Abstract
Purpose
To study the relationship of imprinted gene expression (CDKN1C, H19, IGF2, KCNQ1 and PHLDA2) with human fetal growth.
Methods
RNA was extracted from fetuses with intrauterine growth restriction (IUGR) and from the controls without growth restriction. The gene expression pattern of CDKN1C, H19, IGF2, KCNQ1 and PHLDA2 genes was evaluated using RT-PCR. MS-MLPA was also performed to assess the IC1 and IC2 DNA methylation status on chromosome 11p15.5.
Results
The samples were divided according to their tissue type in placental or fetal tissue. Within each group, IUGR cases and controls were compared. In the IUGR cases, in both fetal and placental tissue groups IGF2 was observed to be down regulated. In another approach, the samples were divided in IUGR and control groups and for each of them placental and fetal tissue was compared. Within the IUGR group up regulation of CDKN1C, KCNQ1, and PHLDA2 was determined in placental samples. IUGR group presented a statistically lower methylation status in both IC1 and in IC2. Regarding differences between fetal and placental samples within this group, methylation status of placental samples was statistically significant down regulated in the imprinting center 1 (IC1).
Conclusions
Genomic imprinting is a phenomenon that plays an important role in fetal and placental development. This study emphasizes the importance of imprinted genes during pregnancy. Differences between tissues could reflect different mechanisms, either compensatory or adverse, that should be investigated in more detail.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-014-0278-0) contains supplementary material, which is available to authorized users.
Keywords: Imprinted genes, RT-PCR, DNA methylation, IUGR
Introduction
Intrauterine growth restriction (IUGR) is a condition in which a fetus is unable to achieve its genetically determined potential size thereby increasing its perinatal risk of morbidity and mortality. IUGR encompasses many different maternal and fetal causes. Concerning fetal problems, chromosomal abnormalities are one of the major causes of growth restriction [1]. Additionally, this phenomenon occurs due to abnormal gene expression in tissues leading to an abnormal fetus growth.
Epigenetics has a central role in the regulation of fetal growth and development. In particular, imprinted genes comprise a small subset of the human genome that has been shown to be essential to fetal, placental and behavioral development. Genomic imprinting is characterized by an epigenetic modification in which one allele is repressed according to the parental origin. In general, paternally expressed genes promote fetal and placental growth while maternally expressed genes limits conceptus growth. This is consistent with the imprinting conflict theory, which postulates that the paternal genome maximizes extraction of maternal resources for the benefit of the paternal offspring while maternal genome limits nutrient provision acting to preserve and distribute such resources more equally between all her potential offspring [2–4]. Currently, there are over 90 known human imprinted genes (www.geneimprint.com). Insulin-like growth factor 2 (IGF2) is a paternally expressed gene, which acts as a growth factor, being associated with several pathologies such as Beckwith-Wiedmann syndrome (BWS, OMIM: #130650), characterized by an overgrowth [5, 6]. With the opposite effect, Cyclin-dependent kinase inhibitor 1C (CDKN1C) [7], Pleckstrin homology-like domain family A member 2 (PHLDA2) and Potassium voltage-gated channel (KCNQ1) are imprinted genes that tend to restrict fetal body’s weight, being associated with restriction growth syndromes such as Silver-Russell syndrome (SRS, OMIM: #180860) [8, 9]. All these genes are located in the same cluster, on chromosome 11 at p15.5. Two independent imprinting control regions or imprinting centers (IC), IC1 and IC2, regulate its expression. IGF2 and the imprinted maternally expressed non coding transcript (H19) are co-localized in the IC1, which contains a methylation-sensitive chromatin insulator that is responsible for controlling the expression of both genes. Therefore, methylation in the IC1 of the paternal chromosome prevents the binding of the transcription factor 11-zinc finger protein (CTCF), resulting in H19 hyper methylation and silencing and allowing the expression of IGF2. On the other hand, in the maternal chromosome, the lack of methylation in this region prevents the activation of IGF2 and the presence of the CTCF insulator facilitates H19 transcription [10].
In humans, deregulation of the IGF2/H19 imprinted region is associated with the overgrowth and tumor predisposition-related BWS and with SRS, mainly characterized by pre- and post-natal growth deficiency.
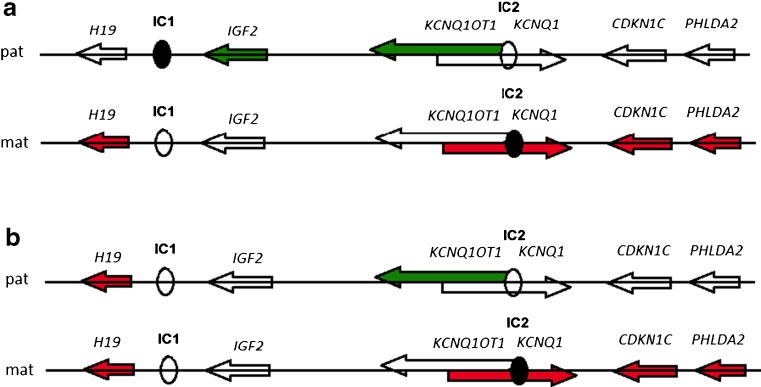
IC2 is much larger and responsible for controlling several maternally expressed genes namely CDKN1C, KCNQ1 and PHLDA2. It also includes a non-coding KCNQ1 overlapping transcript 1 (KCNQ1OT1) gene that has an antisense orientation with respect to the protein-coding gene KCNQ1. In the maternal chromosome, IC2 is methylated, KCNQ1OT1 is not transcribed and the flanking imprinted genes are expressed. On the paternal chromosome the IC2 sequence itself and/or the KCNQ1OT1 transcript mediate the silence of the several genes of the region including the growth inhibitors CDKN1C, PHLDA2 and KCNQ1 genes (Fig. 1a) [10–12].
Fig. 1.
Regulation of imprinted genes expression according to IC1/IC2 methylation, both in paternal (pat) and maternal (mat) chromosomes; (a) – Epigenotypes of normal imprinted gene expression, (b) – Epigenetic abnormalities often detected in SRS patients, representing maternalization of IC1
Genomic DNA methylation in these ICs may exhibit substantial variation among human individuals. This epigenetic variation might contribute either to phenotypic variation, as well as to human disease [10, 13]. Although this subject has been the focus of investigation recently, the role of human imprinted genes in fetal development is not fully understood. With this study we assessed the expression levels of five imprinted genes (CDKN1C, H19, IGF2, KCNQ1 and PHLDA2) in fetal and placental samples from spontaneous abortions or fetal deaths in which IUGR was identified. Additionally, the IC1 and IC2 methylation status on chromosome 11p15.5 in the same samples was evaluated.
Materials and methods
Tissue samples
Two groups were selected: a group with 10 fetuses with IUGR (all of them paired samples of fetal tissue and placenta, corresponding to a total of 20 samples) and a control group with 3 fetuses (from which two of them were paired samples of fetal and placental tissue and one with only fetal tissue corresponding to a total of 5 samples). The evaluation of the cause that determined the fetal loss in both groups (control and with IUGR) was made by the obstetricians (clinical and analytic evaluation) and a pathologist (pathological evaluation, according to the protocol of the Department of Anatomical Pathology), in Centro Hospitalar de São João, Porto. A fetus was considered as having an IUGR when its biometrical parameters were below percentile 10 for gestational age. This analysis was performed by ultrasound or by autopsy, in cases without second trimester ultrasound. In both groups, maternal exposure to cigarette smoking, toxins exposure (as drugs, medication), uterine malformations, inherited thrombophilia, pre-eclampsia and endocrine disorders were excluded. The control group samples were selected from cases with infections or umbilical cord constriction. All samples had a normal karyotype and were obtained from fetuses in the second trimester (gestational age between 14 and 24 weeks). The study was approved by the Ethical Committee of Centro Hospitalar de São João, EPE.
RNA extraction and cDNA synthesis
Selected samples were stored in RNA later at -80 ºC until use. Total RNA was extracted from samples using 1 ml of Tripure Isolation Reagent (Roche, Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s standard protocol. Quantification and purity were determined by NanoDrop 2,000 UV–vis Spectrophotometer (Nanodrop Technologies, Wilmington, USA). For cDNA synthesis, 10 μg of total RNA was subjected to reverse transcription using qScriptTM cDNA Synthesis Kit (Quanta BioSciences, Inc., Gaithersburg, USA) following the manufacturer’s instructions.
Quantitative RT-PCR
RNA expression levels of five imprinted genes (CDKN1C, H19, IGF2, KCNQ1 and PHLDA2) and one housekeeping gene (GADPH) were analyzed by Real-Time PCR on a StepOnePlus™ Real-Time PCR System (Life Technologies Corporation, California, USA). Taqman® Gene Expression Assays (Life Technologies Corporation, California, USA) were used for each target gene CDKN1C (Hs00175938_m1), H19 (Hs00923522_m1), IGF2 (Hs01005963_m1), KCNQ1 (Hs00923522_m1) and PHLDA2 (Hs00169368_m1), and endogenous control GAPDH (Hs99999905_m1). PCR reactions were performed in a 25 μl volume containing 5 μl of cDNA, 12.5 μl of TaqMan® Universal PCR Master Mix System (Life Technologies Corporation, California, USA), 6.25 μl of Rnase-free water and 1.25 μl of 20× TaqMan ® Gene Expression Assay Mix for each gene. PCR was performed in separated wells for each reaction and each sample was run in triplicate. For each gene, cases and controls samples were run in the same RT-PCR plate to minimize intra-plate variations. PCR parameters were as follows: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. In each plate negative controls were included.
DNA extraction and MS-MLPA
DNA extraction was performed according to the manufacturer’s standard protocol using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany).
Afterward, MS-MLPA was performed on genomic DNA with SALSA MS-MLPA kit ME030-C1 BWS/SRS, (MRC-Holland, Amsterdam, The Nederland’s), according to the manufacturer’s instructions. This mix contains 42 probes, 26 of which for the region 11p.15. The methylation status of this region can also be determined by 10 of these probes since they contain a HhaI recognition site. In addition, further 15 probes are added for reference.
After 16 h of hybridization at 60 ºC, samples were equally split into two aliquots. The first aliquot underwent ligation only, whereas the second one underwent ligation followed by enzymatic digestion with HhaI (a restriction enzyme recognizing only unmethylated DNA) and PCR amplification using universal primers. In the latter case, amplification products were obtained and detected by capillary electrophoresis only if the CpG included in the HhaI site was methylated and, therefore, not digested. Ligation, enzymatic digestion, and PCR amplification were performed according to the manufacturer’s instructions. PCR products (1 ml) were mixed with 0.3 ml of internal size standard (GeneScan™ 600 LIZ; Applied Biosystems) and 13.7 ml of deionized formamide, and injected into 3,500 Genetic Analyzer (Life Technologies Corporation, California, USA).
Data analysis
Gene expression values
The results were analysed using the validated Livak method [14].
A mean was calculated between the 3 replicates for the reference gene and the target gene. The value of threshold cycle (CT) of the target gene was deducted to that of reference (ref) or housekeeping gene, for the test samples [ΔCT (sample) = CT (ref) – CT (target gene)]. Later, the expression ratio was considered by calculating the normalized expression (2-ΔCT = normalized expression ratio).
The result obtained for the target gene in the sample was therefore normalized to the expression of a reference gene. Normalizing the expression of the target gene to that of the reference gene compensates for differences in the amount of the cDNA sample.
MS-MLPA
Each probe’s signal was divided by the signal of each reference probe in the sample. After, the median of these ratios was estimated calculating the so called Normalization Constant (NC). This value was divided by the average NC obtained in the undigested reference samples. Values ranging from 0.65 to 1.35 were considered as having normal number copies (diploid); a deletion was suspected for ratio less than 0.65 and duplication was suspected for ratio more than 1.35. Quantification of the methylation status of a CpG site was done by dividing the NC of each MS-MLPA probe obtained on the digested aliquot by the NC of each MS-MLPA probe obtained on the corresponding undigested aliquot. To simplify the interpretation of data, we calculated the average methylation status obtained by the H19 and KCNQ1OT1 probes and indicated them as IC1 and IC2 methylation index, respectively. Due to the reduced number of controls in our series, previous published data was used for the determination of the normal methylation levels ranges, so the mean methylation index for normal samples was 0.52 (range 0.47–0.58) for IC2 and 0.50 (range 0.46–0.55) for IC1 [12].
Results
RT-PCR expression profile
A total of 20 cDNA samples derived from fetuses diagnosed with IUGR and 5 cDNA samples derived from fetuses without growth restriction (control group) were analyzed by RT-PCR for gene expression quantification Analysis of RT-PCR was done using two different approaches: (1) evaluation of the differences between the IUGR group and the control group through the analysis of fetal samples and placental samples, independently; (2) evaluation of the differences between fetal and placental samples within each group. The results are summarized in Figs. 2a, b, 3a, b.
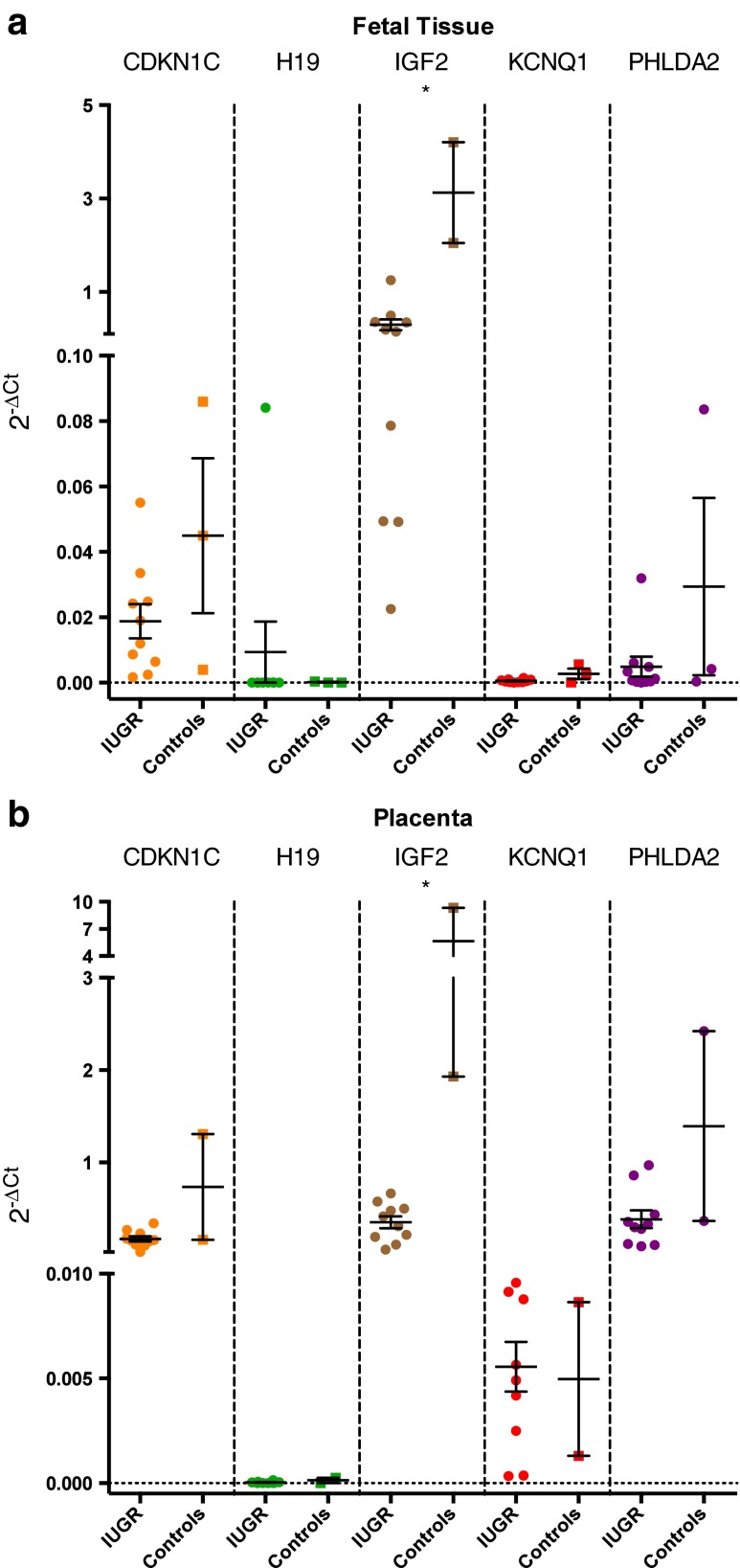
Fig. 2.
(a) Expression levels of the studied genes in fetal tissue samples. (b) Expression levels of the studied genes in placenta samples. cDNA expression was normalized using a housekeeping gene (GAPDH). Error bars represent standard error of the mean (SEM). Significant differences between groups are represented as: * p < 0,05. The data was analysed by Mann–Whitney–Wilcoxon test. To further analysis, the mean, the standard deviation (SD) and the SEM of the respective groups are presented in Table 2 (supplementary material). IUGR – Intrauterine Growth Restriction
Our results showed that in both sample groups, either fetal or placental tissue, IGF2 was down regulated in IUGR cases (p < 0.05 in both). For the other genes, no significant differences between IUGR and control cases were found in both placental and fetal tissue group (Fig. 2a and b).
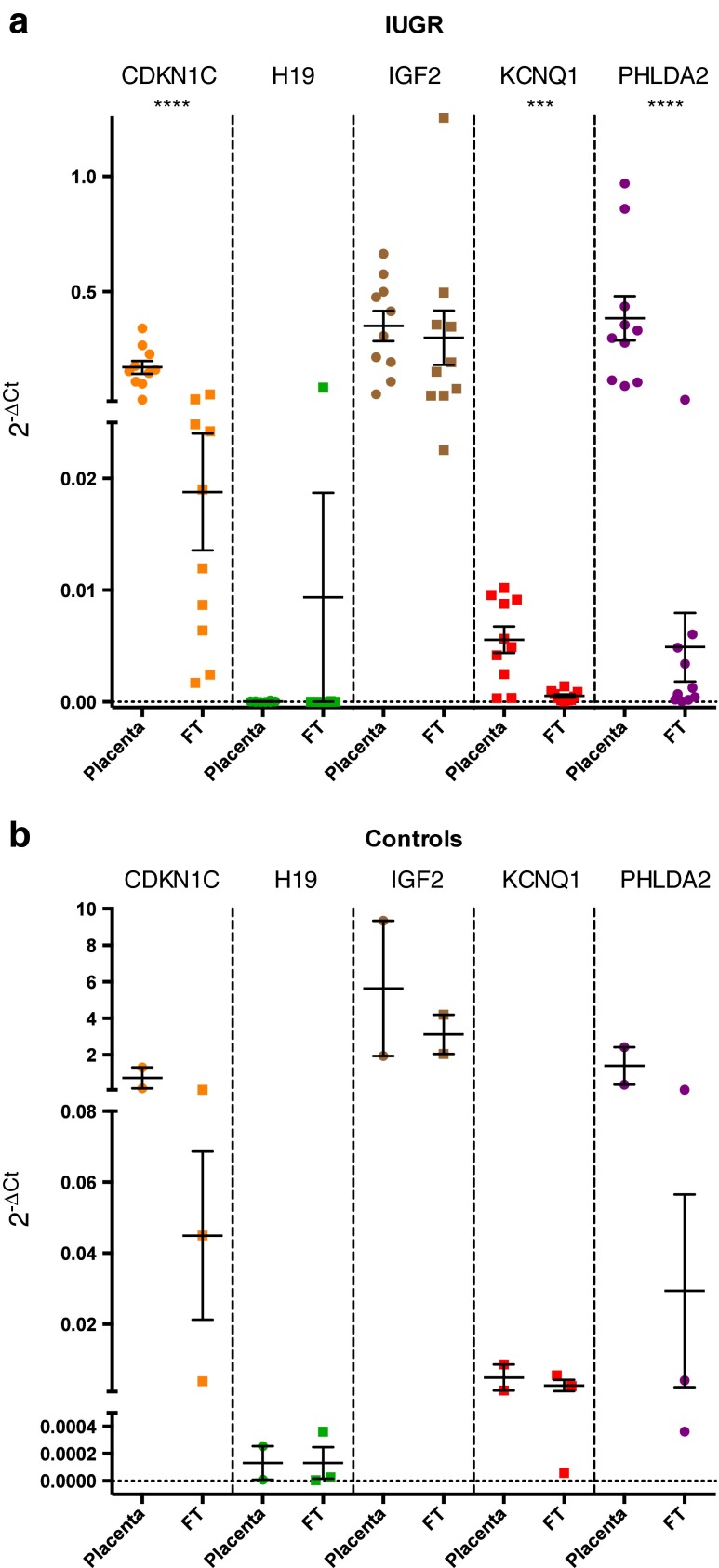
Regarding differences between tissues (fetal versus placental) both groups (IUGR and controls) were evaluated and our results indicated that in the IUGR group, CDKN1C (p < 0.0001), KCNQ1 (p < 0.0005) and PHLDA2 (p < 0.0001) gene expression was upregulated in the placenta. For H19 and IGF2 genes no significant differences between placental and fetal tissue were found in the IUGR group (Fig. 3a). No differences were found in the control group comparing fetal versus placental tissues (Fig. 3b).
Fig. 3.
(a) Expression levels of the studied genes in the growth restriction group. (b) Expression levels of the studied genes in the control group. cDNA expression was normalized using a housekeeping gene (GAPDH). Error bars represent standard error of the mean (SEM). Significant differences between groups are represented as: *** p < 0,0005; **** p < 0,0001. In Fig. 3a the H19, IGF2 and PHLDA2 data was analysed by Mann–Whitney–Wilcoxon test. The CDKN1C and KCNQ1 data was analysed by Student’s t-test. In Fig. 3b data was analysed by Student’s t-test. To further analysis, the mean, the standard deviation (SD) and the SEM of the respective groups are presented in Table 2 (supplementary material). FT – Fetal tissue
DNA methylation analysis
A total of 25 DNA samples derived from fetuses diagnosed with IUGR (n = 20) and a control group (n = 5), were analyzed to evaluate the methylation status. The results are summarized in Table 1. No copy number alterations were found.
Table 1.
Summary of MS-MLPA methylation analysis
| Methylation status | IC1 Nº Samples (M ± SD) | IC2 Nº Samples (M ± SD) | |
|---|---|---|---|
| Growth restriction | Normal | 4 (51.9 ± 1.7) | 11 (51.4 ± 3.3) |
| Hypomethylated | 13 (37.5 ± 4.5) | 8 (40.4 ± 7.7) | |
| Hypermethylated | 3 (60.0 ± 5.7) | 1 (58.2) | |
| Controls | Normal | 1 (51.6) | 1 (56.3) |
| Hypomethylated | 0 (0) | 0 (0) | |
| Hypermethylated | 4 (56.0 ± 0.4) | 4 (62.6 ± 4.6) |
Four out of 20 samples with IUGR presented a normal methylation status in the IC1, 13 out of 20 had IC1 hypo methylation and 3 out of 20 had IC1 hyper methylated. In the control group one sample out of 5 presented a normal methylation status in the IC1, and 4 out of 5 had IC1 hyper methylated.
Regarding IC2 in the IUGR group, a normal methylation status was detected in 11 out of 20 samples, 8 out of 20 had IC2 hypo methylation and 1 out of 20 had IC2 hyper methylation. In the control group 1 out of 5 had a normal methylation status and the other 4 out of 5 had IC2 hypo methylation.
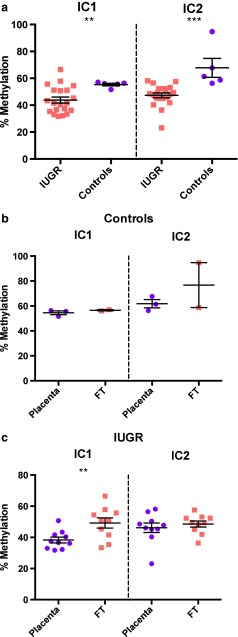
A comparison between the IUGR group and control group was made for IC1 and IC2 methylation status (Fig. 4a). The IUGR group presented a statistical lower methylation status in both IC1 (p < 0.01) and in IC2 (p < 0.0005).
Fig. 4.
(a) Level of methylation in both IC1 and IC2. (b) Level of methylation in both IC1 and IC2 between placenta and fetal samples from control group. (c) Level of methylation in both IC1 and IC2 between placenta and fetal samples from IUGR group. Error bars represent standard error of the mean (SEM). Significant differences between groups are represented as: ** p < 0,01; *** p < 0,0005. In Fig. 4a and b data was analysed by Mann–Whitney–Wilcoxon test. In Fig. 4c data was analysed by Student’s t-test. To further analysis, the mean, the standard deviation (SD) and the SEM of the respective groups are presented in Table 3 (supplementary material). IC – Imprinting Center; IUGR – Intrauterine Growth Restriction; FT – Fetal tissue
Regarding differences between fetal and placental samples within each group (IUGR and control group) no statistical differences were observed in the control group, neither IC1 nor IC2 (Fig. 4b). In the IUGR cases, placental samples methylation status was statistically down regulated in the IC1 (p < 0.01) (Fig. 4c).
Discussion
IUGR comprises one of the leading obstetric complications, suggesting the presence of a pathophysiologic process occurring in utero that is responsible for fetal growth inhibition. Its regulation involves multiple factors with complex mechanisms. In order to better elucidate this process we have studied five imprinted genes, CDKN1C, H19, IGF2, KCNQ1 and PHLDA2, since they play an important role in this biological process. Therefore, mRNA expression of these imprinted genes in IUGR and controls cases was analyzed.
The results from the mRNA expression studies showed that IGF2 gene levels were down regulated in the IUGR group, both in fetal and placental samples. The expression profiles of the other genes evaluated showed no statistically significant differences in fetal and placental samples.
The majority of reports have focused on evaluation of the expression level change in the placenta in association with intrauterine growth defects. In spite of some conflicting evidence regarding down regulation levels in the placenta of IUGR, the majority of authors describe decreased expression in association with intrauterine growth restriction [15–18]. Sibley et al. reported down regulation of IGF2 in cases with IUGR placenta versus normal term placenta, albeit with greater case-to-case variability. In mouse, mutations in Igf2 gene lead to IUGR [19]. IGF2 is considered the major fetal growth factor that regulates fetal/placental growth by stimulating trophoblastic migration and invasion. It also regulates diffusional exchange characteristics of the placenta. Although, intrauterine growth restriction is an extremely complex phenomenon, other genes and processes could contribute for this particular condition [2]. Nevertheless, the biological implications of these differences should be tested in larger series.
In this study, we found statistical differences between the IUGR cases and the control cases in placental tissue samples for CDKN1C and PHLDA2 genes. In a previous study performed by our group, we observed an increase in the PHLDA2 gene expression levels associated with spontaneous abortions, in the first trimester, and with IUGR in second trimester, although not statistically significant [20]. It is important to stress that it has also been suggested that the PHLDA2 gene may have a more profound effect on the placenta at early gestational ages when the placenta is more active, which is also in accordance with our previous report [18, 20]. On the other hand, we found up regulation of these genes, and also for KCNQ1, that are all maternally expressed, in the placenta in comparison with fetal tissue, in the IUGR group. Fetal growth is a complex, dynamic process dependent on the balanced interactions between mother, placenta and fetus. The role of IUGR may be more complex since altered expressions of imprinted genes in the IUGR-associated placenta can be interpreted as causative or protective of fetal growth, some acting to reduce fetal growth, resulting in IUGR (negative effectors), while others may act to enhance fetal growth in a compensatory manner to rescue a pathogenically restricted fetus (positive effectors) [21]. The question if the observed changes in the gene expression in the different tissues, namely fetal versus placental tissue, are compensatory or adverse mechanisms underlying pathology, should be the subject of additional studies.
Additionally to the determination of the mRNA expression levels, the study of the IC1 and IC2 methylation status was carried out in order to evaluate if alteration in the regulation in these imprinting centres could be related with the IUGR.
H19 and IGF2 share common enhancers located downstream of H19. IC1 prevents IGF2 expression from the maternal allele but allows its expression from the paternal allele, due to the DNA methylation mark at IC1. This epigenetic mark is extended to the H19 promoter on the paternal allele, restricting H19 expression to the maternal chromosome. Sixty percent of SRS cases are associated with loss of DNA methylation (LOM) from the paternal IC1 allele [8]. This defect represents a maternalization of IC1; this means that the methylated paternal allele acquires a maternal epigenotype (Fig. 1a and b). Consequently, it is expected a decrease of IGF2 expression and/or an increase in H19 expression and this may be associated with growth restriction, which is one of the main characteristics of the SRS phenotype. The same molecular defect can be applied to IUGR cases.
Our results showed that 65 % of the cases with IUGR were hypo methylated in IC1. This means that IC1 hypo methylation could be one of the possible explanations for the fetal growth restriction. A previous report in mice showed a positive correlation between bacterial infection and IC1 hyper methylation, however in this report the investigators also found a down regulation in the Igf2 expression. Thus, this cause-effect in Igf2 expression remains to be corroborated [22]. It is also important to stress that we also found IC1 hyper methylation in the control group, however, IGF2 expression levels were not down regulated.
Several studies revealed IC1/IC2 epimutations in patients with SRS. In one study MS-MLPA performed in peripheral blood from three patients referred as SRS revealed hypo methylation in both IC1 and IC2 but, when the study was performed using buccal cells, only hypo methylation of the IC1 could be confirmed [23], suggesting that different tissues can have different methylations ranges. This is also corroborated by our results that show statistically significant differences between placental and fetal tissue for IC1 in IUGR group. In the future, it will be important not only to enlarge our series of IUGR cases but also to extend the study to the third trimester of pregnancy.
In the present study, hypo methylation of IC2 was also observed for 40 % of the samples. Previously, Begemann et al. and Azzi et al. found that 3.8 and 4.05 % of SRS cases had IC2 hypo methylation, respectively [23, 24]. The specific reason for the development of IUGR is not clear, nevertheless mosaicism could explain variability, and the loss of paternal or maternal methylation marks could probably not only be attributed to a deficient acquisition of methylation during gametogenesis but be consistent with an incorrect maintenance of methylation after fertilization. The majority of studies performed to evaluate the levels of IC1 and IC2 methylation using MS-MLPA were performed in cases of patients with imprinting disorders, namely SRS or BWS. This is the first study performed in spontaneous abortions cases with IUGR and could reflect more severe cases with extreme methylation variations. Once again, it is important to stress that, so far, it is not possible to exclude that the different methylation levels observed in aborted fetus and placentas were not a cause but a consequence of aberrant development.
In conclusion, several factors can affect fetal growth as maternal nutritional status, diet and exposure to environmental factors. This leads to an alteration in nutrient availability to the fetus and to a modulation of placental gene expression [15]. Imprinted genes can be responsible for IUGR, but additional other non-imprinted genes may also be involved [2].
Since the majority of the studies conducted to date was restricted to the evaluation of the expression level of different imprinted genes in the placenta in cases with IUGR, the present study it is of major importance because differences between tissues could reflect different mechanisms, either compensatory or adverse, that should be investigated in more detail.
It would be interesting to compare our data from the second trimester with data from the third trimester and assess whether there are differences throughout gestation.
More research is needed in this field, because there is not much knowledge about imprinting gene regulation, as well as their proper role in placental and human fetal growth. The possibility of the early determination of differences in the methylation ranges (i.e. hypo methylation of IC1 using MS-MLPA) can be used to identify pregnancies with higher risk of severe IUGR.
Electronic supplementary material
(DOC 120 kb)
(DOC 176 kb)
Footnotes
Capsule Imprinting genes play a very important role in IUGR and methylation or gene expression levels may identify pregnancies at high risk of severe IUGR
Contributor Information
Amilcar Cordeiro, Email: mimed08007@med.up.pt.
Ana Paula Neto, Email: anapaula_neto@hotmail.com.
Filipa Carvalho, Email: filipac@med.up.pt.
Carla Ramalho, Email: carlaramalho@med.up.pt.
Sofia Dória, Phone: + 351 22 5513647, Email: sdoria@med.up.pt.
References
- 1.Snijders RJ, Sherrod C, Gosden CM, Nicolaides KH. Fetal growth retardation: associated malformations and chromosomal abnormalities. Am J Obstet Gynecol. 1993;168(2):547–55. doi: 10.1016/0002-9378(93)90491-Z. [DOI] [PubMed] [Google Scholar]
- 2.Ishida M, Moore GE. The role of imprinted genes in humans. Molecular aspects of medicine. 2012. doi:10.1016/j.mam.2012.06.009. [DOI] [PubMed]
- 3.Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432(7013):53–7. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- 4.Neerhof MG. Causes of intrauterine growth restriction. Clin Perinatol. 1995;22(2):375–85. [PubMed] [Google Scholar]
- 5.St-Pierre J, Hivert MF, Perron P, Poirier P, Guay SP, Brisson D et al. IGF2 DNA methylation is a modulator of newborn’s fetal growth and development. Epigenetics : official journal of the DNA Methylation Society. 2012;7 (10). [DOI] [PMC free article] [PubMed]
- 6.Cerrato F, Sparago A, Di Matteo I, Zou X, Dean W, Sasaki H, et al. The two-domain hypothesis in Beckwith-Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum Mol Genet. 2005;14(4):503–11. doi: 10.1093/hmg/ddi047. [DOI] [PubMed] [Google Scholar]
- 7.Riccio A, Cubellis MV. Gain of function in CDKN1C. Nat Genet. 2012;44(7):737–8. doi: 10.1038/ng.2336. [DOI] [PubMed] [Google Scholar]
- 8.Jacob KJ, Robinson WP, Lefebvre L. Beckwith-Wiedemann and Silver-Russell syndromes: opposite developmental imbalances in imprinted regulators of placental function and embryonic growth. Clin Genet. 2013;84(4):326–34. doi: 10.1111/cge.12143. [DOI] [PubMed] [Google Scholar]
- 9.Ishida M, Monk D, Duncan AJ, Abu-Amero S, Chong J, Ring SM, et al. Maternal inheritance of a promoter variant in the imprinted PHLDA2 gene significantly increases birth weight. Am J Hum Genet. 2012;90(4):715–9. doi: 10.1016/j.ajhg.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19(3):281–9. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Chiesa N, De Crescenzo A, Mishra K, Perone L, Carella M, Palumbo O, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21(1):10–25. doi: 10.1093/hmg/ddr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priolo M, Sparago A, Mammi C, Cerrato F, Lagana C, Riccio A. MS-MLPA is a specific and sensitive technique for detecting all chromosome 11p15.5 imprinting defects of BWS and SRS in a single-tube experiment. European journal of human genetics : EJHG. 2008;16(5):565–71. doi: 10.1038/sj.ejhg.5202001. [DOI] [PubMed] [Google Scholar]
- 13.Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38(12):3880–90. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, et al. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol. 2008;320(1):79–91. doi: 10.1016/j.ydbio.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 16.McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27(6–7):540–9. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Antonazzo P, Alvino G, Cozzi V, Grati FR, Tabano S, Sirchia S, et al. Placental IGF2 expression in normal and intrauterine growth restricted (IUGR) pregnancies. Placenta. 2008;29(1):99–101. doi: 10.1016/j.placenta.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Apostolidou S, Abu-Amero S, O'Donoghue K, Frost J, Olafsdottir O, Chavele KM. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. Journal of molecular medicine (Berlin, Germany) 2007;85(4):379–87. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]
- 19.Sibley CP, Coan PM, Ferguson-Smith AC, Dean W, Hughes J, Smith P, et al. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc Natl Acad Sci U S A. 2004;101(21):8204–8. doi: 10.1073/pnas.0402508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doria S, Sousa M, Fernandes S, Ramalho C, Brandao O, Matias A, et al. Gene expression pattern of IGF2, PHLDA2, PEG10 and CDKN1C imprinted genes in spontaneous miscarriages or fetal deaths. Epigenetics : official journal of the DNA Methylation Society. 2010;5(5):444–50. doi: 10.4161/epi.5.5.12118. [DOI] [PubMed] [Google Scholar]
- 21.Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth defects research Part A, Clinical and molecular teratology. 2011;91(8):682–92. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobetsis YA, Barros SP, Lin DM, Weidman JR, Dolinoy DC, Jirtle RL, et al. Bacterial infection promotes DNA hypermethylation. J Dent Res. 2007;86(2):169–74. doi: 10.1177/154405910708600212. [DOI] [PubMed] [Google Scholar]
- 23.Begemann M, Spengler S, Kanber D, Haake A, Baudis M, Leisten I, et al. Silver-Russell patients showing a broad range of ICR1 and ICR2 hypomethylation in different tissues. Clin Genet. 2011;80(1):83–8. doi: 10.1111/j.1399-0004.2010.01514.x. [DOI] [PubMed] [Google Scholar]
- 24.Azzi S, Rossignol S, Steunou V, Sas T, Thibaud N, Danton F, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet. 2009;18(24):4724–33. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 120 kb)
(DOC 176 kb)