Abstract
This study aimed to investigate a computer-aided system for detecting breast masses using dynamic contrast-enhanced magnetic resonance imaging for clinical use. Detection performance of the system was analyzed on 61 biopsy-confirmed lesions (21 benign and 40 malignant lesions) in 34 women. The breast region was determined using the demons deformable algorithm. After the suspicious tissues were identified by kinetic feature (area under the curve) and the fuzzy c-means clustering method, all breast masses were detected based on the rotation-invariant and multi-scale blob characteristics. Subsequently, the masses were further distinguished from other detected non-tumor regions (false positives). Free-response operating characteristics (FROC) curve and detection rate were used to evaluate the detection performance. Using the combined features, including blob, enhancement, morphologic, and texture features with 10-fold cross validation, the mass detection rate was 100 % (61/61) with 15.15 false positives per case and 91.80 % (56/61) with 4.56 false positives per case. In conclusion, the proposed computer-aided detection system can help radiologists reduce inter-observer variability and the cost associated with detection of suspicious lesions from a large number of images. Our results illustrated that breast masses can be efficiently detected and that enhancement and morphologic characteristics were useful for reducing non-tumor regions.
Keywords: Breast, Magnetic resonance imaging, Detection, Morphologic, Hessian
Introduction
Breast cancer is one of the most common cancers in women [1], and treatment of breast cancer in an earlier stage will reduce the mortality rate [2]. If a tumor is detected in its earlier stages, the chance of successful treatment is higher with proper diagnosis and treatment [2]. Some algorithm on mammography [3] and breast ultrasound (US) [4] have been used for breast cancer detection and diagnosis. However, some limitations might occur using these computer-aided algorithms in different imaging techniques. In mammography, dense fibroglandular tissues can obscure some breast cancers [5], resulting in missed lesions. Mogatadakala et al. [6] proposed a method using order statistic features from multiresolution decompositions of energy-normalized subregions to discriminate surrounding normal and tumor regions in two-dimensional (2-D) US images. However, these algorithms are difficult to be applied to a large three-dimensional (3-D) volume data because the algorithm and feature are designed for 2-D images. Improper US machine settings and scanner operation error may increase the risk of missed lesions.
Compared with US and mammography image, time-intensity curve obtained from dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) can increase the diagnostic accuracy if morphological appearance is indeterminate [7, 8]. Breast magnetic resonance imaging has a moderate specificity but high sensitivity [9, 10]. The detection of early breast cancer in high-risk populations in the preoperative cancer assessment becomes increasingly important [11]. In MRI study, some breast lesion can be identified from maximum intensity projection (MIP) images if manual rotation in different projections is performed [7] while some suspicious lesions are more likely missed in 2-D MIP examination. Because breast lesion is inherent 3-D, 2-D characterization using MIP might be unable to extract the accurate 3-D lesion information due to overlapping between the enhancing fibroglandular tissue and lesion. In viewing of the only 2-D MIP images, the depth of the lesion perpendicular to plane might be lost. Due to the large number of thin-section 3-D DCE-MRI images obtained from the current state-of-the-art MR scanners, manual tumor identification by radiologists is inefficient and subjective to inter-observer variability. To assist in characterizing inherent 3-D lesion and reducing the inter-observer variability, a computer-aided detection (CADe) system for 3-D breast DCE-MRI image data to detect tumor is proposed for clinical practice. In this study, a 3-D multi-scale tumor detection system is developed for detecting not only suspicious-enhanced tissues by time-intensity curve but also whole 3-D mass-like lesions using morphological and textural characteristics.
Materials
Patients Enrollment
This study is approved by the Institutional Review Board, and informed consent is waived for our retrospective study. In our experiment, the dynamic contrast-enhanced magnetic resonance images were obtained by radiologists from August 2006 to August 2009 and consisted of 61 biopsy-proved lesions (21 benign and 40 malignant lesions, size range from 5 to 60 mm, mean 19.3 ± 12.8 mm) in 34 women (age range from 36 to 77 years, mean 51.0 ± 10.4 years) to evaluate the performance of the proposed detection system for breast MRI. The benign lesions included 15 fibrocystic changes, 4 papillomas, and 2 fibroadenomas. The malignant lesions included 35 invasive ductal carcinomas (IDC), 3 ductal carcinoma in situ (DCIS), and 2 invasive lobular carcinomas.
MRI Study
DCE-MRI is a medical imaging technique that traces changes in contrast enhancement of internal tissues, such as breast or brain tissue. In our experiment, DCE-MRI images were acquired using a 1.5T superconductive MR scanner (Signa Excite HD, GE Healthcare, Wauwatosa, WI, USA) with a dedicated eight-channel breast coil from patients in the prone position. The dynamic study was performed with the following parameters: pulse sequence VIBRANT (Volume Imaging for BReast AssessmeNT), fat-suppressed 3-D fast spoiled gradient echo (FSGR), repetition time/echo time/inversion time (TR/TE/TI) = 3.5/1.7/14 ms, flip angle 12°, matrix 256 × 160, image size 256 × 256 pixels, slice thickness 2.5 mm without gap, acquisition 0.75, and field of view 24 × 24 to 30 × 30 cm. There were 56 slices obtained for each acquisition of dynamic study with 14 cm coverage in cranio-caudal distance There were 35 total acquisitions, each lasting 15 s. Intravenous injection of a contrast agent (0.5 mmol/ml, gadodiamide; Omniscan, GE Healthcare AS, Oslo, Norway or gadopentetate dimeglumine; Magnevist, Bayer Healthcare Pharmaceuticals, Montville, NJ, USA) was performed with antecubital venous access through a plastic cannula (20G), with a bolus injection (4 ml/s) at the beginning of fifth acquisition.
Method
The proposed detection algorithm includes four stages. First, the breast region was segmented by template-based segmentation [12]. Based on the area under the curve (AUC) [13], the fuzzy c-means (FCM) clustering method [14] was used to identify suspicious tissue regions. Mass candidates with different sizes were then detected from these suspicious regions using multi-scale detection. The high frequency of false positives (FPs) could lead to unnecessary biopsies. The FPs are further reduced from candidates by blob, enhancement, texture, morphologic, and combined features.
Breast Region Segmentation
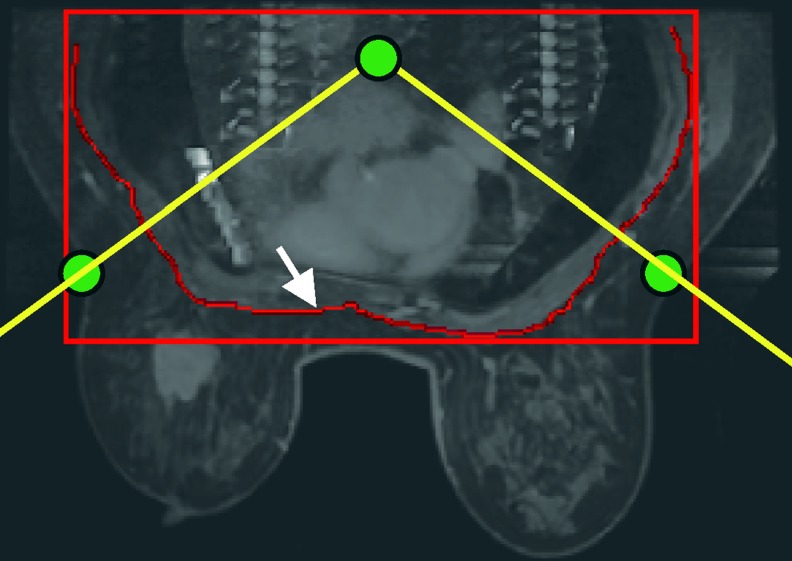
Breast MRI images include not only the breast region but also additional chest regions, such as the lung and the heart. Therefore, the other chest or thorax should be excluded to determine the breast region of interest for tumor detection. However, the shape of the breast and thorax regions is difficult to automatically define due to variation in breast shape and density patterns [12]. Thus, manual inspection of each data is needed to avoid degrading segmentation accuracy from such variation. In the template-based segmentation algorithm [12], the outline between the breast and thorax is manually delineated on one 2-D slice, producing a template to initialize the segmentation outline of other slices. In this study, the manual outline was delineated in the middle transverse slice of the AUC image (Fig. 1). The AUC image was generated from the accumulation of contrast enhancement on time-intensity curve. The intensity of each pixel in AUC image was composed of the area under the curve and similar with the area-under-curve (AUC) color maps in previous study [13]. This outline could exclude most of thorax, chest body region, except for the axillary region. To exclude the axillary region, the thoracic spine and the two breast margins of the bilateral pectoralis muscles were manually identified to define the bilateral margins on the template by a radiologist who had more than 10 years of experience of interpreting breast MR (Yeun-Chung Chang) (Fig. 1). The v-shape cut [12] formed by these three landmarks can exclude the axillary region (Fig. 1). The breast region was determined as the region below the outline inside the v-shape cut (Fig. 1). To determine the breast region in adjacent slices, the outline and landmarks of adjacent slices were used. To evaluate the deformation of chest regions between two adjacent slices, the thorax was defined with a manual bounding box (Fig. 1) to register the chest region inside the same bounding box in the adjacent slice using the demons deformable algorithm [12, 15]. When registering to neighboring slices based on the computed deformation from two adjacent chest regions, the outline and the landmarks were simultaneously deformed and used to extract the breast region of the neighboring slice. Based on the high similarity between the two adjacent slices, the procedure above could be propagated to each two adjacent slices from middle transverse slice to the cranial margin and caudal margin. Figure 2a shows the sequential segmentation results.
Fig. 1.
The solid arrow illustrates the manual outline on the AUC image. Three landmarks were connected to depict the v-shape cut formed by the two oblique lines
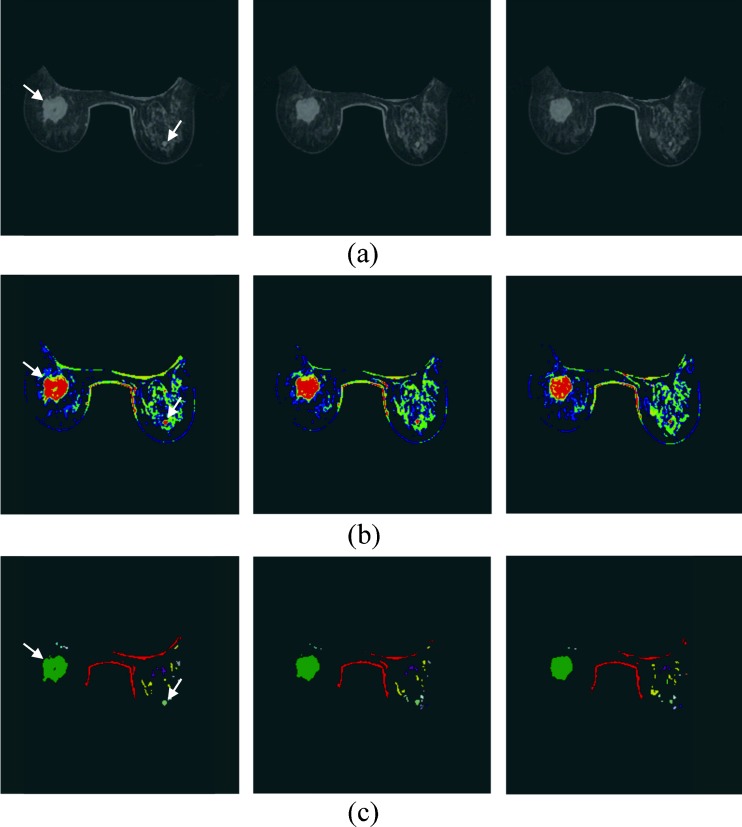
Fig. 2.
The breast segmentation of three continue slices of 3.5 cm IDC and 0.5 cm fibrocystic changes, indicated using white arrows on the AUC image. a AUC image. b The four FCM clusters results are represented by different colors. Non-breast regions and tissues without significant enhancement are colored black. c The connected regions from the two clusters with higher average AUC. The different connected regions are colored by different colors
Finally, the breast region from each slice was segmented using the boundary curve and the v-shape cut (Fig. 1). The suspicious tissues were detected from the segmented breast regions for further mass detection.
Suspicious Tissue Detection
Because diseased tissues usually show more intense enhancement compared to normal tissues [16], the pixels with more enhancement are initially considered as potentially suspected lesions. In this study, the AUC feature computed by the accumulation of contrast enhancement on time-intensity curve [13, 17] was to compute the enhancement degree. The voxels with larger AUCs in the first half of the AUC cumulative histogram [17] computed from the segmented breast region are considered as enhanced tissues. Furthermore, the enhanced tissues were divided into four clusters [14] using FCM clustering method [15, 24], and the average AUC of each cluster was computed. The voxels inside the two clusters with higher average AUC were identified as suspicious (Fig. 2b, c).
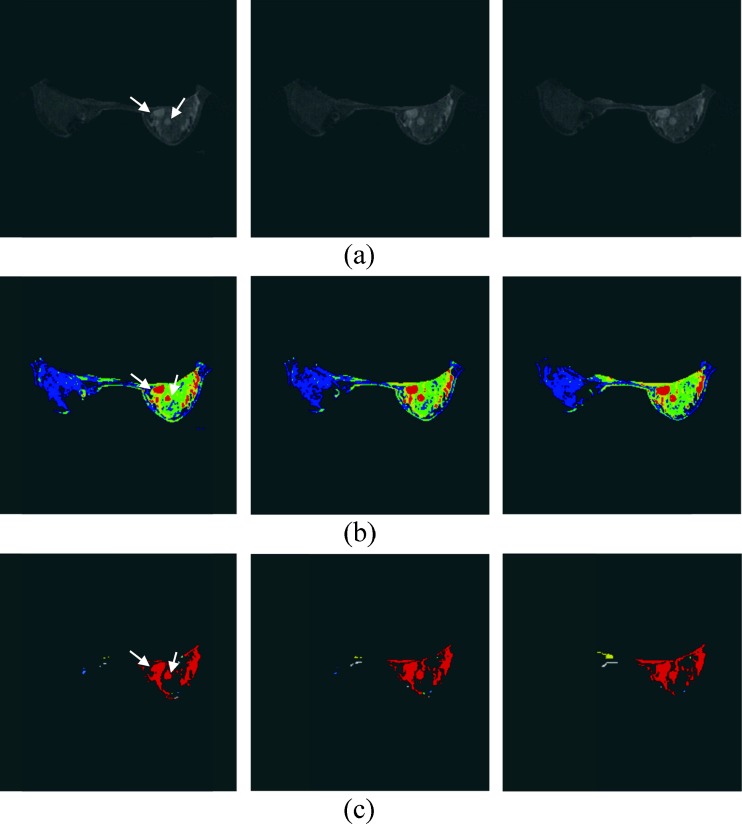
However, the suspicious tissues include some enhanced normal tissues (e.g., fibroglandular tissue, skin, and vessels). Besides, if the tumor is connected with other non-tumor tissues, the real tumor can be obscured within an over-segmented region, as shown in Fig. 3. The over-segmented regions, including tumor tissues and normal tissues, are difficult to diagnose due to their poor shape and synthetic composition. Hence, the tumor-like tissues were further extracted from suspicious tissues using the Hessian method based on the morphologic characteristics.
Fig. 3.
The 1.4 cm IDC and 0.7 cm fibrocystic changes, indicated by white arrows. a AUC image. b The four FCM clusters results are represented by different colors. c The connected regions from the two clusters with higher average AUC. The different connected regions are colored by different colors
Breast Mass Detection
The Hessian method, which has been applied to detect several kinds of geometrical structures [18–20], was used in our study to detect breast masses [21]. The Hessian method calculate the blob-like degree of each suspicious voxel based on the second derivatives along the three-dimensional directions by convoluting the AUC image. The analysis of eigenvalues [18] from the Hessian matrix were defined by
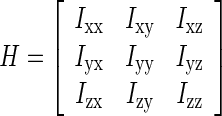 |
1 |
where the elements inside the Hessian matrix H are the partial second derivatives of the image [18], computed by convoluting the AUC image I(x,y,z) with second-order derivatives of 3-D Gaussian kernels G(x,y,z,σ) [19]. The kernels scale σ at each voxel is defined as the radius of the largest blob-like structure centered at this pixel. The eigenvalues of H are defined as λ1, λ2 and λ3 (|λ1| < |λ2| < |λ3|) and the blob-like degree of each suspicious voxel is computed by Oσ(λ) defined as
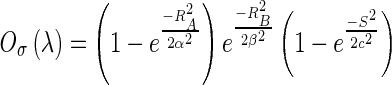 |
2 |
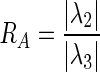 |
3 |
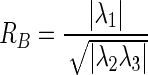 |
4 |
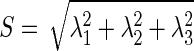 |
5 |
where α, β, and c are user-defined and fixed to 0.5, 0.5, and 20, respectively [18]. If a region is more blob-like or mass-like, the blob-like degrees Oσ(λ) will be higher. By means of blob-like degrees, we can distinguish mass from tubular and plate-like region. The maximum matching response [18], Oσ(λ), of each suspicious voxel evaluated the multi-scale blob-like degree at a range of spatial scales σ of the Gaussian kernel to detect the mass with different sizes, defined by
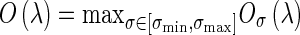 |
6 |
where the kernels scale σmin and σmax are used to control the minimum and the maximum radius of the blob-like structures. Because skin and vessels are not in blob shapes, these enhanced tissues should be of lower blob-like degrees and excluded. The degree was firstly normalized to a range between 0 and 1, and the voxels with the degree lower than 0.2 were eliminated.
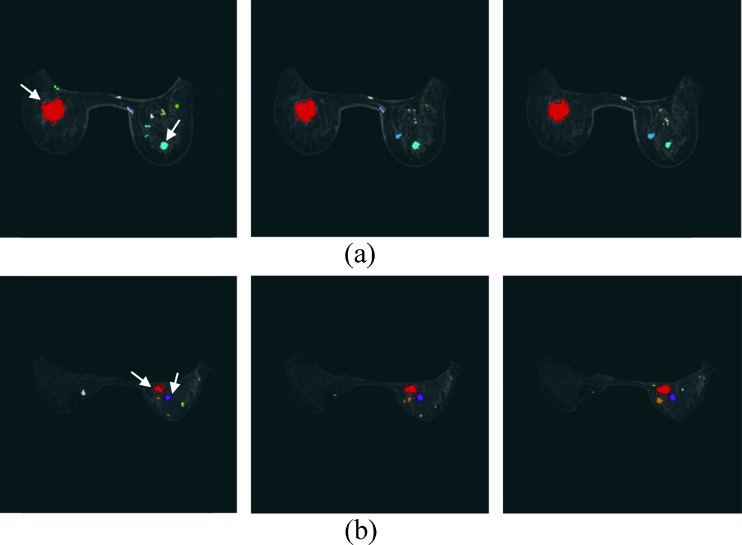
After detecting the mass voxels from the suspicious tissues, the voxels were grouped into regions using the 3-D connected component algorithm [22, 23] (Fig. 4). Then, the mass-like likelihood of each connected region was evaluated to differentiate the mass from other non-tumor regions.
Fig. 4.
Breast mass candidates from a Fig. 2 and b Fig. 3. The different connected regions are displayed by different colors. The tumor is indicated using white arrow
Mass Selection from Candidates
Some false positives were detected as candidates. If all candidates were considered real mass and estimate the tumor-like likelihoods, the processing might be time-consuming and the performance of the CADe system was poor. Before evaluating the likelihoods of these candidates, most false positives should be filtered out to avoid unnecessary evaluation. Because the size of most detected FPs was small, the candidates with smaller volumes [24] were considered the scattered FPs and reduced. Hence, only the 50 largest detected regions per case were considered as mass candidates for further tumor selection.
A probability value, called likelihood, was evaluated for predicting the mass-like likelihood. For evaluating the mass-like likelihood of each candidate, each mass candidate was evaluated for blob, enhancement, texture [25], morphologic [26], and combined features, including previous four feature sets. If the likelihood of a candidate was higher than the threshold value, THreg, this candidate was considered to be a mass.
Mass Selection by Blob and Enhancement Features
To select the masses from the candidates, the blob-like degree of all voxels inside the candidate was averaged as the blob-like degree of candidate. On the other hand, the enhancement degree of each candidate was computed by averaging the AUC characteristics of all the voxels inside the candidate. To select the relatively higher blob-like and enhancement degrees for each case, the rank features were used to sort the 2°, respectively, in descending order. That is, if a candidate is assigned with a smaller rank value, it is comparatively more blob-like or enhanced than the other candidates. Therefore, beside the 2° (Bd and Ed), two rank values (Br and Er) of the blob-like and enhancement degrees were applied (Table 1).
Table 1.
Definition of the features (blob, enhancement, morphological, and texture) for mass selection
| Features | Description | |
|---|---|---|
| Category | Symbol | |
| Blob features | B r | Blob-like degree |
| B d | Blob-like rank | |
| Enhancement features | E r | Enhancement degree |
| E d | Enhancement rank | |
| Morphological features | C 1 | Conventional compactness |
| C 2 | New compactness | |
| R | Radius | |
| R s | Spiculation | |
| ER axis | Axis ratio | |
| ER surface | Surface ratio | |
| V | Volume | |
| Texture features | G 1 | Energy |
| G 2 | Entropy | |
| G 3 | Correlation | |
| G 4 | Inverse difference moment | |
| G 5 | Inertia | |
| G 6 | Cluster shade | |
| G 7 | Cluster prominence | |
| G 8 | Haralick’s correlation | |
Mass Selection by Morphologic Features
Morphological features have been widely used to characterize breast tumors [26, 27]. In our study, seven morphologic features were used to measure the mass-like likelihood of candidates, including two compactness features (C1 and C2), radius (R), spiculation (Rs) [26], surface ratio (ERsurface), axis ratio (ERaxis) [26], and volume (V) features computed by the number of tumor voxels and image resolution (Table 1).
Mass Selection by Texture Features
The texture features computed from the gray level co-occurrence matrix (GLCM) of the first post-contrast acquisition measured not only the intensity distribution but also how correlated spatially a voxel is to its neighbors. GLCM features have been applied for medical image processing, including breast diagnosis [26] and classification [28]. In our study, eight GLCM characteristics [25] were applied to measure the mass-like likelihood of candidates for discrimination between tumors and normal candidates. These GLCM features included energy (G1), entropy (G2), correlation (G3), inverse difference moment (G4), inertia (G5), cluster shade (G6), cluster prominence (G7), and Haralik’s correlation (G8) (Table 1).
Statistical Analysis
To compare the ability to distinguish between mass and FP, statistical analysis was used to analyze features. Each feature was analyzed for normal distribution by the Kolmogorov-Smirnov test [29]. If the p value of Kolmogorov-Smirnov test was greater than or equal to 0.05, it indicated that the feature was normally distributed and the mean and standard deviation of the feature could be further analyzed. For these features, the differences between the masses and FPs were tested by Student’s t test [29]. Otherwise, the median of the feature was given, and the differences between masses and FPs were tested by the Mann-Whitney U test [29]. If the p value of Student’s t test or Mann-Whitney U test was less than 0.05, the features of the masses were considered significantly different from the FP features.
The combined features were selected using the backward elimination algorithm [29]. Two performance indices, detection rate, and FPs per case were used to evaluate the mass detection performance. The detection rate is defined as the number of the selected candidates divided by the number of all true masses confirmed by a radiologist. A higher detection rate means fewer missed masses. FPs per case indicates the number of detected non-tumor regions for each case. A smaller number of FPs indicate that fewer wrongly detected regions needed to be further identified by a radiologist. Using the proposed mass selection features, the mass-like likelihood of each selected region was evaluated by logistic regression [29]. The 10-fold method [30] was adopted for cross validation. All masses were partitioned into 10 groups that were approximately equal in size. In each step of the validation, all candidates in one group were considered as the testing data and the candidates in other nine groups were training data used to train the logistic regression model. The mass-like likelihood of each candidate in the testing data can be predicted by the trained regression model. The procedure was repeated until the likelihoods of all candidates were evaluated. If the evaluated likelihood of the mass region was greater than or equal to the threshold value, THreg, it was considered to be a mass. THreg increased at 0.1 increments from 0.0 to 1.0 to generate the free-response operating characteristics (FROC) [31] curves.
Experiments and Results
Statistical Analysis for Mass Selection
Not all features were normally distributed according to the Kolmogorov-Smirnov test; therefore, the median value and p value from the Mann-Whitney U test were used to analyze the mass selection (Table 2). Except for one texture feature, G4, all blob, enhancement, and morphologic features were significant different between masses and FPs (p < 0.05) (Table 2).
Table 2.
Comparison among different features for selecting masses from candidates
| Feature categories | Features | Type | Median | p value |
|---|---|---|---|---|
| Blob features | B r | Mass | 11 | <0.001* |
| FP | 54 | |||
| B d | Mass | 150 | <0.001* | |
| FP | 136 | |||
| Enhancement features | E r | Mass | 5 | <0.001* |
| FP | 48 | |||
| E d | Mass | 599 | <0.001* | |
| FP | 425 | |||
| Morphological features | C 1 | Mass | 21.80 | <0.001* |
| FP | 29.56 | |||
| C 2 | Mass | 70.45 | <0.001* | |
| FP | 54.19 | |||
| R | Mass | 4.84 | <0.001* | |
| FP | 3.12 | |||
| R s | Mass | 1.37 | <0.001* | |
| FP | 1.21 | |||
| ER surface | Mass | 1.61 | <0.001* | |
| FP | 1.39 | |||
| ER axis | Mass | 0.61 | <0.001* | |
| FP | 0.47 | |||
| V | Mass | 539.03 | <0.001* | |
| FP | 107.67 | |||
| Texture features | G 1 | Mass | 0.36 | 0.036* |
| FP | 0.50 | |||
| G 2 | Mass | 1.78 | <0.001* | |
| FP | 1.15 | |||
| G 3 | Mass | 0.28 | <0.001* | |
| FP | <0.001 | |||
| G 4 | Mass | 0.83 | 0.45 | |
| FP | 0.83 | |||
| G 5 | Mass | 0.35 | <0.001* | |
| FP | 0.27 | |||
| G 6 | Mass | −0.03 | <0.001* | |
| FP | <0.001 | |||
| G 7 | Mass | 0.55 | <0.001* | |
| FP | 0.07 | |||
| G 8 | Mass | 7.59 × 106 | <0.001* | |
| FP | 6.15 × 106 |
*p < 0.05 is considered statistically significant
The median value and p value of Mann-Whitney U test are used to compare each feature between mass and FP
The blob features are the blob-like degree and its rank value (B d and B r). The enhancement features are the enhancement features ant it rank value (E d and E r,). Morphological features are two compactness (C 1 and C 2), radius (R), spiculation (R s), surface ratio (ER surface), axis ratio (ER axis), and volume feature (V). Texture features are energy (G 1), entropy (G 2), correlation (G 3), inverse difference moment (G 4), inertia (G 5), cluster shade (G 6), cluster prominence (G 7), and Haralik’s Correlation (G 8)
Mass Selection by Different Features
After applying the proposed detection method, all masses were included among the candidates. Based on logistic regression, the tumor-like likelihood of each candidate was evaluated for the proposed selection features. In addition to the blob, enhancement, morphologic, and texture features, the combined features were selected from the all features with the backward elimination algorithm to select the masses (Table 3). Based on these feature sets, the detection performance was analyzed by the detection rate and the FPs per case.
Table 3.
Each feature set of the combined features used for mass selection
| Blob features | Enhancement features | Morphologic features | Texture features |
|---|---|---|---|
| B r | E r, E d | C 1, C 2, R, R s, ER surface, V | G 1, G 3, G 5, G 6, G 7, G 8 |
Mass Selection
The 50 largest regions from each patient detected by the Hessian method were considered to be mass candidates. The likelihood of each candidate was evaluated by blob, enhancement, morphologic, texture, and combined features (Table 3). The relation between the detection rate and FPs per case was analyzed using different likelihood thresholds (Table 4). The threshold, THreg, was set at 0.1 and 0.5 to separate masses from FPs based on the combined features. For all masses, the detection rate was 100 % (61/61) with 15.15 FPs and 91.80 % (56/61) with 4.56 FPs based on the combined features and THreg set at 0.1 and 0.5, respectively. For benign masses, the detection rate was 100 % (21/21) with 16.27 FPs and 76.19 % (16/21) with 4.80 FPs. For malignant masses, the detection rate was 100 % (40/40) with 15.27 FPs per case and 100 % (40/40) with 4.68 FPs per case. All masses were detected while THreg set at 0.1. But 5 benign masses were missed using THreg set at 0.5. These five masses (three fibrocystic changes and two papillomas) are considered as more easily missed by our system.
Table 4.
The detection rate and FPs per case for mass detection based on combined feature selection
| All cases | Benign cases | Malignant cases | ||||
|---|---|---|---|---|---|---|
| Detection rate | FPs per case | Detection rate | FPs per case | Detection rate | FPs per case | |
| Th = 0.0 | 100.00 % | 43.74 | 100.00 % | 46.27 | 100.00 % | 43.30 |
| Th = 0.1 | 100.00 % | 15.15 | 100.00 % | 16.27 | 100.00 % | 15.27 |
| Th = 0.2 | 96.72 % | 10.41 | 90.48 % | 11.00 | 100.00 % | 10.57 |
| Th = 0.3 | 91.80 % | 8.00 | 76.19 % | 8.60 | 100.00 % | 8.07 |
| Th = 0.4 | 91.80 % | 6.18 | 76.19 % | 6.80 | 100.00 % | 6.10 |
| Th = 0.5 | 91.80 % | 4.56 | 76.19 % | 4.80 | 100.00 % | 4.43 |
| Th = 0.6 | 81.97 % | 3.53 | 52.38 % | 3.67 | 97.50 % | 3.43 |
| Th = 0.7 | 78.69 % | 2.68 | 42.86 % | 3.00 | 97.50 % | 2.67 |
| Th = 0.8 | 73.77 % | 1.88 | 33.33 % | 2.13 | 95.00 % | 1.90 |
| Th = 0.9 | 62.30 % | 1.12 | 19.05 % | 1.20 | 85.00 % | 1.20 |
| Th = 1.0 | 0.00 % | 0.00 | 0.00 % | 0.00 | 0.00 % | 0.00 |
If the likelihood of detected candidates estimated by the combined features is larger than the logistic regression threshold value, Th, it is selected as a mass. If a case included benign and malignant masses, the case is considered as both benign and malignant case. There are 15 benign cases and 30 malignant cases
Detection Performance Analysis with FROC Curves
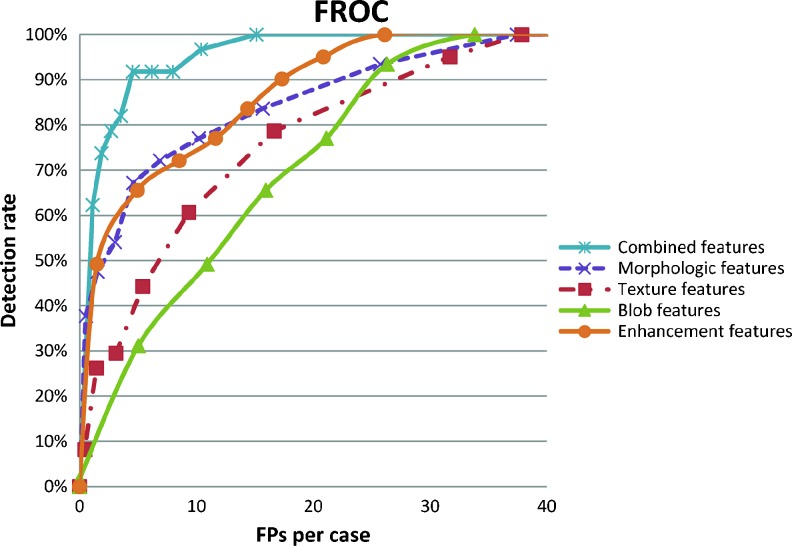
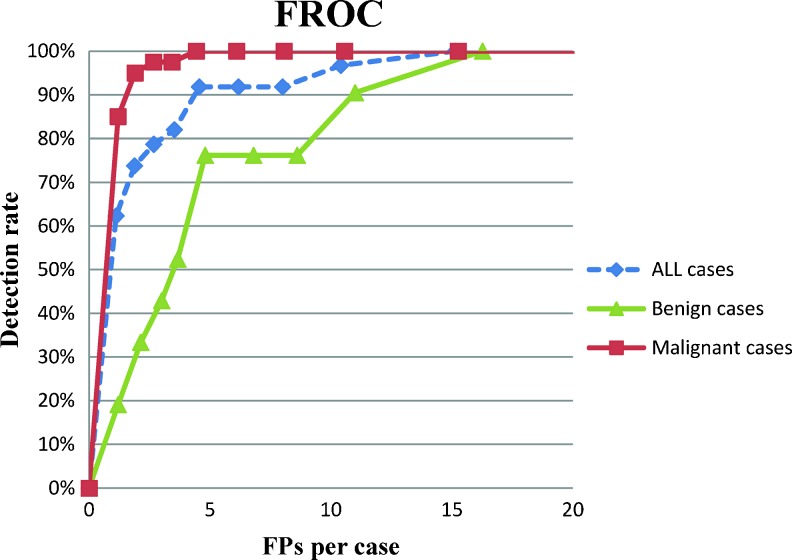
To compare the detection performances of the proposed CADe system using different feature sets, the FROC curve was used to analyze the relationship between the detection rate and the FPs per case (Fig. 5). The combined features were most useful to select masses, which are with higher detection rate and lower FPs. FROC curves for all, benign, and malignant masses using the combined features were shown in Fig. 6. Moreover, the detection performance estimated by the jackknife alternative of FROC-1 (JAFROC) figure of merit (FOM) [32, 33] was applied to analyze the FROC curve of different feature sets statistically (Table 5). The FOM describe whether the mass-like likelihood of tumor is higher than that of FPs. A larger FOM score indicates that the feature set could select mass well. The result reveals that the combined features are most useful to select breast masses and significantly better than other feature sets (p < 0.05).
Fig. 5.
The free-response receiver operating characteristic (FROC) curve of the proposed system for all cases using different feature set
Fig. 6.
The free-response receiver operating characteristic (FROC) curve of the proposed system for all cases, benign cases, and malignant cases using combined features
Table 5.
FOM values of five feature sets for mass selection
| Combined features | Morphological features | Texture features | Blob features | Enhancement features |
|---|---|---|---|---|
| 0.45◎†Φ║ | 0.36Θ‡ | 0.18$ | 0.09# | 0.31 |
All p values of FOM between different pairs of feature set
◎ p < 0.05 while comparison between combined features and morphological features; † p < 0.05 while comparison between combined features and enhancement features; Φ p < 0.05 while comparison between combined features and texture features; ║ p < 0.05 while comparison between combined features and blob features; Θ p < 0.05 while comparison between morphological features and texture features; ‡ p < 0.05 while comparison between morphological features and blob features; $ p < 0.05 while comparison between texture features and enhancement features; # p < 0.05 while comparison between blob features and enhancement features
Detection Performance Analysis According to Different Size Groups
Because small tumors are easily missed [8, 34], the relationship between the detection rate and the tumor size was further analyzed. The detection rate for different size groups is listed in Table 6. Among the five missed masses using THreg 0.5, four masses were smaller than 1 cm and one mass was between 1 and 2 cm (Table 6).
Table 6.
The detection rate for different size groups
| Tumor size | All masses | Benign masses | Malignant masses |
|---|---|---|---|
| ≦1 cm | 77.78 % (14/18) | 71.43 % (10/14) | 100 % (4/4) |
| >1 cm and ≦2 cm | 95.45 % (21/22) | 83.33 % (5/6) | 100 % (16/16) |
| >2 cm | 100 % (21/21) | 100 % (1/1) | 100 % (20/20) |
Discussion
Early tumor detection and proper treatment is important for increasing cancer survival rate [35]. Because MRI is the most sensitive technology for screening high-risk women for breast cancer [36], it has been widely applied by radiologists [8, 34]. Because the amount of three-dimensional (3-D) imaging data with current state-of-the-art MRI techniques is tremendous, the increasing capability of computer assistance was not only to avoid missing tumors but also to reduce interpretation time. To the best of our knowledge, however, multi-scale blob-based detection algorithms have not been applied to detect breast masses for 3-D DCE-MRI. In this study, we proposed a robust algorithm of 3-D tumor detection, including breast segmentation, mass detection, and mass selection to detect differential cancers from everything else in the breast.
In previous MRI detection studies [8, 34], malignant tumors were manually detected from MRI maximum intensity projection plane of enhanced MRI image sets. Nevertheless, the inter-observer inconsistency and variation are inevitable, and performance depends on the experiences of interpreting radiologists. To overcome these issues, automatic detection of benign and malignant breast masses using computer assistance was proposed for 3-D DCE-MRI data in this study. Renz’s study [37] proposed a detection algorithm to detect potential tumors using a hierarchical 3-D Gaussian pyramid to outline regions with high local intensity similarities. However, only enhanced contrast characteristics were used to evaluate tissues. Some normal tissues, such as fibroglandular, skin, and vessels, were also detected to increase the number of FPs. As a result, two malignant lesions were missed and the detection performance was 95.74 % with 17 FPs. In our study, not only enhancement but also blob features characterized the tumor tissues to improve the detection performance.
The whole breast region must be determined before detecting masses. In previous study [38], the border was identified manually by radiologists. However, manual segmentation was usually inconsistent and caused inter-observer variability. A straight line method [39] to segment breast cannot segment the breast tissue near the axilla. In contrast with the previous segmentation methods [38, 39], demons segmentation technique preserves the most of breast region because the outline can deform to segment breast [12, 15]. The semiautomatic breast segmentation initialized by three manual landmarks in this study can segment the precise breast region compared to previous studies. Although the manual landmarks might result in little inter-observer variability, it can improve the accuracy of breast segmentation for clinical practice.
All suspicious tissues from the segmented breast were identified using the FCM clustering method based on AUC feature. However, suspicious tissues identified only using the enhancement characteristic included many flat or lengthy non-tumor tissues. The blob characteristics [40] of masses were applied to further identify the masses from other normal tissues. After mass detection stage, we apply some features, such as the blob, enhancement, texture, morphologic, and combined features, to select the masses from the other normal candidates. Based on our experiment (Table 2), most features show significant difference between masses and FPs (p < 0.05), except for one texture feature, inverse difference moment (G4). In the previous study, the morphologic features [27] of breast MRI are important criteria to characterize breast masses and enhancement-related [13] features are useful to interpret tissues on DCE-MRI. According to the median and p value, the different masses and FPs were more significant in blob, enhancement, and morphological features than texture features. The experiment results indicated the high importance of 3-D morphological and enhancement for tumor detection. The detection performance using the five categories of features is listed in Fig. 5. The combined features performed better than individual features, with higher detection rates and fewer FPs per case. Table 4 and Fig. 6 show that the malignant masses were more easily selected than benign masses. The detection rate of malignant tumors is 100 %, higher than that of benign tumors at 76.19 %. To maintain a high sensitivity for detecting breast cancer, the malignant tumors should not be reduced and at the mass selection process. THreg values set at 0.5 as a suitable threshold to detect all malignant tumors in our experiment, compared to THreg values as 0.1 (Table 4). For the combined features, the detection performances were 100 % (61/61) and 91.80 % (56/61), respectively. Five benign masses were missed using a THreg of 0.5. The JAROC-1 FOM of different feature sets is listed in Table 5. The combined features and morphological features were more capable of detecting breast mass, which are with higher ROM values, 0.45 and 0.36, respectively. The p value for FOM between different pairs of feature sets indicated that the combined features can significantly improve the detection performance from other individual feature set. The morphological features are most used in clinical practice. The differences between morphological and texture features, as well as blob features, are also statically significant.
The p value of all the features for different types of detected regions and the five missed masses were analyzed using a THreg of 0.5. For the clinically used feature, enhancement degree (Ed), the missed masses were more similar with the FPs than malignant and benign masses (p > 0.05). The enhancement degrees (Ed) of the malignant and benign masses (753.78 ± 367.22 and 619.38 ± 209.63) were significantly larger than that of the missed masses (400.00 ± 119.26) (p = 0.018 and 0.032). The tumor volumes (V) of malignant masses (2765.03 ± 3347.59 mm3) were significantly larger than the missed masses (265.37 ± 140.55 mm3) (p = 0.003). Like previous studies [7, 34], masses which are relatively small or have a slight absence of perceptible enhancement could be missed. The miss-detected benign tumors were difficult to be detected using only enhancement and volume features. The morphologic features, C1, C2, R, Rs, ERsurface, ERaxis, and V computed from benign tumors were similar to those from FPs (p > 0.05). The result indicates that morphologic characteristics between the miss-detected masses and benign tumors, as well as normal tissues, are not significantly different. Therefore, they are difficult to be distinguishable only based on the morphologic characteristics. Because all texture features were larger than 0.05, single texture feature was not useful for mass detection. The detection rate analyzed according to different tumor size groups is listed using THreg of 0.5 (Table 6). The malignant masses were detected correctly among all size groups. The missed masses were benign and smaller in size. Prior study [41] with a large cohort of MRI-guided needle localization shows that lesions smaller than 5 mm have low likelihood of being cancer.
Some limitations in this study could be considered further. The voxels with blob-like degrees higher than the 0.2 obtained by experiments were considered to be masses in our study. However, the threshold to determine whether the voxel is a mass tissue can affect the detection performance. Because of poor spatial resolution (image size 256 × 256 pixels and slice thickness 2.5 mm), tumor tissues might seem to connect to enhanced normal tissues on images. Hence, these tumor tissues can be evaluated as lower blob-like degrees and easily filtered out by an automatically defined threshold. Therefore, an automatic algorithm to determine the threshold of blob-like degree on a higher resolution image will be important for future work involving more precise detection of blob-like tissues. Because the missed benign masses contain only small numbers of voxels, a few enhanced voxels can make analysis difficult. The use of higher resolution images might overcome this problem. On the other hand, kinetic features usually analyzed the malignancy likelihood of benign and malignant tumors [17], but the study about using time-intensity curve to distinguish tumor from non-tumor tissues were rare. In addition to the blob, morphological, and texture features, the pharmacokinetic might be incorporated to improve our system in the future.
Conclusion
In the proposed breast DCE-MRI computer-aided detection system, whole breast region is determined using the modified demons deformable algorithm. Masses are detected using multi-scale Hessian method. The mass detection rates are 100 % (61/61) with 15.15 false positives per case and 91.80 % (56/61) with 4.56 false positives per case. To date, this is the first study using a multi-scale blob-based tumor detection algorithm followed by morphologic tumor selection to automatically detect both benign and malignant tumors on 3-D DCE-MRI study. In conclusion, the developed DCE-MRI computer-aided detection (CADe) system might potentially assist physicians to detect mass before clinic diagnosis.
Acknowledgments
The authors would like to thank the National Science Council of the Republic of China (NSC 101-2221-E-002-068-MY3), Ministry of Economic Affairs (102-EC-17-A-19-S1-164), and Ministry of Education (AE-00-00-06) of the Republic of China for the financial support
Contributor Information
Yeun-Chung Chang, Email: ycc5566@ntu.edu.tw.
Ruey-Feng Chang, Phone: +886-2-33661503, FAX: +886-2-23628167, Email: rfchang@csie.ntu.edu.tw.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review, 1975–2008. Bethesda: National Cancer Institute; 2011. [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Roubidoux MA, Helvie MA, Lai NE, Paramagul C. Bilateral breast cancer: early detection with mammography. Radiology. 1995;196:427–431. doi: 10.1148/radiology.196.2.7617856. [DOI] [PubMed] [Google Scholar]
- 4.Kolb TM, Lichy J, Newhouse JH. Comparison of the performance of screening mammography, physical examination, and breast US and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. Radiology. 2002;225:165–175. doi: 10.1148/radiol.2251011667. [DOI] [PubMed] [Google Scholar]
- 5.Kelly KM, Dean J, Comulada WS, Lee SJ. Breast cancer detection using automated whole breast ultrasound and mammography in radiographically dense breasts. Eur Radiol. 2010;20:734–742. doi: 10.1007/s00330-009-1588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mogatadakala KV, Donohue KD, Piccoli CW, Forsberg F. Detection of breast lesion regions in ultrasound images using wavelets and order statistics. Med Phys. 2006;33:840–849. doi: 10.1118/1.2174134. [DOI] [PubMed] [Google Scholar]
- 7.Muradali D, Ghai S, Bukhanov K, Kulkarni S. Nonenhancing breast malignancies on MRI: sonographic and pathologic correlation. Am J Roentgenol. 2005;185:481–487. doi: 10.2214/ajr.185.2.01850481. [DOI] [PubMed] [Google Scholar]
- 8.Teifke A, Hlawatsch A, Beier T, Vomweg TW, Schadmand S, Schmidt M, et al. Undetected malignancies of the breast: dynamic contrast-enhanced MR imaging at 1.0 T. Radiology. 2002;224:881–888. doi: 10.1148/radiol.2243010547. [DOI] [PubMed] [Google Scholar]
- 9.Bluemke DA, Gatsonis CA, Chen MH, DeAngelis GA, DeBruhl N, Harms S, et al. Magnetic resonance imaging of the breast prior to biopsy. JAMA. 2004;292:2735–2742. doi: 10.1001/jama.292.22.2735. [DOI] [PubMed] [Google Scholar]
- 10.Warner E, Plewes DB, Shumak RS, Catzavelos GC, Di Prospero LS, Yaffe MJ, et al. Comparison of breast magnetic resonance imaging, mammography, and ultrasound for surveillance of women at high risk for hereditary breast cancer. J Clin Oncol. 2001;19:3524–3531. doi: 10.1200/JCO.2001.19.15.3524. [DOI] [PubMed] [Google Scholar]
- 11.Yeh ED. Breast magnetic resonance imaging: current clinical indications. Magn Reson Imaging Clin N Am. 2010;18:155–169. doi: 10.1016/j.mric.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Lin M, Chen J-H, Nalcioglu O, Su M-YL: Automatic template-based breast segmentation on MRI using nonrigid registration algorithms. In: International Society for Magnetic Resonance in Medicine (ISMRM) conference, 2012, pp. 2569, City
- 13.Galbraith SM, Lodge MA, Taylor NJ, Rustin GJS, Bentzen S, Stirling JJ, et al. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed. 2002;15:132–142. doi: 10.1002/nbm.731. [DOI] [PubMed] [Google Scholar]
- 14.Su MY, Nie K, Chen JH, Chan S, Chau MKI, Yu HJ, et al. Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI. Med Phys. 2008;35:5253–5262. doi: 10.1118/1.3002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroon D-J, Slump CH: MRI modality transformation in demon registration. In: IEEE International Symposium on Biomedical Imaging, ISBI ‘09. IEEE Signal Processing Society, City, 2009, pp. 963–966
- 16.Hayton P, Brady M, Tarassenko L, Moore N. Analysis of dynamic MR breast images using a model of contrast enhancement. Med Image Anal. 1997;1:207–224. doi: 10.1016/S1361-8415(97)85011-6. [DOI] [PubMed] [Google Scholar]
- 17.Chang YC, Huang YH, Huang CS, Chang PK, Chen JH, Chang RF. Classification of breast mass lesions using model-based analysis of the characteristic kinetic curve derived from fuzzy c-means clustering. Magn Reson Imaging. 2012;20:312–322. doi: 10.1016/j.mri.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Frangi AF, Niessen WJ, Vincken KL, Viergever MA: Multiscale vessel enhancement filtering, Medical Image Computing and Computer-Assisted Intervention - Miccai‘98 1496, 1998, pp. 130–137
- 19.Jiamin L, White JM, Summers RM: Automated detection of blob structures by Hessian analysis and object scale. In: Image Processing (ICIP), 2010 17th IEEE International Conference on, City, 2010, pp. 841–844
- 20.Jia T, Zhao D-Z, Wei Y, Zhu X-H, Wang X: Computer-aided lung nodule detection based on CT images. In: Complex Medical Engineering, 2007 CME 2007 IEEE/ICME International Conference on, City, 2007, pp. 816–819
- 21.Okada K, Akdemir U: Blob segmentation using joint space-intensity likelihood ratio test: application to 3D tumor segmentation. In: Computer Vision and Pattern Recognition, 2005 CVPR 2005 I.E. Computer Society Conference on, vol. 2, City, 2005, pp. 437–444
- 22.Rosenfel A. Connectivity in digital pictures. J ACM. 1970;17:146. doi: 10.1145/321556.321570. [DOI] [Google Scholar]
- 23.Rosenfel A, Pfaltz JL. Sequential operations in digital picture processing. J ACM. 1966;13:471. doi: 10.1145/321356.321357. [DOI] [Google Scholar]
- 24.Chen JH, Chang RF, Chang-Chien KC, Takada E, Huang CS, Chou YH, et al. Rapid image stitching and computer-aided detection for multipass automated breast ultrasound. Med Phys. 2010;37:2063–2073. doi: 10.1118/1.3377775. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med. 2007;58:562–571. doi: 10.1002/mrm.21347. [DOI] [PubMed] [Google Scholar]
- 26.Chang RF, Moon WK, Shen YW, Huang CS, Chiang LR. Computer-aided diagnosis for the classification of breast masses in automated whole breast ultrasound images. Ultrasound Med Biol. 2011;37:539–548. doi: 10.1016/j.ultrasmedbio.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Meinel LA, Stolpen AH, Berbaum KS, Fajardo LL, Reinhardt JM. Breast MRI lesion classification: improved performance of human readers with a backpropagation neural network computer-aided diagnosis (CAD) system. J Magn Reson Imaging. 2007;25:89–95. doi: 10.1002/jmri.20794. [DOI] [PubMed] [Google Scholar]
- 28.Haralick RM. Statistical and structural approaches to texture. Proc IEEE. 1979;67:786–804. doi: 10.1109/PROC.1979.11328. [DOI] [Google Scholar]
- 29.Field AP. Discovering Statistics Using SPSS. London: SAGE Publications; 2009. [Google Scholar]
- 30.Bengio Y, Grandvalet Y. No unbiased estimator of the variance of K-fold cross-validation. J Mach Learn Res. 2004;5:1089–1105. [Google Scholar]
- 31.Yu SY, Guan L. A CAD system for the automatic detection of clustered microcalcifications in digitized mammogram films. IEEE Trans Med Imaging. 2000;19:115–126. doi: 10.1109/42.896785. [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty DP. Validation and statistical power comparison of methods for analyzing free-response observer performance studies. Acad Radiol. 2008;15:1554–1566. doi: 10.1016/j.acra.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty DP, Breatnach ES, Yester MV, Soto B, Barnes GT, Fraser RG. Digital and conventional chest imaging: a modified ROC study of observer performance using simulated nodules. Radiology. 1986;158:35–39. doi: 10.1148/radiology.158.1.3940394. [DOI] [PubMed] [Google Scholar]
- 34.Shimauchi A, Jansen SA, Abe H, Jaskowiak N, Schmidt RA, Newstead GM. Breast cancers not detected at MRI: review of false-negative lesions. Am J Roentgenol. 2010;194:1674–1679. doi: 10.2214/AJR.09.3568. [DOI] [PubMed] [Google Scholar]
- 35.Anderson BO, Shyyan R, Eniu A, Smith RA, Yip CH, Bese NS, et al. Breast cancer in limited-resource countries: an overview of the Breast Health Global Initiative 2005 guidelines. Breast J. 2006;12(Suppl 1):S3–S15. doi: 10.1111/j.1075-122X.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 36.Kuhl CK, Schrading S, Bieling HB, Wardelmann E, Leutner CC, Koenig R, et al. MRI for diagnosis of pure ductal carcinoma in situ: a prospective observational study. Lancet. 2007;370:485–492. doi: 10.1016/S0140-6736(07)61232-X. [DOI] [PubMed] [Google Scholar]
- 37.Renz DM, Bottcher J, Diekmann F, Poellinger A, Maurer MH, Pfeil A, et al. Detection and classification of contrast-enhancing masses by a fully automatic computer-assisted diagnosis system for breast MRI. J Magn Reson Imaging. 2012;35:1077–1088. doi: 10.1002/jmri.23516. [DOI] [PubMed] [Google Scholar]
- 38.Pisano ED, Hendrick RE, Yaffe MJ, Baum JK, Acharyya S, Cormack JB, et al. Diagnostic accuracy of digital versus film mammography: exploratory analysis of selected population subgroups in DMIST. Radiology. 2008;246:376–383. doi: 10.1148/radiol.2461070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JH, Moon WK, Shen YW, Huang CS, Luo SC, Kuzucan A, et al. Comparative study of density analysis using automated whole breast ultrasound and MRI. Med Phys. 2011;38:382–389. doi: 10.1118/1.3523617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorr RT, Alberts DS. Quantitation of ellipsoid tumor areas using a circumferential measuring device. Med Oncol Tumor Pharmacother. 1988;5:249–251. doi: 10.1007/BF03003192. [DOI] [PubMed] [Google Scholar]
- 41.Liberman L, Mason G, Morris EA, Dershaw DD. Does size matter? Positive predictive value of MRI-detected breast lesions as a function of lesion size. AJR Am J Roentgenol. 2006;186:426–430. doi: 10.2214/AJR.04.1707. [DOI] [PubMed] [Google Scholar]