Summary
Prion-like propagation of tau aggregation may underlie the stereotyped progression of neurodegenerative tauopathies. True prions stably maintain unique conformations (“strains”) in vivo that link structure to patterns of pathology. We now find that tau meets this criterion. Stably expressed tau repeat domain indefinitely propagates distinct amyloid conformations in a clonal fashion in culture. Reintroduction of tau from these lines into naïve cells re-establishes identical clones. We produced two strains in vitro that induce distinct pathologies in vivo as determined by successive inoculations into three generations of transgenic mice. Immunopurified tau from these mice re-creates the original strains in culture. We used the cell system to isolate tau strains from 29 patients with 5 different tauopathies, finding that different diseases are associated with different sets of strains. Tau thus demonstrates essential characteristics of a prion. This may explain the phenotypic diversity of tauopathies and could enable more effective diagnosis and therapy.
Introduction
Tauopathies are a diverse group of neurodegenerative diseases defined by accumulation of fibrillar deposits of the microtubule-associated protein tau (MAPT) (Lee et al., 2001). Alzheimer’s disease (AD), the most common tauopathy, affects >30 million people worldwide and will afflict >120 million by 2050 (Holtzman et al., 2011). MAPT mutations cause dominantly inherited tauopathies (Hutton et al., 1998), and most increase the propensity of tau to form amyloids (Barghorn et al., 2000), which are paracrystalline protein assemblies rich in beta-sheet structure (Bonar et al., 1969). Most tauopathy cases are sporadic, with variable clinical and pathological presentation (Lee et al., 2001).
The prion hypothesis posits that pathological aggregates of the mammalian prion protein (PrP) cause infectious, sporadic, and familial neurodegenerative diseases (Prusiner, 1998). In contrast, yeast prions are adaptive and confer phenotypic diversity and rapid evolution of new traits in times of stress (True and Lindquist, 2000). Both yeast and mammalian prions form strains, which are encoded by distinct fibrillar structures (Safar et al., 1998; Toyama et al., 2007). Prion strains determine the incubation periods of disease in humans (Kim et al., 2012) and mice (Legname et al., 2006). In addition, human prion strains are thought to underlie clinical symptoms and pathological presentation (Collinge and Clarke, 2007).
The hypothesis that common neurodegenerative diseases such as AD could be caused by a prion-like mechanism was suggested three decades ago (Prusiner, 1984). Recently, however, experimental work on diverse amyloids has generated new interest (Frost and Diamond, 2010; Guo and Lee, 2014). Human neurodegenerative diseases target unique neural networks (Braak and Braak, 1995; Seeley et al., 2009), an observation most parsimoniously explained by the network-based spread of a toxic agent (Raj et al., 2012; Zhou et al., 2012). Prior studies suggest that tau aggregates spread among cells via templated conformational change (Frost et al., 2009a; Holmes et al., 2013). In vivo work supports this model (Clavaguera et al., 2009; de Calignon et al., 2012; Iba et al., 2013; Kim et al., 2010; Liu et al., 2012) as has similar work with other intracellular amyloids such as α-synuclein (Desplats et al., 2009; Luk et al., 2012) and extracellular amyloids including amyloid β (Meyer-Luehmann et al., 2006).
Bona fide prions are defined by propagation of distinct conformational strains in vivo, and prior studies have hinted at prion-like strain properties of non-PrP human amyloids. For example, amyloid β protein forms at least two distinct, self-propagating fibrillar conformations in vitro (Petkova et al., 2005) and in vivo (Lu et al., 2013). Others have demonstrated propagation in vitro of distinct tau (Frost et al., 2009b; Siddiqua and Margittai, 2010) and α-synuclein conformers (Bousset et al., 2013; Guo et al., 2013; Sacino et al., 2013). While provocative, these prior studies have not demonstrated that non-infectious proteopathic seeds act as true prions. Specifically, it has not been shown that distinct conformations or “strains” are capable of transmission into a living cell/organism, propagation through multiple generations, extraction, and re-introduction to naïve cells/organisms to replicate the same structural phenotype (Collinge and Clarke, 2007). This is important not for semantic reasons, but because if prion mechanisms underlie human disease, only stably propagating strains can account for stereotyped clinical presentation and network spread. In this study we have found that tau acts as a prion by these criteria, and, further, that individual human tauopathies are associated with unique strains.
Results
Homotypic seeding of tau depends on beta-sheet structure
Amino acid sequence disparities impair cross-seeding between PrP moieties from different species, leading to “seeding barriers” (Collinge and Clarke, 2007). To test the fidelity of hetero- vs. homotypic seeding for tau, we expressed several amyloidogenic proteins and exposed them to a variety of fibrillar seeds. Prolonged expression of full-length (FL) tau can be toxic to dividing cells. Thus for tau, we expressed the aggregation-competent core, termed the repeat domain (tau RD; aa 244-372 of the 441 aa FL tau 4R2N) (Wischik et al., 1988). We generated polyclonal HEK293 cell lines stably expressing tau RD-YFP, α-synuclein-YFP, or huntingtin (htt) exon 1(Q25)-YFP (see Figure S1A for construct diagrams). Inclusions did not occur in any line without exposure to exogenous fibrils. However, upon transduction of fibrils (Aβ (1-42), htt exon 1 N17(Q35), α-synuclein, tau RD) using liposomes, we observed homotypic but not heterotypic seeding for each amyloidogenic protein (Figure 1A; Figure S1B for quantification), consistent with sequence-specific templating. Prior reports have indicated that in certain cases, α-synuclein aggregates can cross-seed FL tau (Giasson et al., 2003; Guo et al., 2013; Waxman and Giasson, 2011). Thus, we tested this for both YFP-tagged and untagged versions of FL tau 4R1N P301S. We observed only homotypic seeding and no cross-seeding of tau by α-synuclein or any other amyloid (Figure S1C,D). This is consistent with sequence-specific templating, although we cannot rule out the possibility that different amyloid conformers are capable of heterologous seeding, as has previously been reported (Guo et al., 2013).
Figure 1. Homotypic seeding produces stably propagated tau RD inclusions.

(A) Polyclonal HEK293 lines stably expressing YFP-tagged tau RD, α-synuclein, or htt exon1(Q25) were transduced with buffer, or fibrils of Aβ, Htt, α-syn, or tau RD. Cells were DAPI-stained on Day 6. Only homotypic seeding occurred. See Figure S1A for construct diagrams, Figure S1B for quantification, and Figure S1C,D for similar homotypic seeding using full-length (FL) 4R1N tau P301S.
(B) Polyclonal HEK293 lines stably expressing tau RD-YFP with no mutations (WT), ΔK280 (pro-aggregation), ΔK280/I277P/I308P (2P; anti-aggregation), or P301L/V337M (LM; pro-aggregation) were transduced with either buffer or tau RD fibrils. Upon fibril transduction, all form inclusions, except for 2P.
(C) Tau RD(LM)-YFP cells transduced with either buffer or tau RD fibrils were passaged every two days. On every other passage, the percentage of cells with inclusions was quantified (n=10 fields, each with 150+ cells per condition). Inset highlights inclusion-positive cells at later time points. Error bars represent S.E.M.
(D) At Day 50 following exposure to fibrils, inclusion-positive cells were visible.
(E-F) At Day 3 following exposure to fibrils, tau RD(LM)-YFP cells were diluted sparsely on coverslips and grown for 8 days. Colonies were either 100% inclusion-negative (E) or 100% inclusion-positive (F).
Amyloids typically feature a cross beta-sheet conformation (Bonar et al., 1969). We exploited two proline substitutions (I277P/I308P) in tau that block its ability to enter into this quaternary form (Bergen et al., 2001) to test whether inclusion formation requires this property. Polyclonal HEK293 cell lines stably expressing tau RD-YFP with no mutations (WT); P301L/V337M (LM: pro-aggregation); ΔK280 (pro-aggregation); or ΔK280/I277P/I308P (2P: anti-aggregation) were transduced with tau RD fibrils. All formed inclusions except tau RD(2P)-YFP, confirming that beta-sheet structure is required for tau RD inclusion formation in our model system (Figure 1B).
Stable inheritance of tau RD aggregates
Seeded htt exon 1 (Ren et al., 2009), Sup35NM (Krammer et al., 2009), SOD1 (Münch et al., 2011), and α-synuclein (Bousset et al., 2013) form persistent intracellular inclusions in cultured cells. We tested this for tau RD. We transduced tau RD fibrils or buffer into polyclonal tau RD(LM)-YFP (hereafter, referred to as tau RD) cells, chosen for their superior ability to be seeded relative to tau RD(WT)-YFP, and quantified the percentage of cells with inclusions on every other passage. Transduced fibrils induced tau RD inclusions that persisted >50 days post-exposure (Figure 1C). We hypothesized that the aggregated state was stably inherited, since inclusion-containing cells formed local clusters (Figure 1D). To test this, we sparsely diluted fibril-transduced tau RD cells to isolate individual colonies. These were composed of either 100% inclusion-negative (Figure 1E) or 100% inclusion-positive (Figure 1F) cells, indicating stable inheritance of the aggregated state.
Tau RD propagates conformationally distinct strains
Only prion protein (PrP) (Birkett et al., 2001) and certain fungal prions (e.g. Sup35 [PSI+]) (Derkatch et al., 1996) unequivocally propagate distinct conformational states, or strains, in cell culture. To test the ability of tau RD to propagate distinct conformers, we diluted fibril-transduced monoclonal tau RD cells and isolated individual clones that stably propagated inclusions (Figure 2A). Previous work with the Sup35 protein has indicated that inclusion morphology is a proxy for biochemically distinct yeast prion strains in dividing mammalian cells (Krammer et al., 2009). We thus characterized 20 tau RD clones based on inclusion morphology, numbered in order of isolation. Most (Figure S2A) featured small juxtanuclear inclusions with many nuclear speckles, exemplified by Clone 9 (Figure 2B). Clone 10 alone propagated a single, large juxtanuclear inclusion (Figure 2B). We confirmed that stably propagated tau RD inclusions were amyloids as Clones 9 and 10, but not inclusion-negative Clone 1, bound X-34, a Congo red derivative that stains beta-sheet structures (Figure 2C).
Figure 2. Generation of stably inherited tau RD prion strains.
(A) A monoclonal HEK293 line stably expressing tau RD(LM)-YFP (hereafter referred to as tau RD) was transduced with tau RD fibrils. At Day 3, cells were diluted sparsely in a 10 cm dish. At Day 12, inclusion-positive colonies were identified and picked, amplifying to confluency in separate 10 cm dishes. At Day 30, cells were re-plated for confocal analysis or harvested for subsequent experiments.
(B) Confocal analysis of morphologically distinct tau RD prion strains. Clone 1 does not contain inclusions. Clone 9 contains nuclear speckles and a small juxtanuclear inclusion. Clone 10 features one very large juxtanuclear inclusion and no nuclear speckles. See Figure S2A for other clones.
(C) Clones 1, 9 and 10 were stained with X-34, an amyloid dye. X-34 staining is only observed in Clone 9 and Clone 10, indicating that the propagated aggregates are amyloids.
(D) SDD-AGE demonstrates that Clone 10 features larger aggregates than Clone 9.
(E) Sedimentation analysis was performed on Clones 1, 9, and 10. Pellet (P) was isolated from supernatant (S) by ultracentrifugation. For Clones 9 and 10, supernatant was loaded at a 3:1 ratio to pellet and total (T) to allow clear detection; Clone 1, a 1:1 ratio. Clone 1 has all tau RD in the supernatant, whereas Clone 9 has almost all tau RD in the pellet. Clone 10 has mixed solubility.
(F) Limited proteolysis (pronase) digests all tau RD in Clone 1, but reveals protease-resistant tau RD peptides between 10 and 13 kDa as well as between 20 and 25 kDa in Clone 9 and 10. Unlike Clone 9, Clone 10 digestion produces a doublet, consistent with a distinct conformation.
(G) A split-luciferase assay reports differential seeding efficiency of tau RD prion strains. A polyclonal HEK293 line expressing both tau RD-CLuc and tau RD-Nluc was transduced with lysate from the three clones. Clone 1 does not seed aggregation. Clone 9 seeds robustly, whereas Clone 10 seeds significantly less. Averages of four separate experiments are shown, each read in quadruplicate 48 hours post-transduction (error bars = S.E.M, * = p<0.05, **** = p<0.0001). See Figure S2B for evidence that differences in cell confluency do not account for differences in luminescence.
(H) Inclusion elimination rates differ between clones. After transduction with lysate from Clone 9 or 10, the percentage of cells containing inclusions was quantified on days 4, 17, and 30 (n=10 fields, each with 150+ cells per condition). Cells with inclusions derived from Clone 9 are eliminated more rapidly from the population. Error bars = S.E.M, **** = p<0.0001.
(I) Clone 9-transduced cells grow more slowly. After transduction of stable cells, colonies with inclusions derived from Clone 9 have fewer cells than colonies with inclusions derived from Clone 10. Colonies without inclusions have identical cell numbers (error bars = S.E.M, **** = p<0.0001). See Figure S2C for differences in cell growth rate in tau RD(LM)-HA cells and Figure S2D for LDH toxicity assay in tau RD(LM)-HA background.
(J) Clones 1, 9, and 10 maintain distinctive morphologies after 6 months in culture. See also Figure S2E-H for data indicating that juxtanuclear Clone 10 but not Clone 9 inclusions are aggresomes.
(K) Structural characteristics (limited proteolysis digestion patterns) of strains are propagated with high fidelity over six months.
To characterize the clones biochemically, we first used semi-denaturing detergent agarose gel electrophoresis (SDD-AGE), a method that differentiates strains based on aggregate size (Kryndushkin et al., 2003). Tau RD species from Clone 10 were larger than those propagated by Clone 9 (Figure 2D). Thus, the Clone 10 fibrils may not be as readily fragmented into smaller species (Tanaka et al., 2006). Next, we used sedimentation analysis to differentiate the strains (Tanaka et al., 2006). Clone 1 had entirely soluble tau RD, whereas Clones 9 and 10 had insoluble tau RD (Figure 2E). Clone 10 featured more soluble tau RD than Clone 9. To probe for structural differences, we used limited proteolysis as has been used previously for differentiating PrP conformers (Bessen and Marsh, 1994). Cell lines propagating aggregates (Clones 9, 10) featured pronase-resistant species between 10 and 13 kDa as well as between 20 and 25 kDa in size (Figure 2F). Clone 9 produced a smear between 10-13 kDa, while Clone 10 produced a clear doublet. These studies indicated clear differences in biochemical characteristics of the clones, consistent with distinct strain conformations.
Prion strains often have different seeding efficiencies, which can result in variable incubation times in vivo (Legname et al., 2006). Thus we compared the clones, modifying a pre-existing split-luciferase complementation assay (Naik and Piwnica-Worms, 2007) for use as a tau aggregation sensor (Figure S1A). Clone 1 contained no seeding activity. However, inclusion-containing lines seeded robustly, especially Clone 9, which seeded more than Clone 10 (Figure 2G). Differences in seeding were not an artifact of cell confluency, as determined by normalizing to cell number in seeding experiments (Figure S2B).
Next, we compared the toxicities of Clone 9 and 10. Although Clone 9 lysate initially seeded a greater number of cells, these were rapidly eliminated relative to those induced by Clone 10 (Figure 2H). Furthermore, cells containing Clone 9-derived inclusions grew more slowly than those derived from Clone 10 (Figure 2I). While growth rate of non-transfected HEK293 cells was not affected by inoculation with Clone 9 lysate, growth of tau RD cells was impaired following the same treatment (Figure S2C). This was not seen for Clone 10. Finally, an LDH assay suggested that Clone 9 lysate is toxic to tau RD cells relative to a sham treatment (Figure S2D).
A previous study reported that tau from human brain can induce aggresome structures in vitro (Santa-Maria et al., 2012). Thus, we examined the subcellular localization of inclusions associated with Clones 9 and 10. Based on anti-vimentin stains (Figure S2E), electron microscopy (Figure S2F), and anti-γ-tubulin stains (Figure S2G), we conclude that juxtanuclear Clone 10 inclusions are canonical aggresomes, unlike the inclusions of Clone 9. Intranuclear Clone 9 inclusions did not colocalize with PML bodies (Figure S2H). Thus, Clones 9 and 10 propagate conformationally distinct tau prion strains, with different consequences for the cell. To test the fidelity of strain inheritance, we passaged them continuously for 6 months. Inclusion morphologies (Figure 2J) and limited proteolysis patterns (Figure 2K) associated with Clones 9 and 10 were unaltered. Thus, tau RD prion strains are robust, maintaining their phenotypes indefinitely in cell culture.
Transfer of strain phenotype to naïve cells
To rule out an effect of cell background on strain formation, we transduced Clone 9 and 10 lysates into naïve monoclonal tau RD cells, isolating 6 colonies (A-F) for each (Figure 3A). We evaluated derivative clones (9C was lost in passage) by inclusion morphology (Figure 3B); SDD-AGE (Figure 3C); sedimentation analyses (Figure 3D, S3B); seeding activity (Figure 3E); and limited proteolysis (Figure 3F). In all cases, derivative clones matched their associated progenitors, indicating that tau RD prion strains are encoded by conformation, independent of cell background. Faithful templating into naïve cells also occurred after passive addition of lysates to media (Figure S3A), thus indicating that bypassing physiological uptake is not necessary for templating. With a Tet Off line to control tau RD expression, we demonstrated that the aggregate-positive phenotype can be cured by stopping expression for 7 days and then restarting it (Figure S3C,D).
Figure 3. Tau RD aggregates transfer strain conformations into naïve cells.
(A) Lysates from Clones 9 and 10 were transduced into naïve tau RD-YFP cells and monoclonal inclusion-containing cells were isolated and amplified. Six secondary clones were generated for each condition, but one (Clone 9C) failed to amplify.
(B) Morphologies of primary clones are maintained in secondary cell lines. See also Figure S3A, which demonstrates that this templating of morphology is not dependent on liposome-mediated transduction of lysate.
(C) SDD-AGE of lysates from both primary and secondary clones demonstrates similar aggregate sizes in secondary clones relative to the primary ones. A line separates gels run separately.
(D) Sedimentation analysis was performed as described in Figure 2E. Secondary clones feature similar sedimentation patterns to the clones from which they were derived. For original blots, see Figure S3B.
(E) Split-luciferase complementation demonstrates similar seeding efficiencies in secondary lines vs. parental lines. Averages of four separate experiments are shown, each read in quadruplicate 48-hours post-transduction of lysate (error bars = S.E.M, **** = p<0.0001).
(F) Limited proteolysis shows that all Clone 10 derivatives feature a doublet whereas Clone 9 derivatives are associated with an unresolvable band between 10 and 13 kDa. Clone 9 derivatives feature a more resistant band between 20 and 25 kDa. See Figure S3C,D for reversibility of aggregated state.
(G) Lysates from Clones 9 and 10, but not Clone 1, induce detergent-resistant FL tau P301S-YFP species, which co-localize with AT8 (red) in primary cortical neurons. Clone 9 induces tangle-like structures throughout the soma and neuritic processes. Clone 10 primarily seeds punctate-like structures in the soma. See Figure S3E for data showing that Clone 9 seeds more widespread inclusion formation, Figure S3F for similar results in neurons expressing untagged FL tau P301S, and Figure S3G for images of tangles throughout processes of Clone 9-inoculated neurons.
(H) Clone 9 and Clone 10 lysates containing tau RD(P301L/V337M)-YFP, do not seed inclusion formation in neurons expressing WT FL tau. For evidence that this is due to an asymmetric seeding barrier between FL tau with and without P301 mutations, see Figure S3H.
To rule out an artifact of using artificial truncated tau RD and dividing cell model systems, we examined FL tau inclusion formation in primary cortical neurons. Neurons expressing FL tau P301S-YFP formed detergent-resistant inclusions following treatment with Clone 9 or 10 but not Clone 1 or PBS (Figure 3G). Clone 9 seeded very robustly relative to Clone 10 (Figure S3E). Clone 9 lysate created inclusions throughout the soma and processes of neurons with untagged and YFP-tagged FL P301S tau, whereas Clone 10 lysate primarily seeded inclusion bodies confined to the soma (Figure 3G,S3F,G). Corroborating prior studies (Aoyagi et al., 2007; Miyasaka et al., 2001), we observed a seeding barrier between WT tau and P301 mutants (P301L, P301S). Specifically, aggregates from Clones 9 and 10, which feature both the P301L and V337M mutations, never seeded aggregation in neurons expressing FL tau WT-YFP (Figure 3H) and FL tau WT (no tag) (data not shown). This seeding barrier was confirmed to be asymmetric by using a panel of split-luciferase tau RD mutant pairs (Figure S3H), which demonstrated that WT tau RD can seed all forms of RD (WT, P301L, P301S, P301L/V337M), whereas P301 mutants cannot seed WT.
Tau strains induce unique pathologies in transgenic tau P301S mice
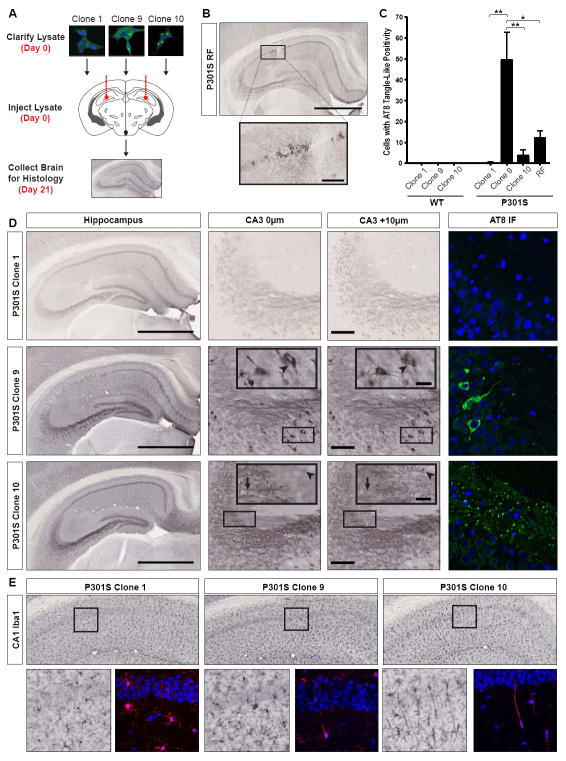
Inoculation of recombinant fibrils into transgenic P301S mice (Yoshiyama et al., 2007), which express a form of mutant tau associated with dominantly inherited tauopathy, rapidly induces pathology within weeks (Iba et al., 2013). Thus, we tested whether tau strains formed in cell culture would have similar effects. We inoculated equivalent amounts of lysate from Clones 1, 9, and 10, as well as recombinant tau RD fibrils (RF) bilaterally into the hippocampi of 3-month old mice (Figure 4A). For all experiments, conditions were gender-matched (Table S1). After 3 weeks, RFs induced tangle-like pathology when assessed by AT8 (Figure 4B,C), an antibody against FL phospho-tau, as previously reported (Iba et al., 2013). Clone 9 and 10 induced unique pathologies, whereas Clone 1 did not cause any detectable abnormality (Figure 4C,D). While Clone 9 induced tangle-like inclusions throughout CA1 and CA3, Clone 10 induced AT8-positive puncta in mossy fiber tracts (Figure 4D). Staining with MC1, an antibody against conformationally-altered tau (Jicha et al., 1997) confirmed these differences (Figure S4A). X-34, an amyloid dye, primarily recognized Clone 9 pathology (Figure S4A), although light staining was observed in CA1 of Clone 10-inoculated mice. Pathological differences could not be explained by differences in the amount of total or insoluble tau RD inoculated (Figure S4E,F). Injected WT mice never developed pathology (Figure S4B), possibly due to a seeding barrier between inoculated tau RD and WT murine tau (Figure 3H, S3E,H).
Figure 4. Clone 9 and 10 induce unique tau and microglia pathology in P301S mice.
(A) Lysates (10 μg total protein) were injected bilaterally into the hippocampi of 3-month P301S and wild-type (WT) mice. 21 days post-injection, left hemispheres were collected for histology; right hemispheres for homogenization. See Figure S1 for description of mice used in all experiments.
(B) Recombinant tau RD fibrils (RF) induce tangle-like, AT8-positive tau pathology near the injection site in CA1 (scale bars: hippocampus – 1 mm; inset – 100 μm).
(C) Quantification of tangle-like, AT8-positive cell bodies within the hippocampus (CA1 and CA3) of WT and P301S mice. P301S mice injected with Clone 9 lysate have significantly more AT8-positive cell bodies than those injected with Clone 1, Clone 10, or RF (error bars = S.E.M., * = p<0.05, ** = p<0.01). WT mice do not develop pathology after injection.
(D) P301S mice were inoculated with Clone 1, Clone 9, or Clone 10 lysate. Representative whole hippocampus images are shown with the corresponding CA3 z-stacks. Arrowheads in Clone 9 CA3 insets highlight an AT8-positive cell body that can be seen throughout both z-stack images. The arrow and arrowhead in Clone 10 CA3 insets each represent a different AT8-positive puncta that is visible in only one z-stack plane (scale bars: hippocampus – 1 mm; CA3 – 100 μm; CA3 inset and AT8 IF – 25 μm; n=3-4 per clone). See Figure S4A for MC1 and X-34 staining, Figure S4B for lack of pathology in inoculated WT mice.
(E) Iba1 staining of microglia in CA1 of inoculated P301S mice indicates that only Clone 10 induces the formation of rod microglia, which extend highly polarized processes into CA1. See Figure S4C for columns of rod microglia in these animals and Figure S4D for absence of these microglia in Clone 10- inoculated WT animals. See Figure S4E and Figure S4F for data indicating that identical amounts of total and insoluble tau were used in inoculations.
P301S mice inoculated with Clone 10 uniquely accumulated elongated Iba1-positive rod microglia (Figure 4E), which aligned end-to-end parallel to CA1 pyramidal axons (Figure S4C). Such unique coupling of rod microglia has been observed in a rodent traumatic brain injury model, and may be protective for injured axons (Ziebell et al., 2012). WT mice inoculated with Clone 9 and 10 did not feature this pathology, indicating that endogenous human P301S tau is required for this induced microglial phenotype (Figure S4D).
Tau strains are stably propagated through multiple generations in mice
Prions can be stably passaged in vivo (Bruce et al., 1994). Thus, we performed serial inoculation of brain homogenates into naïve P301S mice (Figure 5A). Brain homogenate from WT or P301S mice inoculated with Clone 1, 9 or 10 (termed generation G0) was inoculated into naïve P301S mice (generation G1). After 28 days, brains were collected for histology and biochemistry, and the process was repeated in a second round of P301S mice (generation G2). Immunohistochemistry demonstrated identical pathology for each generation of mice: Clone 9 serial propagation induced AT8- positive, tangle-like pathology in CA1 and CA3 regions, whereas Clone 10 serial propagation induced AT8-positive puncta in the mossy fiber tracts of the hippocampus (Figure 5B,C, S5A). Clone 1 induced no pathology in any generation (Figure 5B, S5B). Homogenate from WT mice inoculated with the original cell tau strains (G0) did not produce pathology upon passage into P301S mice (Figure S5C). Therefore, pathology observed in G1 and G2 cannot be due to residual tau RD seeds from the original inoculum, and tau prions propagate unique phenotypes for multiple passages in vivo.
Figure 5. Tau strains passage stably through multiple generations of P301S mice.
(A) Lysates (10 μg protein) were injected bilaterally into the hippocampi of 3-month P301S mice (Generation 0/G0). 21 days post-injection, brains were collected for histology and homogenization. Hippocampal homogenate (10 μg) was then bilaterally inoculated into a new round of 3-month P301S mice (Generation 1/G1) followed by a 28-day incubation before the process was repeated for a new cohort of 3-month P301S mice (Generation 2/G2). At G0 and G2, hippocampal homogenates were IPed (anti-tau 8.5; epitope = aa 25-30; outside RD region) and inoculated into the original tau RD-YFP line to test the fidelity of strain inheritance (G0 and G2 clones). For each cohort, n=3-4 animals.
(B-C) AT8 staining (DAB = B and immunofluorescence = C) reveals that the morphological phenotypes of phosphorylated tau inclusions breed true through multiple generations of tau P301S mice. See Figure S5A for images of whole hippocampi, Figure S5B for images of Clone 1 AT8 immunofluorescence, and Figure S5C for data indicating that strain passage is not due to residual tau RD seeds remaining in diluted inoculate.
In vivo tau strains maintain phenotypes upon passage back into cells
To conclusively test whether tau strains are biochemically stable after passage in vivo, we isolated FL P301S tau from micro-dissected hippocampi of injected mice (G0), using a monoclonal antibody (HJ8.5) that binds an epitope present in FL tau but not tau RD (Yanamandra et al., 2013). We assessed seeding activity in G0 samples by split-luciferase complementation and inclusion counts. Only hippocampi from P301S mice (G0) injected with tau RD aggregates contained seeding activity (Figure 6A, S6A). This did not correlate with the amount of immunoprecipitated tau (Figure S6D). WT mouse hippocampi never seeded, regardless of the inoculum. We next tested whether the strains introduced into G0 mice could be re-isolated in tau RD cells. Scoring of single colonies based on morphology (containing or lacking nuclear speckles) suggested that strains were unaltered following a single passage through mice (Figure S6B,C). To further confirm this, a single representative colony associated with each mouse was blindly selected and amplified. All G0-Clone 9 and G0-Clone 10 samples recapitulated the morphologies of the original Clones 9 and 10 (Figure 6B). Limited proteolysis patterns (Figure 6C) and seeding propensities were also identical (Figure 6D).
Figure 6. Strains transfer faithfully to cell culture after passage through Generation 0 and Generation 2 mice.
(A) IPed material was transduced into tau RD-YFP cells prior to passage onto coverslips. At 96 hours, cells were fixed. Only Clone 9-, Clone 10-, and RF-inoculated mice seed inclusions robustly. WT mouse homogenates never seed aggregation. Ten fields, each with 100+ cells, were analyzed per brain, and averages were collapsed within cohorts (error bars = S.E.M., **** = p<0.0001). See also Figure S6A for split-luciferase complementation data.
(B) Inclusion morphologies are maintained following passage through P301S mice (G0). IPed FL tau from individual P301S mice inoculated with Clone 9 or Clone 10 was transduced into tau RD-YFP cells, and a single representative clone per mouse was isolated and amplified. All G0-derived clones continue to propagate the original phenotypes. See also Figure S6B,C for quantification of colony morphologies prior to monoclonal cell line isolation and Figure S6D for quantification of total IPed tau used in G0 experiments.
(C) Limited proteolysis reveals that G0 clones feature similar banding patterns to the original parental lines, with G0-Clone 10 featuring a doublet between 10-13 kDa (vs. smear for G0-Clone 9) and a band between 20 and 25 kDa that is slightly smaller than G0-Clone 9 bands.
(D) Split-luciferase complementation demonstrates similar seeding efficiencies in G0 clones relative to original parental lines. Averages of four separate experiments are shown, each read in quadruplicate 48- hours post-transduction of lysate (error bars = S.E.M, **** = p<0.0001).
(E). IPed material from pooled G2 mice was transduced into naïve tau RD-YFP cells prior to passage onto coverslips. At 96 hours, cells were fixed. Seeding of inclusion formation is significantly greater for G2-Clone 9 and G2-Clone 10 mice than G2-Clone 1 mice. G2-Clone 1 tau induces inclusions on rare occasions (~1% of cells). Seeding is specific to tau as IgG-precipitated material never seeds. Ten fields, each with 150+ cells, were analyzed per condition (error bars = S.E.M., * = p<0.05, **** = p<0.0001). See also Figure S6E for split-luciferase complementation data.
(F) Inclusion morphologies are maintained following passage through three generations of mice. IPed full-length tau from pooled G2 homogenates was transduced into tau RD-YFP cells, and 12 clones per cohort were isolated. Representative examples are shown. The two clones boxed in red feature similar limited proteolysis digestion patterns and seeding ratios to each other, which are unique from all 22 other clones. See Figure S6F for quantification of colony morphologies prior to monoclonal cell line isolation and Figure S6G for images of all 24 clones.
(G) Limited proteolysis reveals that G2 clones feature similar banding patterns to their parental lines, with G2-Clone 10 featuring a doublet between 10-13 kDa (vs. smear for G2-Clone 9) and a band between 20 and 25 kDa that is slightly smaller than G2-Clone 9 digests. Two clones (boxed in red), one for each cohort, are unique in featuring bands at 15 and 25 kDa.
(H) Split-luciferase complementation demonstrates similar seeding efficiencies in G2 clones relative to original parental lines. Seeding ratios were averaged across clones, each of which was read in quadruplicate 48-hours post-transduction of lysate (error bars = S.E.M, **** = p<0.0001). Boxed in red are two outlier clones (9G and 10D), which also feature unique inclusion morphologies and limited proteolysis digestion patterns.
Similar experiments were performed following the third passage (G2). IPed tau from pooled G2-Clone 9 and G2-Clone 10 homogenates seeded far more strongly than G2-Clone 1 homogenates (Figure 6E). IgG-precipitated material did not seed (Figure 6E, S6E), and IPed tau seeded as strongly as crude homogenate in a split-luciferase assay (Figure S6E ). Tau alone thus accounts for the seeding activity reported in these assays.
Next, we introduced IPed material from G2 mice into tau RD reporter cells and scored colonies based on morphology prior to isolation of monoclonal lines. G2-Clone 9 colonies almost exclusively featured nuclear inclusions, whereas virtually all G2-Clone 10 colonies lacked them (Figure S6F). The rare inclusion-positive colonies associated with G2-Clone 1 also featured nuclear inclusions, which suggested that some of the G2-Clone 10 colonies containing nuclear inclusions could arise from an intrinsic P301S-derived strain. Monoclonal strains (n=12) were blindly selected for each G2 cohort. In all but one case (G2-Clone 10D), inclusion morphologies matched that of the original inoculate (Figure 6F, S6G). For both Clone 9 and 10 cohorts, 11 of 12 clones matched their parental counterpart based on both limited proteolysis (Figure 6G) and seeding activity (Figure 6H). Intriguingly, the two outliers (G2-Clone 9G, G2-Clone 10D) had identical proteolysis patterns and seeding ratios, which were unique from those of all other clones. We speculate that these clones result from an intrinsic strain within 4-month P301S mice. We conclude that tau prion strains are stable across numerous passages through cells and animals.
Spread of tau pathology to distant, synaptically connected regions
After inoculation with recombinant tau fibrils, pathology can develop in synaptically connected regions (Iba et al., 2013). Our preliminary observations indicated that serial inoculations with Clone 9 induced pathology in the ipsilateral entorhinal cortex (EC) (data not shown). To test for spread, we performed a final inoculation (G3) into the left hippocampus of P301S mice. After five weeks, G3-Clone 9 mice had AT8-positive pathology in regions that project to or from the hippocampus (Figure 7A) including ipsilateral and contralateral EC, retrosplenial cortex (RSp), and contralateral hippocampus (Figure 7B-D, S7A) (Andersen, 2007; van Groen et al., 2003). Ipsilateral EC had robust pathology in layers II/III, whereas contralateral EC pathology occurred in deeper cortical layers, suggesting spread along defined anatomical connections (van Groen et al., 2003). Furthermore, pathology was observed in ipsilateral subiculum and dentate gyrus (Figure S7B). In contrast, G3-Clone 1 brain did not show AT8-positivity above baseline (Figure 7B,D). Overt spread was not observed in G3-Clone 10 mice (data not shown), perhaps due to its decreased seeding ability (Figure 2G,S3E). A heat-map summarizes the brain regions with enhanced AT8-positive pathology in G3-Clone 9 mice (Figure 7C). These results agree with previous work suggesting that seeded intracellular amyloids spread along discrete neural networks (de Calignon et al., 2012; Iba et al., 2013; Liu et al., 2012; Luk et al., 2012; Zhou et al., 2012). We cannot completely exclude the possibility that this was due to trans-synaptic spread of inoculum.
Figure 7. Anterograde and retrograde spread of pathology to synaptically connected regions in Generation 3-Clone 9 (G3-Clone 9) mice.
(A) Schematic of known projections to and from the hippocampus (DG = dentate gyrus; MEC/LEC = medial and lateral entorhinal cortices; MF = mossy fibers; RSp = retrosplenial cortex; Sub = subiculum).
(B) Representative images of AT8 staining in the hippocampi of G3 mice inoculated with 10 μg of G2 brain homogenate. Spread of Clone 9 pathology to the contralateral hippocampus is evident. See Figure S7A for whole brain slices.
(C) Summary of pathology present in G3-Clone 9 mice. Gradient represents semi-quantitative analysis of neurofibrillary tangle-like AT8 cell body positivity observed in each region (PPA = posterior parietal association area) both 2.5 and 3.0 mm posterior to bregma.
(D) AT8 histopathology observed in brain regions with known projections to/from the hippocampus. Ipsilateral AT8 pathology is observed in the EC and appears in cortical layers II-III, whereas contralateral pathology is observed in deeper layers of the EC. Pathology is also observed in the retrosplenial cortex, especially ipsilateral to the injection site. See Figure S7B for subiculum and dentate gyrus images.
Intra-patient and inter-disease phenotypic diversity in the tauopathies
It has been hypothesized that conformationally distinct tau prion strains may be associated with individual tauopathies (Clavaguera et al., 2013b; Frost and Diamond, 2010), and a recent study found that inoculation of transgenic human tau mice with brain homogenates from patients with different tauopathies recapitulates certain pathological features of the diseases (Clavaguera et al., 2013a). To examine whether inclusion morphology is a reasonable indicator of distinct strains, we first used our cell model to examine brain homogenates from three individuals with clinically distinct, pathologically verified tauopathies (all patient samples obtained from the Neurodegenerative Disease Brain Bank at UCSF): Alzheimer’s disease (AD), corticobasal degeneration (CBD), and argyrophilic grain disease (AGD). We transduced immunoprecipitated (IP) or crude (CR) homogenate into the monoclonal Tet Off HEK293 cell line (Figure 8A), used for its relatively high tau RD expression and greater seeding efficacy, and characterized resulting colonies morphologically and biochemically (Figure S8A-I). Each brain induced a unique inclusion morphology, independent of the transduction method (IP vs. CR) (Figure S8A-D). By analyzing three representative clones derived from each brain by sedimentation analysis (Figure S8E), seeding (Figure S8F,G), and limited proteolysis (Figure S8H,I), we concluded that morphology reliably differentiates biochemically distinct strains. Next, we expanded our analysis to include IPed tau from patients with AD (n=6); AGD (n=6); CBD (n=6); Pick’s disease (PiD, n=5); and progressive supranuclear palsy (PSP; n=6) (Table S2). Excepting PiD, a three-repeat tauopathy, these are predominantly four-repeat (AGD, CBD, PSP) or mixed-repeat (AD) tauopathies that differ in the morphology and distribution of neuronal and glial tau inclusions (Lee et al., 2001). We transduced IPed tau from each sample into the monoclonal Tet Off cell line and isolated clones with inclusions (Figure 8A). We identified six morphological phenotypes: 1) No seeding; 2) Toxic (all cells with inclusions died and clones could not be isolated); 3) Mosaic (unstable prion strain); 4) Ordered; 5) Disordered; 6) Speckles (Figure 8B). We blindly scored all clones based on tau RD inclusion morphology. This revealed distinct strain compositions across the diseases (Figure 8C). AD patient samples revealed remarkable homogeneity, suggesting a predominant strain. Other disorders revealed inter-patient variation. Some patients featured homogeneous strain composition (e.g. certain patients with AGD, PSP), whereas others exhibited considerable heterogeneity. With few exceptions (e.g. AD1-AD4; AGD2; CBD5; PiD3), most patient samples produced two or more strains. The range of phenotypes associated with single patients suggests a diversity of patient-derived tau prion strains. Since the cell-based strain isolation system can likely amplify only a subset of strains, these data suggest that a disease-associated ensemble or “cloud” of conformations exists within individual patients. Nevertheless, certain tauopathies can be differentiated by their strain composition.
Figure 8. Diverse tau prion strains within patients and across diseases.

(A) Schematic illustrating methods used to generate patient-derived tau RD prion strains in a monoclonal Tet Off-tau RD-YFP line. See Figure S8 for data indicating that different inclusion morphologies are associated with different biochemical and seeding properties.
(B) Morphological phenotypes associated with tau RD prion strains induced by patient material: no seeding, toxic, mosaic, ordered, disordered, speckles. Representative examples are shown.
(C) IPed tau from 29 patient samples (AD = Alzheimer’s disease, AGD = argyrophilic grain disease, CBD = corticobasal degeneration, PiD = Pick’s disease, PSP = progressive supranuclear palsy) was transduced into tau RD-YFP cells (Tet Off) and as many inclusion-positive clones as could be identified for each patient sample were blindly picked and amplified. Once confluent in 10 cm dishes, morphological phenotypes were scored by a separate blinded experimenter. See Table S2 for numerical values, patient-related information, and tissue origin.
Discussion
Many papers describing “prion-like” behavior of proteins associated with neurodegenerative diseases have been published in the last several years. In the case of tau, fibrils transmit its aggregated state from the outside to the inside of a cell (Frost et al., 2009a; Holmes et al., 2013), suggesting that this mechanism could account for the stereotyped progression of tauopathies. This model of disease was subsequently supported in vivo with reports of trans-synaptic spread of pathology (de Calignon et al., 2012; Kim et al., 2010; Liu et al., 2012) and protein-only induction of tau inclusions (Iba et al., 2013). Work with other intracellular amyloids (Desplats et al., 2009; Holmes and Diamond, 2012; Münch et al., 2011; Ren et al., 2009) has suggested that prion-like transmission can explain the progression of many neurodegenerative diseases.
Whether or not various non-infectious amyloids are “true” prions has become a contentious subject of debate. Some define prions as being capable of inter-organism transmission of pathology and by the ability to survive freely in the environment (Aguzzi and Rajendran, 2009). To date, there exists no evidence that this definition can be applied to proteins other than PrP. This restrictive definition, based on early research into prion diseases such as kuru and scrapie, potentially ignores a rich biology that mechanistically unites many common diseases. Importantly, we now know that the vast majority of human prion diseases have non-infectious etiology, and that their great phenotypic heterogeneity can be attributed to strains (Collinge and Clarke, 2007). With respect to prion-like intracellular amyloids in humans, recent data indicate that homogenates from distinct tauopathies may reproduce certain pathological features of the diseases in transgenic mice, which is consistent with strain behavior (Clavaguera et al., 2013a). Other studies explicitly suggest the existence of α-synuclein strains, based on the production of different α-synuclein conformers in vitro (Bousset et al., 2013; Guo et al., 2013; Sacino et al., 2013). However, to account for phenotypic diversity at a systems level, a prion strain must replicate with remarkable reliability for extended periods of time. A stringent test of this is to ensure that the strain is stable, isolatable, and replicates its phenotype through living systems with high fidelity (Bruce et al., 1994; Prusiner, 1998). Until now, these characteristics have not been linked to a mammalian protein other than PrP. Based on these criteria, our data strongly suggest that tau should be considered a bona fide prion. Fittingly, we also find that different tauopathies are associated with different strains. This has direct implications for understanding the phenotypic diversity of tauopathies.
Tau as a prion in cell culture and mice
We began this work by establishing a monoclonal HEK293 cell line that stably expresses the tau repeat domain fused to YFP. In the absence of tau aggregate exposure, these cells propagate only tau RD monomer (“naïve” cells). Induction of aggregation with recombinant fibrils, however, created clonal lines (Clone 9 and Clone 10) that indefinitely propagate unique aggregate structures, or strains, from mother to daughter cells. These strains differ with respect to inclusion morphology, aggregate size, sedimentation profile, seeding capacity, protease digestion patterns, toxicity, and subcellular localization. Importantly, these properties are cell-independent, as we re-created the strains by protein transfer into naïve cells. Furthermore, the distinct inclusion morphologies we observed may represent specific cellular responses to different aggregate conformations, consistent with their unique patterns of compartmentalization. The cell culture system established here may thus prove useful to detect, propagate and characterize additional tau prion strains, as well as to understand the cellular mechanisms that govern strain replication, subcellular localization, degradation, and toxicity.
In vivo, we found that strains 9 and 10 induce unique pathological phenotypes in transgenic P301S mice. Moreover, Clone 10 lysate uniquely results in the formation of rod-shaped microglia, which indicates that distinct tau conformers initiate different physiological responses in vivo. More remarkably, we report that the morphological phenotypes breed true through multiple generations of mice, the first time this has been observed for an amyloid other than PrP. We recognize that pathological phenotypes can be prone to bias in detection. Thus, we passaged strains back to naïve tau RD-YFP cells, conclusively demonstrating the robust inheritance of tau conformations. This data also indicates that the repeat domain is sufficient to encode strains that are unaltered by templating of their structure to FL tau. Therefore, the reported cell model is useful for detecting and propagating physiologically relevant tau prion strains. Finally, using unilateral inoculation of Clone 9 lysate, we show that tau aggregation propagates along known anatomical connections, supporting conclusions of previous studies (de Calignon et al., 2012; Iba et al., 2013; Liu et al., 2012). More importantly, however, these cellular and in vivo studies indicate for the first time that a mammalian protein amyloid other than PrP templates itself with high fidelity through living systems.
Tau prion strains in human tauopathy brains
Knowing that tau acts as a prion in experimental models, we examined whether this concept could explain phenotypic diversity observed in tauopathies. Brain samples from three patients with distinctive tauopathies induced diverse self-propagating tau prion strains in culture. Our initial work with these novel strains indicated that inclusion morphology is a reliable surrogate for more labor-intensive biochemical characterization. This led us to assess the morphological phenotypes of tau strains derived from numerous patients (n=29) across a spectrum of tauopathies (AD, AGD, CBD, PiD, PSP). Each of the diseases was associated with several cellular inclusion morphologies, although certain diseases (AD, CBD, PiD) are more homogeneous than others (PSP, AGD). It is noteworthy that AD pathology is characteristically more uniform than other tauopathies (Duyckaerts et al., 2009; Feany et al., 1996), and the tau strains isolated from AD brains were by far the most homogeneous. The isolation of multiple conformers from individuals suggests that a tau aggregate ensemble exists within each person, and that standard methodologies (e.g. histopathology, inoculation into mice, protease digestion) will be insufficient for a nuanced understanding of this conformational complexity. Similar to what has been reported for PrP amyloids (Collinge and Clarke, 2007; Li et al., 2010), we speculate that these clouds of tau conformers are prone to selective pressures at the cellular level, which may have implications for therapies that target extracellular tau (Holmes et al., 2013; Yanamandra et al., 2013).
Although we have now succeeded in categorizing multiple distinct strains, the cell-based isolation method can only detect those that successfully template to tau RD-YFP and propagate without overt cellular toxicity. The inability to re-select Clone 9 derivatives in the Tet-Off background illustrates this problem. On the other extreme, strains that do not propagate with high fidelity may be lost prior to clonal selection. For example, the strains present in several AGD and PiD samples were not stable in cell culture, making detailed characterization of these strains difficult using our model system. Furthermore, it is likely that seeding barriers between tau from patient brain (consisting of various tau isoforms and post-translationally modified species) and tau RD in cell culture limits the strains we can detect. Our observation of an asymmetric seeding barrier between P301 mutants and WT tau underscores this limitation, as does recent work indicating similar barriers between three-repeat (3R) and four-repeat (4R) tau (Dinkel et al., 2011). Despite some limitations, the model system presented here has many advantages over standard animal inoculations, as it is less resource-intensive and can parse multiple conformations from a single isolate. Finally, knowledge of the existence of multiple strains in vivo may allow us to characterize them on molecular terms and diagnose patients with much greater precision, possibly by determining structures and conformational epitopes specific to individual diseases. This could help facilitate therapeutic strategies tailored toward the underlying protein pathology.
Expanding the spectrum of prion diseases
Our data, along with cell culture (Holmes and Diamond, 2012), pathological (Clavaguera et al., 2013b) and imaging (Greicius and Kimmel, 2012) studies are consistent with the model of cell-cell “transmission” of neurodegenerative diseases throughout the nervous system. Our finding of multiple self-propagating conformations in experimental and patient-derived tau preparations suggests that tau should be defined as a prion, because it encodes self-catalyzing conformational information that it propagates indefinitely with high fidelity. Importantly, however, there is no evidence to suggest that AD or other tauopathies are infectious in the classical sense, as they are not known to be communicable between individuals. The infectious property of PrPSc might reflect its anomalous biochemical stability or expression profile, whereas a host of other cell biological and biophysical properties, especially the ability to encode self-propagating conformers, will more appropriately unify the growing family of “prionlike” proteins. Indeed, the vast majority (>95%) of human prion diseases appear to be genetic or sporadic, indicating that infectivity should not be a restrictive criterion. We predict that strains associated with distinct clinical phenotypes will also be identified for synucleinopathies and ALS/FTLD spectrum disorders, both of which feature diversity in pathological presentation (Halliday et al., 2011; Van Langenhove et al., 2012). Understanding disparate amyloid neurodegenerative diseases in light of this model should create new possibilities for common diagnostic and therapeutic approaches.
Experimental Procedures
Statistical Analysis
Unless explicitly stated, all statistical analyses used one-way analysis of variance with Bonferroni’s multiple comparison test.
Liposome-mediated transduction of fibrils, lysate, brain homogenate
Cell lines were plated at 250,000 cells/well in 12-well plates. 24 hours later, fibrils or lysate were combined with OptiMEM (Gibco) to a final volume of 100 μL. 96 μL OptiMEM and 4 μL lipofectamine-2000 (Invitrogen) was then added to a final volume of 200 μL. After 20-minutes, liposome preparations were added to cells. 18 hours later, cells were re-plated in wells of a 6-well plate. For more details, see Extended Experimental Procedures.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
SDD-AGE was performed as previously described (Kryndushkin et al., 2003) with minor modifications. Cell pellets lysed in 0.05% Triton X were clarified by sequential centrifugations (500xg, 1000xg). Low-SDS 1.5% agarose gels were prepared by dissolving agarose in buffer G (20 mM Tris-Base, 200 mM glycine, in ddH2O) with 0.02% SDS. For each condition, 5 μg of clarified cell lysate was incubated with 0.02% SDS sample buffer for 7 minutes prior to loading. SDD-AGE was run in Laemmli buffer (Buffer G with 0.1% SDS). Protein was transferred to Immobilin P (Millipore). Membranes were probed for tau using rabbit polyclonal anti-tau ab64193 (1:4000, AbCam) and counter-probed with goat anti-rabbit HRP (1:4000, Jackson Immunotherapy). For more details, see Extended Experimental Procedures.
Sedimentation analysis
Clarified cell lysate was centrifuged at 100,000xg for 1 hour. Supernatant was placed aside and the pellet was washed with 1.5 mL PBS prior to ultracentrifugation at 100,000xg for 30 minutes. The supernatant/wash was aspirated and the pellet was re-suspended by boiling in RIPA buffer with 4% SDS and 100 mM DTT. Bradford assay (Bio-Rad) with BSA standard curve was used to normalize all protein concentrations. Samples were ran on 4-15% SDS-PAGE gels (Bio-Rad) and protein was transferred to Immobilin P (Millipore). Membranes were probed for tau as described above. For more details, see Extended Experimental Procedures.
Split-luciferase complementation assay
Polyclonal HEK293 cells stably expressing tau RD-Cluc and tau RD-Nluc were plated at 240,000 cells/well in 12-well plates 24-hours prior to cell lysate transduction. Clarified cell lysate was prepared as described above. 20 μg (10 μL) of each cell lysate was diluted with 90 μL OptiMEM (Gibco) and incubated with 96 μL OptiMEM and 4 μL lipofectamine-2000 (Invitrogen) for 20 minutes. Liposome preparations were then added to cells and 18 hours later, cells were re-plated in quadruplicate in a 96-well plate. 24 hours later, media was aspirated from wells and replaced with luciferin solution (150 μg/mL D-luciferin potassium salt, Gold Biosciences, in Dulbecco’s phosphate-buffered saline, Gibco). Cells were incubated with luciferin solution for 3 minutes at 37°C prior to reading luminescence with a Tecan M1000 fluorescence plate reader. For more details, see Extended Experimental Procedures.
Protease digestion
Pronase (Roche) was diluted in PBS to a final concentration of 1 mg/mL and single-use aliquots were stored at -80°C. Clarified cell lysate was pre pared as previously described (see SDD-AGE) and protein concentrations were normalized to 1.7 μg/μL. 17 μg (10 μL) of cell lysate was added to 10 μL of pronase at a concentration of 100 μg/mL (diluted in PBS) for a final volume of 20 μL. Cell lysates were digested at 37°C for one hour. Reactions were quenc hed by addition of 20 μL 2x sample buffer (final SDS concentration of 1%) and boiling for 5 minutes. 15 μL of each sample was ran on a 10% Bis-Tris NuPAGE gel (Novex by Life Technologies) and protein was transferred to Immobilin P (Millipore). Membranes were probed for tau as described above. For more details, see Extended Experimental Procedures.
Animals and hippocampal injections
Transgenic mice expressing FL human tau 4R1N P301S under the murine prion promoter (Yoshiyama et al., 2007) were maintained on a B6C3 background. P301S and non-transgenic littermates were anesthetized with isoflurane and were bilaterally injected into the hippocampus (from bregma: −2.5 mm posterior, +/-2 mm lateral, −1.8 mm ventral) with either 2 μL of 5 μg/μL clarified lysate/homogenate or 2 μL of 2.5 μg/μL recombinant tau RD fibrils as previously described (Devos and Miller, 2013). For all experiments, conditions were gender-matched. (Table S1).Unilateral injections were used for G3 spread experiments. For more details, see Extended Experimental Procedures.
Histology and Immunohistochemistry
50 μm sections were taken through the entire left hemisphere using a freezing microtome. For DAB stains, brain slices were incubated with indicated antibodies overnight at 4°C. Slices were then counter-stained with the appropriate secondary. Using the VECTASTAIN Elite ABC Kit (Vector Labs), slices were then incubated at room temperature for 30 minutes, followed by DAB development using the DAB Peroxidase Substrate Kit with the optional nickel addition (Vector Labs). Histological images and z-stacks were captured using the Olympus Nanozoomer 2.0-HT (Hamamatsu) and analyzed with the NDP viewer software (Hamamatsu). For immunofluorescence stains, slices were incubated in blocking solution with indicated primary antibody overnight at 4°C, f ollowed by appropriate secondary labeling. For more details, see Extended Experimental Procedures.
Supplementary Material
Highlights.
Tau stably propagates multiple amyloid conformations (strains) in dividing cells
Strains induce unique pathologies in transgenic mice
Strains can be passaged through multiple generations of mice and back to cells
Different tauopathies are associated with different sets of strains
Acknowledgments
We thank John Cirrito, Peter Davies, David Holtzman, Paul Kotzbauer, Jeffrey Milbrandt, David Piwnica-Worms for reagents. We thank Dorrie Young for help with figures and Jan Bieschke, William Dauer, Jen Dulle, Bess Frost, Brandon Holmes, Suzanne Schindler, Kevin Stein, and Heather True for critiques. Our work was funded by the Tau Consortium, Muscular Dystrophy Association, American Health Assistance Foundation, Ruth K. Broad Foundation, Harrington Discovery Institute, and the NIH (1F31NS086251, DWS; 1R01NS071835, MID; 1R01NS078398, TMM). This work was supported by the Hope Center Alafi Neuroimaging Lab and a P30 Neuroscience Blueprint Interdisciplinary Center Core award to Washington University (P30NS057105). Human tissue samples were provided by the Neurodegenerative Disease Brain Bank at the University of California, San Francisco, which receives funding support from NIH grants P01AG019724 and P50AG023501, the Consortium for Frontotemporal Dementia Research, and the Tau Consortium.
Footnotes
Conflict of Interest: M.I.D. acknowledges a potential conflict of interest in that he is a co-inventor of antibodies used in this study (HJ9.3 and HJ8.5) that have been licensed by C2N Diagnostics through an agreement with Washington University in St. Louis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Andersen P. The Hippocampus Book. Oxford University Press; 2007. [Google Scholar]
- Aoyagi H, Hasegawa M, Tamaoka A. Fibrillogenic nuclei composed of P301L mutant tau induce elongation of P301L tau but not wild-type tau. J Biol Chem. 2007;282:20309–20318. doi: 10.1074/jbc.M611876200. [DOI] [PubMed] [Google Scholar]
- Barghorn S, Zheng-Fischhöfer Q, Ackmann M, Biernat J, Bergen von M, Mandelkow EM, Mandelkow E. Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry. 2000;39:11714–11721. doi: 10.1021/bi000850r. [DOI] [PubMed] [Google Scholar]
- Bergen von M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow EM, Mandelkow E. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J Biol Chem. 2001;276:48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkett CR, Hennion RM, Bembridge DA, Clarke MC, Chree A, Bruce ME, Bostock CJ. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. Embo J. 2001;20:3351–3358. doi: 10.1093/emboj/20.13.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonar L, Cohen AS, Skinner MM. Characterization of the amyloid fibril as a cross-beta protein. Proc Soc Exp Biol Med. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- Bousset L, Pieri L, Ruiz-Arlandis G, Gath J, Jensen PH, Habenstein B, Madiona K, Olieric V, Böckmann A, Meier BH, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. doi: 10.1038/ncomms3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–8. doi: 10.1016/0197-4580(95)00021-6. discussion278–84. [DOI] [PubMed] [Google Scholar]
- Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Transmission of bovine spongiform encephalopathy and scrapie to mice: strain variation and the species barrier. Philos Trans R Soc Lond, B, Biol Sci. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci U S a. 2013a doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Lavenir I, Falcon B, Frank S, Goedert M, Tolnay M. “Prionlike” templated misfolding in tauopathies. Brain Pathol. 2013b;23:342–349. doi: 10.1111/bpa.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suárez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Lee H-J, Bae E-J, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee S-J. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S a. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos SL, Miller TM. Direct intraventricular delivery of drugs to the rodent central nervous system. J Vis Exp. 2013:75. doi: 10.3791/50326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel PD, Siddiqua A, Huynh H, Shah M, Margittai M. Variations in filament conformation dictate seeding barrier between three- and four-repeat tau. Biochemistry. 2011;50:4330–4336. doi: 10.1021/bi2004685. [DOI] [PubMed] [Google Scholar]
- Duyckaerts C, Delatour B, Potier M-C. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 2009;118:5–36. doi: 10.1007/s00401-009-0532-1. [DOI] [PubMed] [Google Scholar]
- Feany MB, Mattiace LA, Dickson DW. Neuropathologic overlap of progressive supranuclear palsy, Pick’s disease and corticobasal degeneration. J Neuropathol Exp Neurol. 1996;55:53–67. doi: 10.1097/00005072-199601000-00006. [DOI] [PubMed] [Google Scholar]
- Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009a;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Ollesch J, Wille H, Diamond MI. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem. 2009b;284:3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM-Y. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300:636–640. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Kimmel DL. Neuroimaging insights into network-based neurodegeneration. Curr Opin Neurol. 2012;25:727–734. doi: 10.1097/WCO.0b013e32835a26b3. [DOI] [PubMed] [Google Scholar]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct α-Synuclein Strains Differentially Promote Tau Inclusions in Neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday GM, Holton JL, Revesz T, Dickson DW. Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 2011;122:187–204. doi: 10.1007/s00401-011-0852-9. [DOI] [PubMed] [Google Scholar]
- Holmes BB, Diamond MI. Cellular mechanisms of protein aggregate propagation. Curr Opin Neurol. 2012;25:721–726. doi: 10.1097/WCO.0b013e32835a3ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Devos SL, Kfoury N, Li M, Jacks R, Yanamandra K, Ouidja MO, Brodsky FM, Marasa J, Bagchi DP, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci U S a. 2013;110:E3138–E3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Iba M, Guo JL, McBride JD, Zhang B, Trojanowski JQ, Lee VM-Y. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J Neurosci. 2013;33:1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jicha GA, Bowser R, Kazam IG, Davies P. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J Neurosci Res. 1997;48:128–132. doi: 10.1002/(sici)1097-4547(19970415)48:2<128::aid-jnr5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Kim C, Haldiman T, Surewicz K, Cohen Y, Chen W, Blevins J, Sy M-S, Cohen M, Kong Q, Telling GC, et al. Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrP(C) PLoS Pathog. 2012;8:e1002835. doi: 10.1371/journal.ppat.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee S, Jung C, Ahmed A, Lee G, Hall GF. Interneuronal transfer of human tau between Lamprey central neurons in situ. J Alzheimers Dis. 2010;19:647–664. doi: 10.3233/JAD-2010-1273. [DOI] [PubMed] [Google Scholar]
- Krammer C, Kryndushkin D, Suhre MH, Kremmer E, Hofmann A, Pfeifer A, Scheibel T, Wickner RB, Schätzl HM, Vorberg I. The yeast Sup35NM domain propagates as a prion in mammalian cells. Proc Natl Acad Sci U S a. 2009;106:462–467. doi: 10.1073/pnas.0811571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Legname G, Nguyen H-OB, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S a. 2006;103:19105–19110. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K. Transsynaptic spread of tau pathology in vivo. PLoS ONE. 2012;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J-X, Qiang W, Yau W-M, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Kehm VM, Zhang B, O’Brien P, Trojanowski JQ, Lee VM-Y. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J Exp Med. 2012;209:975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- Miyasaka T, Morishima-Kawashima M, Ravid R, Kamphorst W, Nagashima K, Ihara Y. Selective deposition of mutant tau in the FTDP-17 brain affected by the P301L mutation. J Neuropathol Exp Neurol. 2001;60:872–884. doi: 10.1093/jnen/60.9.872. [DOI] [PubMed] [Google Scholar]
- Münch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc Natl Acad Sci U S a. 2011;108:3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Piwnica-Worms D. Real-time imaging of beta-catenin dynamics in cells and living mice. Proc Natl Acad Sci U S a. 2007;104:17465–17470. doi: 10.1073/pnas.0704465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Some speculations about prions, amyloid, and Alzheimer’s disease. N Engl J Med. 1984;310:661–663. doi: 10.1056/NEJM198403083101021. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S a. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P-H, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacino AN, Thomas MA, Ceballos-Diaz C, Cruz PE, Rosario AM, Lewis J, Giasson BI, Golde TE. Conformational templating of α-synuclein aggregates in neuronalglial cultures. Mol Neurodegener. 2013;8:17. doi: 10.1186/1750-1326-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- Santa-Maria I, Varghese M, Ksiezak-Reding H, Dzhun A, Wang J, Pasinetti GM. Paired helical filaments from Alzheimer disease brain induce intracellular accumulation of Tau protein in aggresomes. J Biol Chem. 2012;287:20522–20533. doi: 10.1074/jbc.M111.323279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqua A, Margittai M. Three- and four-repeat Tau coassemble into heterogeneous filaments: an implication for Alzheimer disease. J Biol Chem. 2010;285:37920–37926. doi: 10.1074/jbc.M110.185728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- van Groen T, Miettinen P, Kadish I. The entorhinal cortex of the mouse: organization of the projection to the hippocampal formation. Hippocampus. 2003;13:133–149. doi: 10.1002/hipo.10037. [DOI] [PubMed] [Google Scholar]
- Van Langenhove T, van der Zee J, Van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann Med. 2012;44:817–828. doi: 10.3109/07853890.2012.665471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman EA, Giasson BI. Induction of intracellular tau aggregation is promoted by α-synuclein seeds and provides novel insights into the hyperphosphorylation of tau. J Neurosci. 2011;31:7604–7618. doi: 10.1523/JNEUROSCI.0297-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischik CM, Novak M, Thøgersen HC, Edwards PC, Runswick MJ, Jakes R, Walker JE, Milstein C, Roth M, Klug A. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci U S a. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K, Kfoury N, Jiang H, Mahan TE, Ma S, Maloney SE, Wozniak DF, Diamond MI, Holtzman DM. Anti-tau antibodies that block tau aggregate seeding in vitro markedly decrease pathology and improve cognition in vivo. Neuron. 2013;80:402–414. doi: 10.1016/j.neuron.2013.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama Y, Higuchi M, Zhang B, Huang S-M, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM-Y. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell JM, Taylor SE, Cao T, Harrison JL, Lifshitz J. Rod microglia: elongation, alignment, and coupling to form trains across the somatosensory cortex after experimental diffuse brain injury. J Neuroinflammation. 2012;9:247. doi: 10.1186/1742-2094-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.