided by 22.5 (Matthews, et al., 1985).
2.4. Quantitative real-time, reverse-transcription PCR
The left hippocampus, which had been placed at the time of euthanasia in RNAlater Solution (Life Technologies), was retrieved and total RNA was prepared using the TRI Reagent (Life Technologies). Reverse transcription was performed on total RNA (1µg) using an iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad Laboratories, Hercules, CA). Quantitative real-time, reverse-transcription PCR (qPCR) was performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and ready-to-use TaqMan Gene Expression Assays (Applied Biosystems). We quantified mRNA levels for PGC-1α, PGC-1β, PGC-1 related co-activator (PRC), nuclear respiratory factor 1 (NRF-1), NRF-2, mitochondrial transcription factor A (TFAM), cytochrome c oxidase subunit 2 (COX2), cytochrome oxidase subunit 4 isoform 1 (COX4), citrate synthase (CS), doublecortin (DCX), VEGF-A, BDNF, TNF-α, and CCL11. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal loading control. Our qPCR amplifications were performed using an Applied Biosystems StepOnePlus Real-Time PCR System (Applied Biosystems).
2.5. MtDNA copy number
To quantify mtDNA, total DNA was extracted from the left hemisphere cortex using a phenol-chloroform based method as previously described (Guo, et al., 2009). DNA concentration and purity were determined spectrophotometrically by determining the absorbance at 260 nm and the 260 nm/280 nm ratio. Each real-time PCR reaction was performed using 10 ng of DNA as the template. We used TaqMan Gene Expression Assays (Applied Biosystems) to quantify the amounts of three targets present on mtDNA, NADH dehydrogenase subunit 1 (ND1), COX2, and ATP synthase F0 subunit 6 (ATP6), as well as the amount of a nuclear target, the 18s rRNA gene. The relative mtDNA to nuclear DNA copy number ratio was determined using the comparative ΔΔCT method, in which ND1: 18s rRNA, COX2: 18s rRNA, and ATP6: 18s rRNA ratios were calculated.
2.6. Immunochemistry
Brain protein lysates from left hemisphere cortex were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Rockford, IL) according to the manufacturer’s instructions. Protein concentrations were measured using a BCA protein assay reagent kit (Thermo Scientific). Primary antibody binding was detected using horseradish peroxidase-conjugated secondary antibodies (1:2000 dilution; Cell Signaling Technology, Beverly, MA) and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific). Densitometry was performed using a ChemiDoc XRS with Quantity One software (Bio-Rad).
Several energy-sensitive proteins and mitochondrial biogenesis related proteins were analyzed by Western blot. Primary antibodies purchased from Cell Signaling Technology included antibodies to Akt (1:1000 dilution; #5373), phospho-Ser473 Akt (1:1000 dilution; #4060), glycogen synthase kinase 3 beta (GSK3β) (1:1000 dilution; #9315), phospho-Ser9 GSK3β (1:1000 dilution; #9322), mammalian target of rapamycin (mTOR) (1:1000 dilution; #2983), phospho-Ser2448 mTOR (1:1000 dilution; #2976), TFAM (1:500 dilution; #7495), forkhead box protein O1 (FOXO1) (1:500 dilution; #2880), forkhead box O3a (FOXO3a) (1:500 dilution; #2497) , and GAPDH (1:2000 dilution; #2118). Primary antibodies purchased from Abcam (Cambridge, MA) included antibodies to PGC-1β (1:500 dilution; #ab61249) and citrate synthase (1:500 dilution; #ab96600). Antibody to NRF1 (1:200 dilution; #sc-33771) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to PGC-1α (1:1000 dilution; #PA5–22958) and histone deacetylase 1 (HDAC1) (1:1000 dilution; #PA1–860) were purchased from Thermo Scientific. An antibody to COX4 (1: 2000 dilution; #A21348) was purchased from Life Technologies.
2.7. Plasma CCL11 levels
Non-fasting plasma samples were used to evaluate plasma CCL11 levels using a commercially available Mouse CCL11/Eotaxin Quantikine ELISA Kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s instruction. Plasma CCL11 levels were measured in both CT and EX groups at baseline and at the end of the 8-week study.
2.8. Statistical analysis
Data were summarized by means and standard errors. Mean values were compared by independent samples t-tests or paired t-tests using SPSS 18.0 (SPSS Inc., Chicago, IL, USA). Pearson’s correlation analysis and Spearman’s rank-order correlation analysis were used to test inter-parameter relationships. p-values less than 0.05 were considered statistically significant.
3. Results
3.1. Effect of treadmill training on blood lactate levels
Blood lactate levels in the EX group were measured every week immediately after an exercise session. The treadmill speed was adjusted based on these measurements.
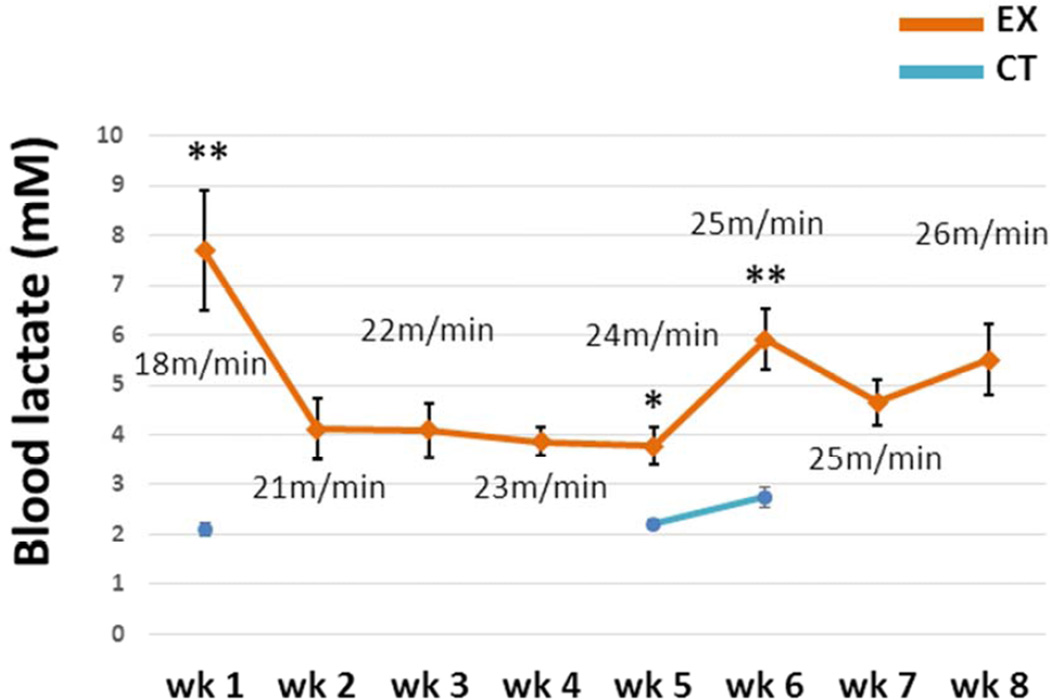
Blood lactate levels in the CT group were measured at week 1, 5, and 6. Lactate levels in the CT group ranged from 2.1–2.7 mM, while post-exercise lactate levels in the EX group ranged from 3.8–7.7 mM (Fig. 1). At each comparable time point EX group lactate levels exceeded CT group lactate levels.
Figure 1. Blood lactate levels.
Blood lactate levels were determined in the EX mice immediately after a treadmill running session on a weekly basis. In the CT mice, blood lactate levels were measured at the end of weeks 1, 5, and 6. Blood lactate levels were always significantly higher in the EX mice after a treadmill running session than they were in the CT group. The treadmill speed for each week is indicated. Wk=week. * p < 0.05, compared to CT group; ** p < 0.001, compared to CT group.
3.2. Effect of treadmill training on weight, glucose, and insulin
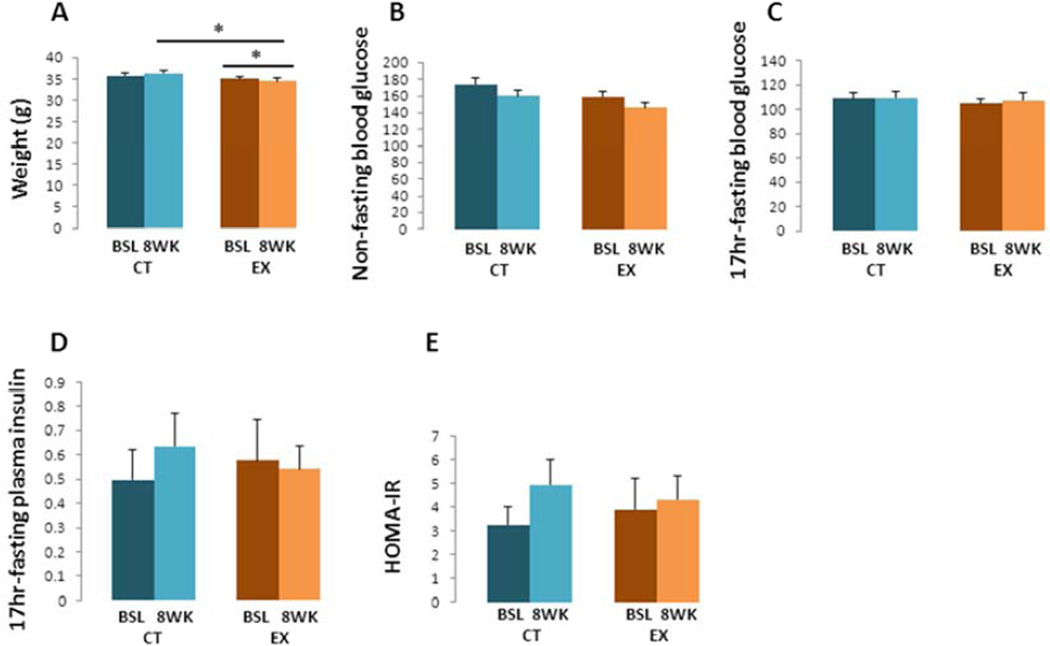
At the beginning of the study body weights were comparable between the CT and EX groups. After 8 weeks, the EX group mean weight was 5 % lower than it was in the CT group (p < 0.05, Fig. 2A). Paired t test analysis also showed EX mice lost 2 % of their starting body weight (p < 0.05, Fig. 2A), while the CT group weight did not change.
Figure 2. Body weights, blood glucose, plasma insulin, and HOMA-IR.
(A) After 8 weeks of treadmill exercise the EX group weighed less than the CT group. A paired t test analysis also showed that the EX group weighed less at the end of the study than it did at the start. (B) Significant differences in non-fasting blood glucose levels were not observed. (C) No inter- or intragroup differences were detected in 17-hour fasting blood glucose levels. (D) No inter- or intragroup differences were detected in 17-hour fasting plasma insulin levels. (E) Significant differences in HOMA-IR were not observed. Bsl=baseline; wk=week. * p < 0.05, compared to CT group.
Non-fasting blood glucose levels were comparable between the CT and EX groups at the beginning of the study, and levels did not significantly change over the 8 week study duration (Fig. 2B). Similarly, no inter- or intra-group differences were detected in fasting blood glucose or plasma insulin levels (Fig. 2C – D). HOMA-IR values did not change significantly over the 8 week study duration (Fig. 2E).
3.3. Effect of treadmill training on bioenergetics-relevant genes and proteins
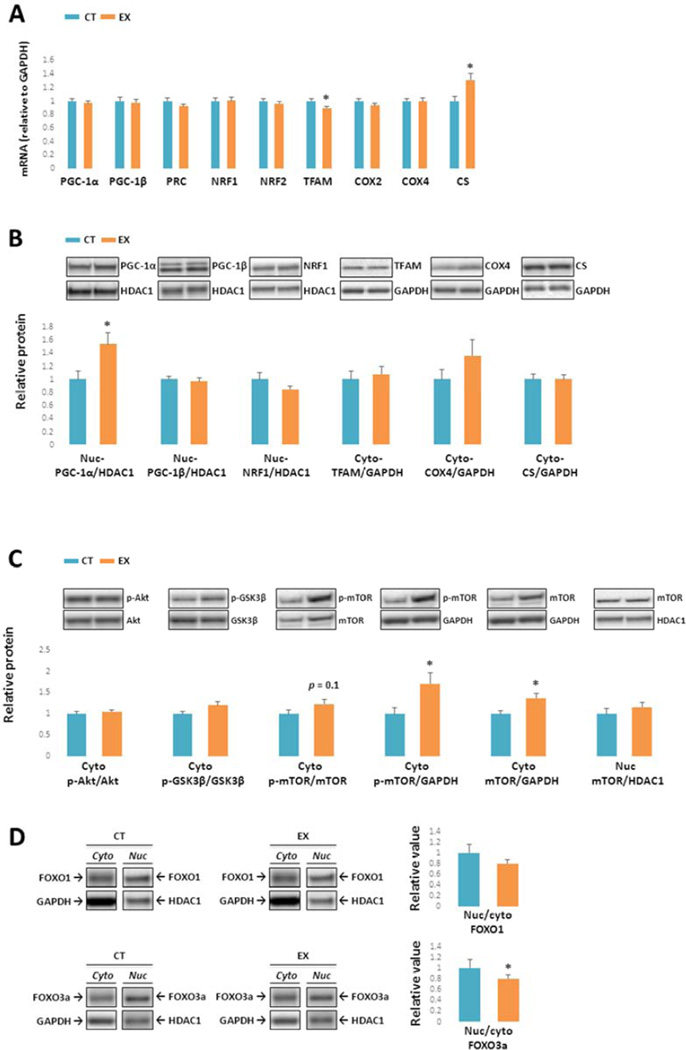
The PGC-1 family, a group of transcriptional coactivators, plays a central role in a regulatory network that governs the transcriptional control of mitochondrial biogenesis and respiratory function (Scarpulla, 2011). Three members have been identified: PGC-1α, PGC-1β and PRC. These coactivators target NRF-1 and NRF-2, which regulate TFAM expression (Scarpulla, 2008,Vina, et al., 2009). TFAM initiates replication and transcription of mitochondrial DNA (mtDNA). We found PGC-1α, PGC-1β, PRC, COX2, COX4, NRF1, and NRF2 mRNA levels were comparable between CT and EX group brains (Fig. 3A). Compared to the CT group, TFAM mRNA decreased by 10% in the EX group (p < 0.05), while CS mRNA levels increased by 30% (p < 0.05).
Figure 3. Mitochondrial biogenesis-relevant gene expression, proteins, and pathways.
(A) mRNA levels of PGC-1α, PGC-1β, PRC, COX2, COX4I1, NRF1, and NRF2 were comparable between the EX and CT groups. TFAM mRNA was slightly lower in the EX group, while CS mRNA was higher. (B) Nuclear PGC-1α protein levels were significantly higher in the EX group. No inter-group differences were observed in nuclear PGC-1β, NRF1,TFAM, COX4I1, or CS protein levels. (C) Cytoplasmic Akt Ser473 phosphorylation (normalized to total Akt protein), and cytoplasmic GSK3β Ser9 phosphorylation (normalized to total GSK3β protein) were comparable between the CT and EX groups. Although cytoplasmic mTOR Ser2448 phosphorylation, when normalized to total mTOR protein, was not significantly increased it was higher in the EX group when referenced to GAPDH, and cytoplasmic total mTOR was also elevated. Total mTOR in the nucleus was comparable between the CT and EX groups. (D) No inter-group differences were detected in FOXO1 protein distributions. The FOXO3a nucleus: cytoplasm ratio was lower in the EX group. Cyto: cytoplasmic; nuc: nuclear. * p < 0.05, compared to CT group.
Nuclear PGC-1α protein levels were significantly higher in EX brains (50 % higher than in CT brains, p < 0.05, Fig. 3B). No inter-group differences were detected in nuclear PGC-1β, NRF1, or TFAM protein levels. COX4 and CS protein levels were comparable between the groups.
Levels of other proteins that monitor or respond to cell energy states are shown in Fig. 3C. Cytoplasmic Akt phosphorylation at Ser 473 (normalized to total Akt protein) and GSK3β Ser 9 phosphorylation (normalized to total GSK3β protein) were comparable between groups. When corrected for total mTOR protein levels, cytoplasmic mTOR phosphorylation at Ser 2448 in EX brains trended higher (25 % increase, p = 0.1), and normalizing mTOR Ser 2448 levels to GAPDH suggested mTOR phosphorylation was indeed higher in the EX group (70 % increase, p < 0.05). Total mTOR protein in the cytoplasm was higher in the EX group (40 % increase, p < 0.05), while its level in the nucleus was comparable between groups. FOXO1 nuclear and cytoplasmic protein levels, as well as the FOXO1 nuclear: cytoplasm ratio, did not change across groups (Fig. 3D). The FOXO3a nuclear: cytoplasm protein ratio was significantly lower in EX brains (20 % decrease, p < 0.05, Fig. 3D).
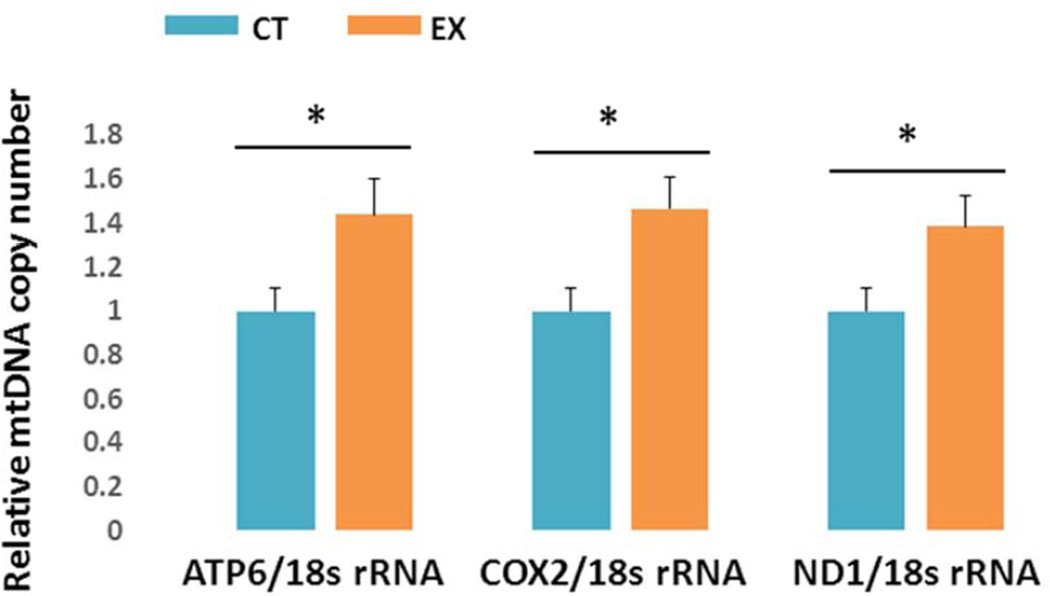
Brain mtDNA copy number, as determined by qPCR, demonstrated exercise-related increases. Ratios of ATP6: 18s rRNA, COX2: 18s rRNA, and ND1: 18s rRNA were all significantly higher in EX brains than they were in CT brains (40–50 % increase, p < 0.05, Fig. 4).
Figure 4. mtDNA copy number.
mtDNA copy numbers were higher in the EX group when measured using primers to ATP synthase F0 subunit 6 (ATP6) (A), COX2 (B), and NADH dehydrogenase subunit 1 (ND1) (C). * p < 0.05, compared to CT group.
3.4. Effect of treadmill training on neurogenesis-related factors
DCX is a microtubule-associated protein expressed specifically in migrating central nervous system neuronal precursors. Its presence in the brain is widespread during early development (Gleeson, et al, 1999), and in the adult brain DCX expression is retained within the hippocampus and the subventricular zone/olfactory bulb axis. DCX expression reportedly reflects levels of adult neurogenesis (Couillard-Despres, et al, 2005). In rodents, both VEGF-A and BDNF reportedly contribute to exercise-induced neurogenesis. VEGF-A activates angiogenesis, which indirectly facilitates neurogenesis. BDNF directly supports neurogenesis (Fabel, et al, 2003,Jin, et al, 2002,Rossi, et al., 2006).
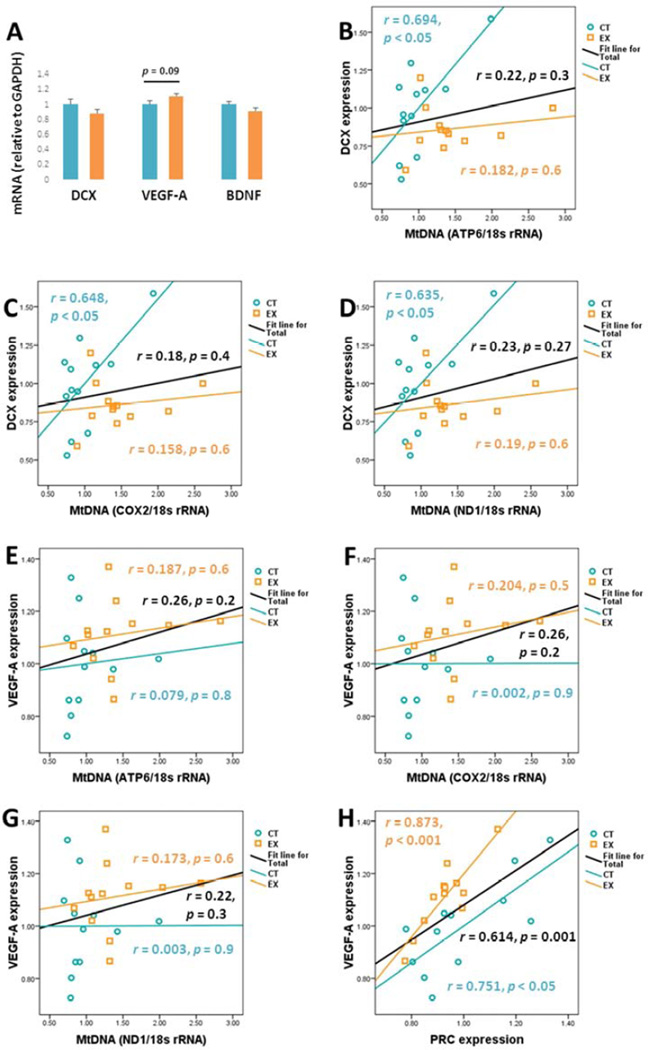
DCX mRNA levels were comparable between hippocampus-derived total RNA preparations (Fig. 5A). There was perhaps a trend towards increased VEGF-A mRNA in the EX group, but this was not a statistically significant change (12 % higher mean absolute value, p = 0.09). BDNF mRNA levels were comparable between the two groups.
Figure 5. Non-inflammatory neurogenesis-related factors and correlations.
(A) mRNA levels for BDNF, DCX, VEGF-A were each normalized to GAPDH mRNA. BDNF and DCX mRNA was comparable between groups. VEGF-A mRNA levels trended higher in the EX group, although this difference was not significant. (B–D) shows the relationship between brain DCX mRNA and mtDNA copy numbers. DCX mRNA levels in the CT group, but not the EX group, positively correlated with mtDNA copy number. (E–G) shows the relationship between brain VEGF-A mRNA and mtDNA copy numbers. No significant correlations were detected. (H) VEGF-A and PRC mRNA levels positively correlated, both in the individual groups and when the groups were combined.
We assessed relationships between neurogenesis-related gene expression and bioenergetics-relevant parameters. For the combined CT and EX groups, no significant relationship between DCX mRNA levels and ATP6, COX2, or ND1 -determined mtDNA copy numbers was observed (DCX and ATP6 mtDNA: r = 0.22, p = 0.3; DCX and COX2 mtDNA: r = 0.18, p = 0.4; DCX and ND1 mtDNA: r = 0.23, p = 0.27; Fig. 5B–D). When analyzed by group, in the CT group positive correlations were observed between DCX mRNA levels and mtDNA content (DCX and ATP6: r = 0.694, p < 0.05; DCX and COX2: r = 0.648, p < 0.05; DCX and ND1: r = 0.635, p < 0.05; Fig. 5B – D). No correlations, however, were observed between DCX mRNA and EX group mtDNA levels (Fig. 5B – D).
Significant correlations were not seen between hippocampal VEGF-A mRNA levels and mtDNA copy numbers, either in the combined CT and EX groups or when groups were analyzed separately (Fig. 5E–G). A strong positive correlation was observed between hippocampal VEGF-A and PRC mRNA expression, when the CT and EX groups were combined (r = 0.614, p < 0.05, Fig. 5H) and when the groups were analyzed separately (CT: r = 0.751, p < 0.05; EX: r = 0.873, p < 0.001; Fig. 5H).
3.5. Effect of treadmill training on inflammatory factors
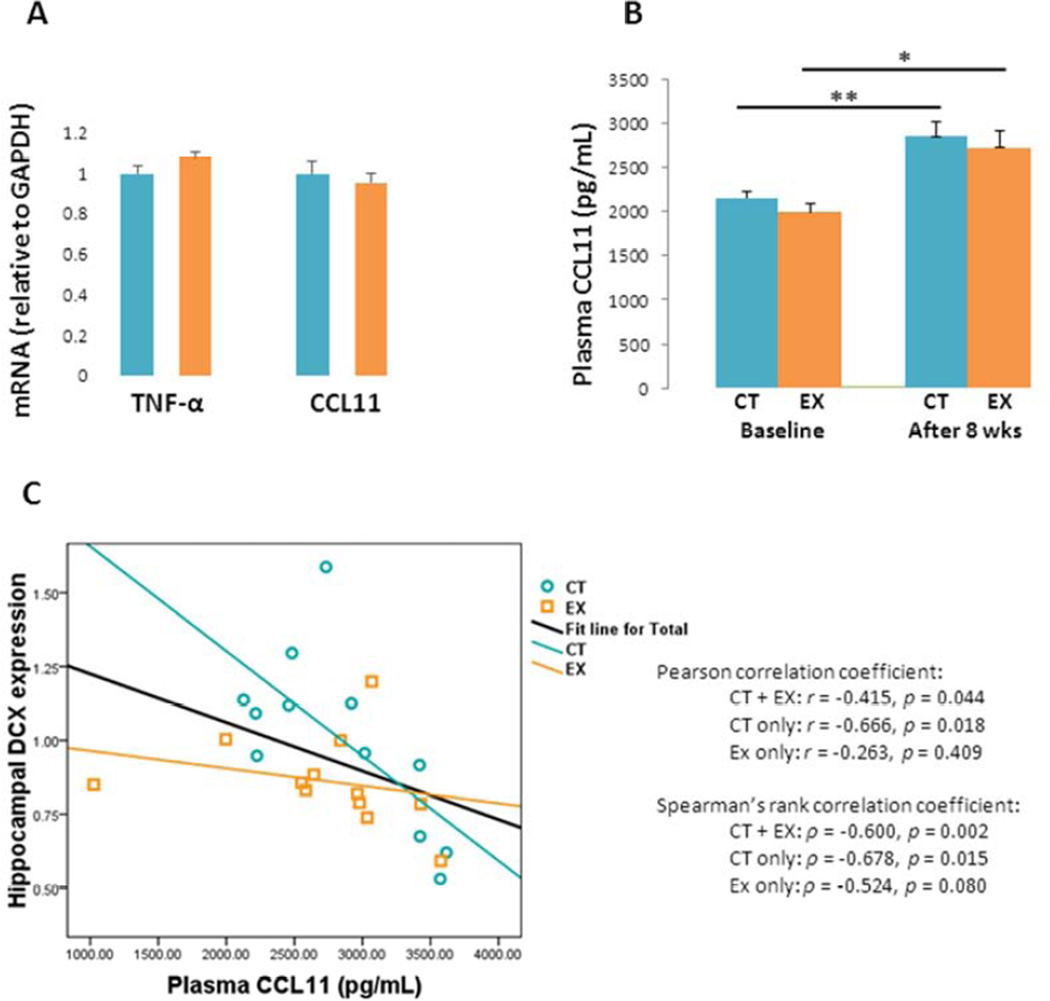
Previous studies suggest exercise lowers brain TNF-α expression in young adult mice (E, et al., 2013a,E, et al., 2013b). In the aged mice we studied, though, exercise did not reduce TNF-α mRNA levels (Fig. 6A). Hippocampal CCL11 mRNA levels were also comparable between the CT and EX groups. Plasma CCL11 protein levels were comparable between the CT and EX groups at baseline and after the 8 week study period, although over the 8 week study period significant increases in plasma CCL11 protein occurred in both groups (CT: 30% higher after 8 weeks compared to baseline, p < 0.001; EX: 40% higher after 8 weeks compared to baseline, p < 0.05; Fig. 6B).
Figure 6. Inflammatory factors.
(A) Hippocampal TNF-α and CCL11 mRNA levels were equivalent between the CT and EX groups. (B) Plasma CCL11 protein levels were comparable between the CT and EX groups. Plasma CCL11 protein levels rose in both groups over the 8-week duration of the study. (C) shows the relationship between brain DCX mRNA levels and plasma CCL11 levels. For the CT and combined groups a negative correlation between brain DCX mRNA and plasma CCL11 protein levels was observed. This relationship was not significant within the EX group.
No correlation was seen between TNF-α mRNA or plasma CCL11 plasma protein and mtDNA copy number (data not shown). Pearson’s correlation analysis detected a moderate negative association between hippocampal DCX mRNA expression and plasma CCL11 protein, when data from the CT and EX groups were pooled (r = −0.415, p < 0.05, Fig. 6C), as well as when just the CT group was considered (r = −0.666, p < 0.05, Fig. 6C). This negative correlation did not hold when the EX group was analyzed separately (r = −0.263, p = 0.4, Fig. 6C). Spearman’s rank-order correlation analysis, which is less sensitive to potential outliers than Pearson’s analysis, showed similar results (CT + EX: ρ = −0.600, p < 0.005; CT only: ρ = −0.678, p < 0.05; EX only: ρ = −0.524, p > 0.05; Fig. 6C).
4. Discussion
We found that in aged mice, exercising above the lactate threshold affects bioenergetics-relevant parameters. Brain PGC-1α protein, mTOR and phospho-mTOR protein, CS mRNA, and mtDNA copy number increased. Our data also suggest that in aged mice, exercise may mitigate CCL11-mediated neurogenesis suppression (Villeda, et al., 2011).
Our study shows it is feasible to exercise aged mice at an intensity that exceeds their lactate threshold, even for a sustained period of up to eight weeks. As is the case with younger mice (E, et al., 2013b), because aged mice become increasingly fit over time the exercise intensity required to surpass the lactate threshold rises. An incremental exercise protocol is therefore indicated.
Although exercise resulted in a small decline in weight, it did not reduce blood glucose, plasma insulin, or HOMA-IR values in our aged mice. Based just on these data we would not conclude exercise did not enhance insulin sensitivity. Prior research indicates exercise enhances the liver’s ability to perform gluconeogenesis (E, et al., 2013a,E, et al., 2013b), which may elevate blood glucose and confound the HOMA-IR calculation.
To our knowledge, only a small number of studies have explored the nexus between exercise, brain aging, and brain bioenergetics. Boveris and Navarro found that in 19-month old mice, moderate intensity treadmill exercise blunted an age-associated decline in brain COX activity (Boveris and Navarro, 2008). In mtDNA POLG mutator mice, which experience accelerated aging and age-related declines in mitochondrial function (Kujoth, et al., 2005,Trifunovic, et al., 2004), moderate-intensity treadmill training over five months preserved brain mitochondrial function even though PGC-1α gene expression did not change (Safdar, et al., 2011). In 12-month old rats, 15 weeks of daily 30-minute treadmill exercise sessions (at 18 m/min) increased brain PGC-1α and NRF1 protein levels, but mtDNA copy number or levels of other bioenergetics-relevant genes or proteins were not reported (Marosi, et al., 2012). Our current study, in which mice were 21 months of age at completion, contributes to this literature.
mtDNA copy number is commonly used to assess mitochondrial biogenesis and mass (Medeiros, 2008). Increased mtDNA copy numbers in the brains of our EX mice argues that even in aged mice, exercise induces at least a partial increase in mitochondrial biogenesis or at least an increase in some mitochondrial components. Supra-lactate threshold exercise also appears to increase brain mtDNA copy number in younger mice (E, et al., 2013b). As PGC-1α co-activates the expression of mitochondrial biogenesis-relevant genes, increased PGC-1α protein is also consistent with an induction of mitochondrial biogenesis. In our prior study of young mice, exercise induced neither PGC-1α protein nor gene expression, but did associate with increased PRC gene expression (E, et al., 2013b). Whether this divergence reflects differences between young and old mice, or differences in the exercise protocols used between studies, is unclear.
Another group, though, did report vigorous exercise increased mouse brain PGC-1α mRNA levels; protein levels were not reported (Steiner, et al., 2011). Increased CS gene expression in our study is also consistent with a mitochondrial mass increase.
On the other hand, we did not observe increased CS protein or any increases in ETC subunit mRNA or protein. TFAM mRNA was also lower in EX mice. It is therefore important to stress that the functional relevance of exercise-induced mtDNA copy number and PGC-1α protein increases remains to be determined.
Akt regulates multiple proteins that monitor or respond to bioenergetic states. Its activity positively correlates with the extent of its Ser473 phosphorylation (Alessi, et al., 1996). Akt directly phosphorylates GSK3β at its Ser9 amino acid (which inhibits activity), and indirectly contributes to mTOR Ser2448 phosphorylation (which enhances activity) (Cole, 2012,Nave, et al., 1999). In our aged mice, exercise changed neither Akt Ser473 nor GSK3β Ser9 phosphorylation, yet mTOR Ser2448 phosphorylation increased. We did not determine the functional relevance of increased mTOR Ser2448 phosphorylation or exactly how it occurred. Our data are nevertheless consistent with a previous report that found inhibiting mTOR directly with rapamycin, but not indirectly via Akt inhibition, reduces PGC-1α expression (Cunningham, et al., 2007). In addition to potentially supporting the authors’ conclusion that mTOR promotes mitochondrial biogenesis in an Akt-independent manner, increased mTOR phosphorylation in our current study implies mTOR may contribute to exercise-induced increases in brain mtDNA copy number and PGC-1α protein (Cunningham, et al., 2007).
Exercise increases neurogenesis in the hippocampal dentate gyrus of both young and old mice (Fabel, et al., 2003,van Praag, et al., 2005). VEGF-A, which induces angiogenesis, also appears to support neurogenesis (Fabel, et al., 2003,Font, et al., 2010,Jin, et al., 2002). We previously found that in young mice, both supra-lactate threshold exercise and lactate itself induced brain VEGF-A gene expression (E, et al., 2013b). In our aged mice, mean VEGF-A gene expression perhaps trended higher in the EX mice, although this was not a statistically significant increase. We believe this trend is worth noting because of an observed correlation between VEGF-A and PRC gene expression; our earlier study in young mice found a similar relationship (E, et al., 2013b).
In addition, our current study found FOXO3a, which suppresses VEGF-A expression (Karadedou, et al., 2012), experienced a subtle nuclear to cytoplasmic shift. This would predictably reduce FOXO3a nuclear activity and facilitate VEGF-A expression. Interestingly, FOXO3a was also recently shown to reduce mtDNA copy number and mitochondria-related nuclear gene expression (Ferber, et al., 2012). Translocation of FOXO3a to the cytoplasm, therefore, is consistent with an upregulation of mitochondrial biogenesis pathways.
We did not directly assess the effects of exercise on hippocampal neurogenesis, although measurements of hippocampal BDNF, DCX, and CCL11 gene expression, as well as plasma CCL11 protein, may provide additional insight into the relationship that may exist between exercise, hippocampal neurogenesis, and brain aging. We did not detect an increase in hippocampal BDNF gene expression, which argues either a dissociation between hippocampal BDNF expression and BDNF protein may exist, or that BDNF produced outside the hippocampus may play a contributory role (Katoh-Semba, et al., 1997). Hippocampal mRNA levels of the DCX gene, which is expressed by immature neurons and is used as a marker of new neuron formation (Wojtowicz and Kee, 2006), did not show inter-group differences. Exercise did not induce obvious inter-group differences in hippocampal gene expression or plasma protein levels of CCL11, a chemokine recently shown to inhibit hippocampal neurogenesis (Villeda, et al., 2011).
Despite these negative findings, correlative analysis suggests exercise may still subtly influence hippocampal neurogenesis by affecting these genes and proteins. To this point our observation that hippocampal DCX gene expression and plasma CCL11 protein levels negatively correlated in CT mice, but not in EX mice, is pertinent. If plasma CCL11 levels in mice indeed rise with advancing age (a phenomenon we were able to validate) (Villeda, et al., 2011), and CCL11 suppresses hippocampal neurogenesis, then an inverse correlation between CCL11 plasma protein and hippocampal DCX would be expected. Our correlational analysis, therefore, suggests that in aged mice supra-lactate threshold exercise may subtly mitigate CCL11’s anti-neurogenesis effects, even if it fails to alter CCL11 levels. In other words, exercise may “uncouple” the previously reported inverse relationship between CCL11 and hippocampal neurogenesis.
CT mice showed a positive correlation between hippocampal DCX gene expression and brain cortex mtDNA copy number. This is consistent with the possibility that robust bioenergetic function enhances neurogenesis, that bioenergetic function is more robust in immature neurons, or both. Why or how supra-lactate threshold exercise negated this relationship is unclear.
The impact of potential confounding factors, such as changes in plasma glucocorticoid levels, is worth considering. Because we were specifically interested in testing the impact of supra-lactate threshold exercise on brain bioenergetic infrastructures, we utilized a treadmill exercise protocol rather than a voluntary wheel-running approach. We would expect mice would find forced treadmill exercise more stressful than voluntary wheel running, which could raise glucocorticoid production, and chronically elevated glucocorticoids alters mouse hippocampal neuron morphology (Woolley, et al., 1990). On the other hand, at least in human studies it appears that even though forced exercise acutely increases plasma cortisol, resting levels remain unchanged (Duclos, et al., 2003). Partly because we did not measure plasma glucocorticoids, and partly because it was beyond the scope of our hypothesis (that supra-lactate threshold exercise alters brain bioenergetic infrastructures), we cannot say whether or not a stress-induced cortisol increase impacted our data (for example, by preventing an exercise-induced increase in hippocampal BDNF expression).
The lactate threshold is typically exceeded in untrained human subjects when the VO2 reaches 60–70% of its maximum, and in trained human subjects when the VO2 reaches 75–80% of its maximum (Kenney, et al., 2012). From a translational perspective, the ability of aged humans to perform supra-lactate threshold (high intensity) exercise is worth considering. To this point, several studies have shown in elderly subjects, aerobic exercise at an intensity of 60–80% of the VO2max is possible (Baker, et al., 2010,Belman and Gaesser, 1991). In addition, long-term, high-intensity training improves memory and executive function test performance in young subjects, as well as in aged subjects with mild cognitive impairment (Baker, et al., 2010,Griffin, et al., 2011).
Overall, our data support the view that physical exercise can impact brain bioenergetics, neurogenesis, and inflammation-related parameters in aged mice. Although other studies find physical exercise functionally benefits neurologic endpoints in aged mice (van Praag, et al., 2005), it remains to be shown whether the specific changes we now describe affect nervous system performance. Lastly, what mediates the exercise-associated changes we observed remains unresolved. Previous work by our group argues lactate, a byproduct of exercising muscle that accesses the brain and which is used by the brain as an energy source, may play a role (E, et al., 2013b).
Acknowledgements
This project was supported by the University of Kansas Alzheimer’s Disease Center (P30 AG035982), the Frank and Evangeline Thompson Alzheimer’s Treatment Program fund, and a Mabel Woodyard Fellowship Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement: The authors declare no competing financial interests.
REFERENCES
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15(23):6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Baker LD, Frank LL, Foster-Schubert K, Green PS, Wilkinson CW, McTiernan A, Plymate SR, Fishel MA, Watson GS, Cholerton BA, Duncan GE, Mehta PD, Craft S. Effects of aerobic exercise on mild cognitive impairment: a controlled trial. Arch Neurol. 2010;67(1):71–79. doi: 10.1001/archneurol.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belman MJ, Gaesser GA. Exercise training below and above the lactate threshold in the elderly. Med Sci Sports Exerc. 1991;23(5):562–568. [PubMed] [Google Scholar]
- Billat VL, Mouisel E, Roblot N, Melki J. Inter- and intrastrain variation in mouse critical running speed. J Appl Physiol. 2005;98(4):1258–1263. doi: 10.1152/japplphysiol.00991.2004. [DOI] [PubMed] [Google Scholar]
- Boveris A, Navarro A. Systemic and mitochondrial adaptive responses to moderate exercise in rodents. Free Radic Biol Med. 2008;44(2):224–229. doi: 10.1016/j.freeradbiomed.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Cole AR. GSK3 as a Sensor Determining Cell Fate in the Brain. Front Mol Neurosci. 2012;5:4. doi: 10.3389/fnmol.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard-Despres S, Winner B, Schaubeck S, Aigner R, Vroemen M, Weidner N, Bogdahn U, Winkler J, Kuhn HG, Aigner L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur J Neurosci. 2005;21(1):1–14. doi: 10.1111/j.1460-9568.2004.03813.x. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Duclos M, Gouarne C, Bonnemaison D. Acute and chronic effects of exercise on tissue sensitivity to glucocorticoids. J Appl Physiol (1985) 2003;94(3):869–875. doi: 10.1152/japplphysiol.00108.2002. [DOI] [PubMed] [Google Scholar]
- E L, Lu J, Burns JM, Swerdlow RH. Effect of exercise on mouse liver and brain bioenergetic infrastructures. Exp Physiol. 2013a;98(1):207–219. doi: 10.1113/expphysiol.2012.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E L, Lu J, Selfridge JE, Burns JM, Swerdlow RH. Lactate Administration Reproduces Specific Brain and Liver Exercise-Related Changes. J Neurochem. 2013b doi: 10.1111/jnc.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl CT, Claasen JH, Bonde S, Kokaia Z, Lindvall O. Inflammation is detrimental for neurogenesis in adult brain. Proc Natl Acad Sci U S A. 2003;100(23):13632–13637. doi: 10.1073/pnas.2234031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabel K, Fabel K, Tam B, Kaufer D, Baiker A, Simmons N, Kuo CJ, Palmer TD. VEGF is necessary for exercise-induced adult hippocampal neurogenesis. Eur J Neurosci. 2003;18(10):2803–2812. doi: 10.1111/j.1460-9568.2003.03041.x. [DOI] [PubMed] [Google Scholar]
- Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012;19(6):968–979. doi: 10.1038/cdd.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font MA, Arboix A, Krupinski J. Angiogenesis, neurogenesis and neuroplasticity in ischemic stroke. Curr Cardiol Rev. 2010;6(3):238–244. doi: 10.2174/157340310791658802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23(2):257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Griffin EW, Mullally S, Foley C, Warmington SA, O’Mara SM, Kelly AM. Aerobic exercise improves hippocampal function and increases BDNF in the serum of young adult males. Physiol Behav. 2011;104(5):934–941. doi: 10.1016/j.physbeh.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9(4):261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2002;99(18):11946–11950. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadedou CT, Gomes AR, Chen J, Petkovic M, Ho KK, Zwolinska AK, Feltes A, Wong SY, Chan KY, Cheung YN, Tsang JW, Brosens JJ, Khoo US, Lam EW. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2012;31(14):1845–1858. doi: 10.1038/onc.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh-Semba R, Takeuchi IK, Semba R, Kato K. Distribution of brain-derived neurotrophic factor in rats and its changes with development in the brain. J Neurochem. 1997;69(1):34–42. doi: 10.1046/j.1471-4159.1997.69010034.x. [DOI] [PubMed] [Google Scholar]
- Kenney WL, Wilmore JH, Costill DL, Wilmore JH. 5th ed. Human Kinetics: Champaign, IL; 2012. Physiology of sport and exercise. [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lira VA, Benton CR, Yan Z, Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;299(2):E145–E161. doi: 10.1152/ajpendo.00755.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marosi K, Bori Z, Hart N, Sarga L, Koltai E, Radak Z, Nyakas C. Long-term exercise treatment reduces oxidative stress in the hippocampus of aging rats. Neuroscience. 2012;226:21–28. doi: 10.1016/j.neuroscience.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Medeiros DM. Assessing mitochondria biogenesis. Methods. 2008;46(4):288–294. doi: 10.1016/j.ymeth.2008.09.026. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Horvath B, Zsengeller Z, Batkai S, Cao Z, Kechrid M, Holovac E, Erdelyi K, Tanchian G, Liaudet L, Stillman IE, Joseph J, Kalyanaraman B, Pacher P. Mitochondrial reactive oxygen species generation triggers inflammatory response and tissue injury associated with hepatic ischemia-reperfusion: therapeutic potential of mitochondrially targeted antioxidants. Free Radic Biol Med. 2012;53(5):1123–1138. doi: 10.1016/j.freeradbiomed.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344:427–431. Pt 2. [PMC free article] [PubMed] [Google Scholar]
- Rossi C, Angelucci A, Costantin L, Braschi C, Mazzantini M, Babbini F, Fabbri ME, Tessarollo L, Maffei L, Berardi N, Caleo M. Brain-derived neurotrophic factor (BDNF) is required for the enhancement of hippocampal neurogenesis following environmental enrichment. Eur J Neurosci. 2006;24(7):1850–1856. doi: 10.1111/j.1460-9568.2006.05059.x. [DOI] [PubMed] [Google Scholar]
- Rothman SM, Griffioen KJ, Wan R, Mattson MP. Brain-derived neurotrophic factor as a regulator of systemic and brain energy metabolism and cardiovascular health. Ann N Y Acad Sci. 2012;1264(1):49–63. doi: 10.1111/j.1749-6632.2012.06525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108(10):4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. Tumor necrosis factor alpha inhibits oxidative phosphorylation through tyrosine phosphorylation at subunit I of cytochrome c oxidase. J Biol Chem. 2008;283(30):21134–21144. doi: 10.1074/jbc.M801954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813(7):1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler J, Bentz BG, Harbrecht BG, Di Silvio M, Curran RD, Billiar TR, Hoffman RA, Simmons RL. Tumor necrosis factor alpha inhibits hepatocyte mitochondrial respiration. Ann Surg. 1992;216(5):539–546. doi: 10.1097/00000658-199211000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Murphy EA, McClellan JL, Carmichael MD, Davis JM. Exercise training increases mitochondrial biogenesis in the brain. J Appl Physiol. 2011;111(4):1066–1071. doi: 10.1152/japplphysiol.00343.2011. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25(38):8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, Lucin KM, Czirr E, Park JS, Couillard-Despres S, Aigner L, Li G, Peskind ER, Kaye JA, Quinn JF, Galasko DR, Xie XS, Rando TA, Wyss-Coray T. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477(7362):90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vina J, Gomez-Cabrera MC, Borras C, Froio T, Sanchis-Gomar F, Martinez-Bello VE, Pallardo FV. Mitochondrial biogenesis in exercise and in ageing. Adv Drug Deliv Rev. 2009;61(14):1369–1374. doi: 10.1016/j.addr.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Wojtowicz JM, Kee N. BrdU assay for neurogenesis in rodents. Nat Protoc. 2006;1(3):1399–1405. doi: 10.1038/nprot.2006.224. [DOI] [PubMed] [Google Scholar]
- Woolley CS, Gould E, McEwen BS. Exposure to excess glucocorticoids alters dendritic morphology of adult hippocampal pyramidal neurons. Brain Res. 1990;531(1–2):225–231. doi: 10.1016/0006-8993(90)90778-a. [DOI] [PubMed] [Google Scholar]