Abstract
Cancer screening with magnetic resonance imaging (MRI) is currently recommended for very high risk women. The high variability in the diagnostic accuracy of radiologists analyzing screening MRI examinations of the breast is due, at least in part, to the large amounts of data acquired. This has motivated substantial research towards the development of computer-aided diagnosis (CAD) systems for breast MRI which can assist in the diagnostic process by acting as a second reader of the examinations. This retrospective study was performed on 184 benign and 49 malignant lesions detected in a prospective MRI screening study of high risk women at Sunnybrook Health Sciences Centre. A method for performing semi-automatic lesion segmentation based on a supervised learning formulation was compared with the enhancement threshold based segmentation method in the context of a computer-aided diagnostic system. The results demonstrate that the proposed method can assist in providing increased separation between malignant and radiologically suspicious benign lesions. Separation between malignant and benign lesions based on margin measures improved from a receiver operating characteristic (ROC) curve area of 0.63 to 0.73 when the proposed segmentation method was compared with the enhancement threshold, representing a statistically significant improvement. Separation between malignant and benign lesions based on dynamic measures improved from a ROC curve area of 0.75 to 0.79 when the proposed segmentation method was compared to the enhancement threshold, also representing a statistically significant improvement. The proposed method has potential as a component of a computer-aided diagnostic system.
Keywords: Computer-aided diagnosis, Magnetic resonance imaging, Breast, Cancer, Supervised learning, Pattern recognition
Introduction
Regular breast cancer screening has been identified as a critical component towards improving breast cancer survival rates [1]. Genetic mutations on the BRCA1/2 genes can result in up to an 85 % lifetime risk of developing breast malignancies [2]. Dynamic contrast-enhanced magnetic resonance imaging (MRI) has been shown to be the most sensitive screening method for the detection of breast cancer in high risk women [3]. The American Cancer Society has recommended that women with a lifetime breast cancer risk of 20–25 % or greater should receive MRI-based screening starting at age 25 to 30 [4]. MRI screening has also been shown to detect cancers missed by mammography and ultrasound in women at moderately elevated cancer risk who have dense breasts [5]. Magnetic resonance imaging-based breast screening is likely to play an increasing clinical role in the future.
It has been shown that there is a high degree of variability between trained radiologists in their ability to correctly diagnose lesions from breast MRI examinations [6]. Breast MRI examinations typically involve the acquisition of hundreds of images compared with just four images for typical x-ray mammogram-based breast cancer screening. Variable radiologist performance and the time consuming nature of radiologic analysis of large breast MRI examinations provides motivation for the research, design, and development of computer-aided diagnosis systems to assist breast MRI radiologists in identifying very early stage malignancies.
When analyzing a contrast-enhanced breast MRI examination, a radiologist will visually inspect the images for a number of markers of malignancy. Patterns in the changes in lesion signal intensity over time (i.e., rapid uptake followed by a washout phase) can be indicative of cancer, and these dynamics constitute one of the main features that a radiologist looks for when reading a breast MRI examination. Radiologists also look for spiculated lesions (or generally irregularly shaped lesions), tumor margins that are not sharp and heterogeneous tissue vascularization, all of which are suggestive of cancer and together influence their final diagnosis according to the Breast Imaging-Reporting and Data System (BI-RADS) lexicon. Assessing tumor characteristics based on the visual assessment of a radiologist is susceptible to human error, which highlights the need for automated methods for characterizing potentially malignant lesions. This has motivated substantial research towards the development of computer-aided diagnostic (CAD) systems [7–12]. CAD systems for breast MRI often focus on the dynamic information (how a lesion’s signal intensity changes over the course of the examination after the injection of a contrast agent). However, fully characterizing suspicious lesions should incorporate additional measurements such as assessing a lesion’s margin. A semi-automatic segmentation algorithm has the potential to clearly delineate a lesion from surrounding breast tissue and thus to facilitate the extraction of measurements that might assist in the diagnosis of radiologically suspicious lesions.
Segmentation of suspicious lesions from breast MRI examinations has been the subject of considerable research. Research in breast MRI lesion segmentation has been performed using the fuzzy c-means algorithm [7–9], the Markov Random Field Model [10] as well as combining the k-means algorithm with the Markov Random Field [11]. Lesion segmentation has also been performed manually [12] which is very time consuming and subject to human error. The c-means and k-means algorithms are seeded with the results of a random number generator which can cause reproducibility problems because the algorithm’s random initialization is different each time the algorithm runs. The Markov random field method has multiple parameters which make it challenging to produce optimal segmentations as the researcher has several parameters to tune in order to configure the system for optimal operation in any given application. This paper presents an alternative technique for the segmentation of suspicious lesions from breast MRI examinations based on a proposed supervised learning method that is easy to use and typically requires no parameter tuning.
Materials and Methods
Screening Study
Women at high risk for breast cancer based on family history or a known genetic mutation were recruited for a high risk cancer screening study which included annual breast MRI in addition to mammography. Participation in screening was offered to all eligible women in the context of genetic counseling. Informed consent was obtained from all participants. The data was collected at Sunnybrook Health Sciences Centre in Toronto, Canada as part of an imaging study that recruited 550 high risk women.
The screening protocol used is as follows. Simultaneous bilateral magnetic resonance imaging was performed using a 1.5 T magnet (GE Signa, version 11.4). Sagittal images were obtained with a phased-array coil arrangement using a dual slab interleaved bilateral imaging method [13]. This provided 3D volume data over each breast obtained with an RF spoiled gradient recalled sequence (SPGR, scan parameters: TR/TE/angle = 18.4/4.3/30°, 256 × 256 × 32 voxels, FOV: 18 × 18 × 6–8 cm). Imaging is performed before and after a bolus injection of 0.1 mmol/kg of contrast agent (Gd-DTPA). Each bilateral acquisition was obtained in 2 min and 48 s. Slice thickness was 2 to 3 mm. The screening study that produced the data used in this paper has been the subject of many other studies [14–19].
Current Study
Ethics approval for this retrospective study was obtained from the institutional review board of Sunnybrook Health Sciences Centre. This retrospective analysis includes 49 malignant lesions and 184 benign lesions. Ground truth for malignant lesions is based on the analysis of tissue biopsies by a histopathologist. When the histopathologist determines a tissue sample to be non-cancerous, a benign diagnosis is accepted. In cases where a suspicious mass did not receive a biopsy but returned to screening without observed changes to the lesion for greater than 1 year, then a benign diagnosis is also accepted.
Image registration is the process of aligning images that vary in position over time. This is performed to compensate for any patient motion that occurs during the examination which can obscure acquired lesion measurements and in extreme cases hide a small lesion completely. For this study, we used a three-dimensional non-rigid registration technique for magnetic resonance breast images that was applied globally to the breast examination [14], in order to help ensure that breast tissues are spatially aligned at each time point.
Proposed Semi-Automatic Segmentation Method
The proposed method for semi-automatic segmentation of suspicious breast MRI lesions is evaluated in the context of a computer-aided diagnostic system. This system involves having the user draw an ellipse to define examples of a lesion deemed suspicious (see Fig. 1 middle frame red ellipse). A second ellipse is drawn defining tissue deemed not suspicious (see Fig. 1 middle frame blue ellipse). The semi-automatic lesion segmentation method described here is formulated as a supervised learning problem. The samples contained within the first (red) ellipse are assigned as the positive training samples. The samples contained within the second (blue) ellipse are assigned as the negative training samples. The proposed method for performing semi-automatic lesion segmentation is formulated as a supervised learning problem and defined as:
 |
1 |
where, 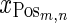 is the positive training data with m samples and n measurements
is the positive training data with m samples and n measurements
Fig. 1.
An example malignant lesion (left pane), with user-defined ellipses marking suspicious tissue in red and non-suspicious tissue in blue (center pane). The resultant region-of-interest (ROI) is computed as defined in equation 1 (right pane—red lines mark the edges of the defined ROI)
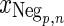 is the negative training data with p samples and n measurements
is the negative training data with p samples and n measurements
sign(x) = 1, x > = 0; sign(x) = ‐ 1, x < 0
α is the input bias parameter ranging from 0 to 1
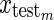 is a single test vector of n measurements replicated in m rows
is a single test vector of n measurements replicated in m rows
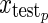 is a single test vector of n measurements replicated in p rows
is a single test vector of n measurements replicated in p rows
This equation was formulated to be capable of solving the supervised learning problem without a tightness of fit parameter (such as gamma in the radial basis function [15, 16]). This formulation was developed to modify an existing equation [16] in order to make it easy to use, not requiring the user to tune a parameter that controls the tightness of the separating classification function. The input test vector and the positive and negative training sets consist of the relative MRI signal intensity values scaled in the range 0 to 1.
For each lesion in this study, a 25 × 25 voxel bounding square was extracted around each suspicious lesion for further analysis (see blue squares on Fig. 2). The testing samples are each voxel within the 25 × 25 voxel bounding square. A 25 × 25 patch was chosen as we have many small lesions detected in our highly sensitive screening program and a 25 × 25 patch allows visual analysis of our smallest lesions (about 2 to 3 mm across). The alpha term in equation 1 is a biasing parameter that allows the user to modify the final segmentations to be inclined to group more samples as part of the positive group (alpha >0.5) or to group more samples as part of the negative non-suspicious group (alpha <0.5). However, the algorithm is quite reliable at the default setting of 0.5 providing no bias between the two user-defined groups. All the results presented in this study were obtained at an alpha setting of 0.5. This parameter is discussed in more detail in the discussion.
Fig. 2.
Final regions-of-interest produced by the proposed technique demonstrated on malignant lesions (above the green line) and benign lesions (below the green line). Red lines mark the border between a lesion and its surrounding non-suspicious tissue. Blue lines separate pixel patches extracted from each of the examinations included in this study
Validation/Statistical Analyses
The above proposed procedure was run individually on each lesion. Comparative segmentation was also performed using the enhancement threshold set to 60 % based on the results of a previous study [17]. A margin measure was computed for each segmented lesion [18] based on both the proposed method and the enhancement threshold set at 60 % [17]. This measure assesses the margin at the contrast peak and divides it by the margin at the final time point; thus, the measure highlights lesions whose margins become less sharp over the course of the examination [18]. Segmentations provided by the proposed method and the comparative enhancement threshold method are used to compute the margin measure. The receiver operating characteristic (ROC) curve area was computed assessing the separation obtained between our malignant and benign lesions based on the margin measure extraction from the proposed method’s regions-of-interest as well as from the enhancement threshold generated regions-of-interest. The Wilcoxon sign rank paired statistical test was run to determine the existence of statistically significant improvements when using the proposed technique relative to the enhancement threshold.
We are interested in determining if the lesion segmentation method is producing accurate regions-of-interest. However, we do not have per voxel ground truth on which to evaluate the results of each segment (region-of-interest). As such, we are evaluating the technique based on its ability to support the computation of a margin measure indicative of malignancy. It is believed that improved separation between malignant and benign lesions as assessed by their margins [18] is an effective way of evaluating segmentation performance in this context. The ROC area produced for margin measurements for each method was computed and the paired Wilcoxon sign rank test was used to compare the margin measurements obtained after the use of either segmentation technique.
We have also elected to evaluate the segmentation method based on the established signal enhancement ratio (SER) method for assessing vascular dynamics. The SER method is defined as SER = (SIpeak − SIpre)/(SIfinal − SIpre), where SIpeak is the signal intensity closest to the peak of the bolus injection, SIpre is the signal intensity of the pre-contrast volume, and SIfinal is the signal intensity of the final post contrast acquisition. For this analysis, the average SER values across each segmented lesion were computed for both the proposed method and the enhancement threshold method. The ROC area produced for the average SER values for each method was computed, and the paired Wilcoxon sign rank test was used to compare the average SER results obtained after the use of either segmentation technique.
Results
An example of the procedure for defining the training samples is provided in Fig. 1 where the red ellipse was drawn first and defines tissue of interest (in this case a malignant lesion) and the second ellipse (drawn in blue) defines non-suspicious tissue. The resultant segmentation produced by the proposed method is provided in Fig. 1 (right pane). The resultant delineated regions-of-interest produced by the proposed supervised learning method for each lesion included in this study are provided in Fig. 2 with red lines marking the final lesion delineations. Blue squares delineate the local neighborhood of relative signal intensity values around each lesion (each square is 25 × 25 voxels or 17.5 × 17.5 mm). The first three rows plus the first four samples of the fourth row of the montage in Fig. 2 represent the 49 malignant lesions included in this study (all of the samples above the green line). The remaining samples below the green line consist of benign lesions.
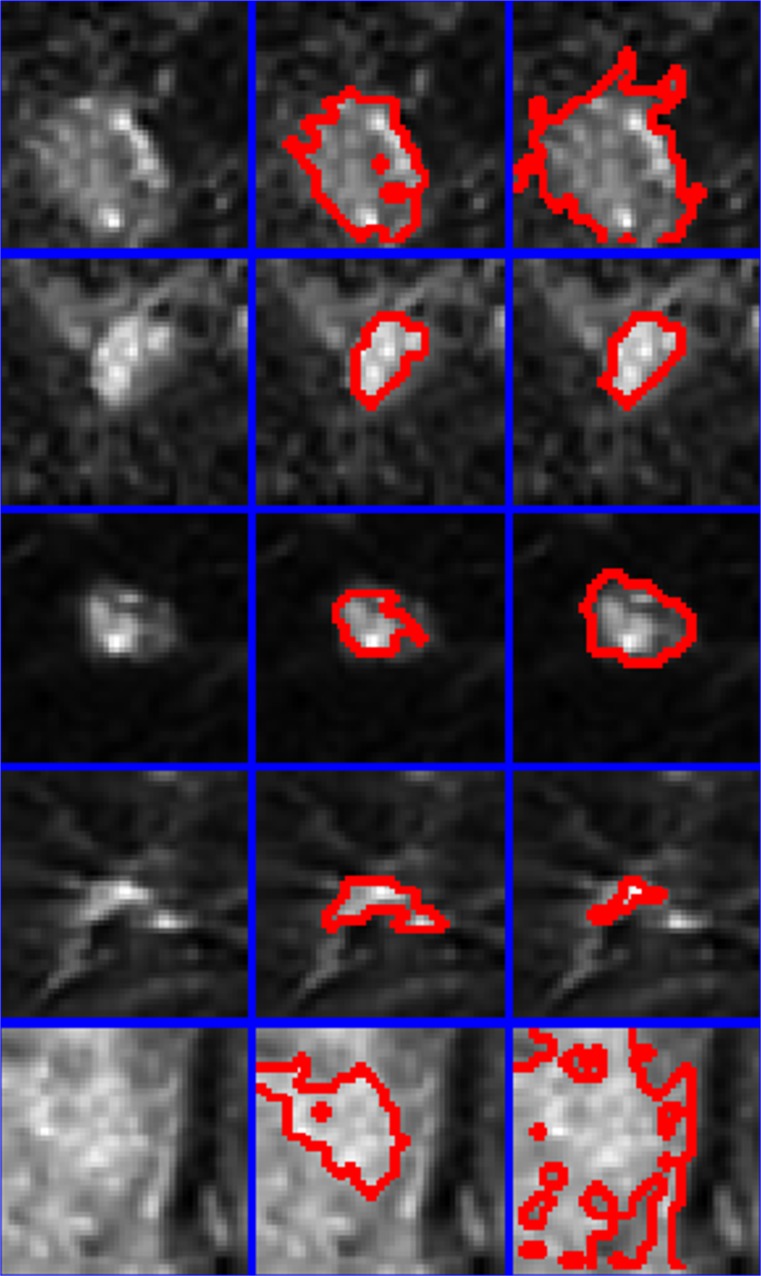
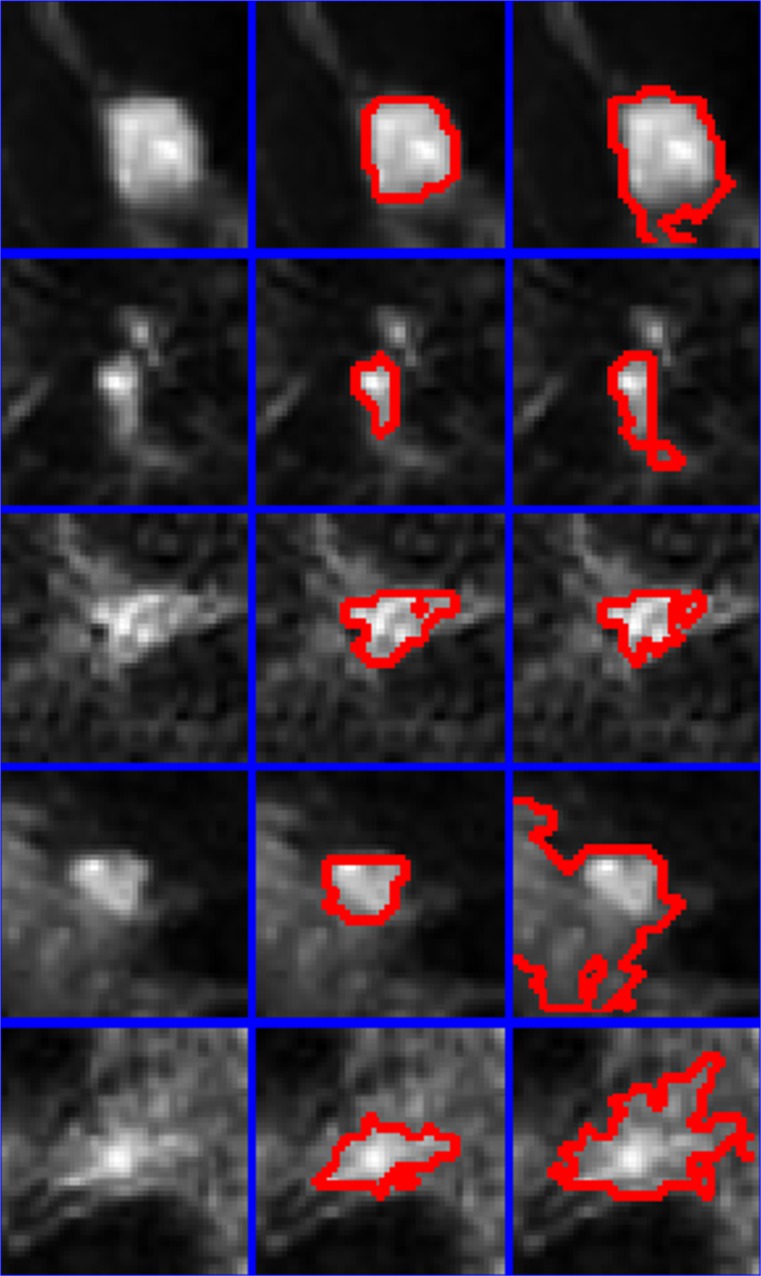
Semi-automatic segmentation using equation 1 was performed on all of the lesions in the dataset (results provided in Fig. 2). The enhancement threshold was also used to segment lesions at a setting of 60 % [17] for comparative purposes. Figures 3 and 4 provide magnified images of malignant and benign lesions, respectively in the left column, the results of the proposed technique is provided in the middle column and compared with the enhancement threshold in the right column.
Fig. 3.
Five malignant lesions are presented (left column) along with comparative regions-of-interest produced by the proposed technique (center column) and the 60 % enhancement threshold (right column). Red lines mark the border between a lesion and its surrounding non-suspicious tissue. Blue lines separate pixel patches extracted from examinations included in this study
Fig. 4.
Five benign lesions are presented (left column) along with comparative regions-of-interest produced by the proposed technique (center column) and the 60 % enhancement threshold (right column). Red lines mark the border between a lesion and its surrounding non-suspicious tissue. Blue lines separate pixel patches extracted from examinations included in this study
We have elected to compare these two lesion delineation techniques by extracting a known measure of malignancy and comparing how much separation is obtained between our malignant and benign lesions from either segmentation approach. We have computed the receiver operating characteristic (ROC) curve areas for both the signal enhancement ratio and a margin assessment metric [18] as computed for each of the two segmentation approaches addressed. The enhancement threshold produced a ROC area based on the margin measure [18] of 0.6309. The proposed segmentation method produced regions-of-interest from which a margin measure [18] was computed yielding a ROC area of 0.7315. This represents a statistically significant improvement over the results obtained from enhancement threshold-based segmentation as measured by the Wilcoxon sign rank test (p < 0.0001). The enhancement threshold segmentation resulted in regions-of-interest from which mean signal enhancement ratio (SER) measurements were computed yielding a ROC area of 0.7510. The proposed segmentation method produced regions-of-interest from which mean SER measurements were computed yielding a ROC area of 0.7857. This represents a statistically significant improvement over the results obtained by the enhancement threshold based on the Wilcoxon sign rank test (p < 0.001). SER computations were performed by averaging all of the computed SER values within each region-of-interest.
Discussion
The proposed semi-automatic region-of-interest segmentation algorithm presented was evaluated on an extremely challenging breast MRI screening dataset consisting of many lesions that are just 2 to 5 mm across (see Fig. 2 where each blue box is 17.5 by 17.5 mm). The proposed method yielded improvements in the separation between malignant and benign lesions as assessed by a margin measurement [18] and the established signal enhancement ratio when compared with enhancement threshold-based lesion segmentation. The enhancement threshold method has a tendency to overestimate the final region-of-interest in circumstances where the suspicious lesion is located within fibroglandular tissue undergoing gross physiological enhancement as demonstrated in Fig. 3, bottom row. The enhancement threshold method also has a tendency to underestimate small early stage malignancies as demonstrated in Fig. 3, fourth row, where the enhancement threshold not only appears to underestimate the lesion volume, but misses a secondary enhancement site immediately adjacent to the main tumor site.
In the standard use of supervised learning algorithms (when a CAD system’s final prediction is based on measurements acquired from many examinations), testing on our training data is correctly discouraged as it can easily overestimate the actual separation available between our cancers and benign samples through overfitting. This is quite dangerous in the context of a standard supervised machine learning-based CAD system which is trained on many malignant and benign samples simultaneously. It should be noted that the approach presented here is quite different. Although testing and training are performed on the same data, this is performed individually on each examination, thus it will not cause the typical problems associated with overestimating the separation between malignant and benign lesions as it is merely testing a lesion and its surrounding tissue as being more like the user provided tissue of interest (Fig. 1, red ellipse) or more like the less-suspicious tissue (Fig. 1, blue ellipse). Any error that this paradigm introduces is limited to the segmentation of each individual lesion. Thus, if a given segmentation is deemed overly biased due to the selected training samples, the user can simply redefine the training samples or make use of the alpha parameter in order to bias the results towards what is qualitatively deemed correct. In this study, the proposed technique performed quite well, each sample in Fig. 2 was computed at an alpha setting of 0.5, indicating no bias between the two groups defined by user drawn ellipses.
The main limitation of this study is that it was performed on a single screening dataset. Future work will involve testing this approach in the context of multiple independent challenging screening datasets. An additional limitation of the study is that it included measures for assessing the lesion’s margin as well as its standard vascular dynamics; future work will look at adding measures of vascular heterogeneity and irregularity of the lesion shape as potential additional computable markers of malignancy. This study is also limited by an inability to assess false positives and false negatives on a per voxel basis as we do not have ground truth regarding the true diagnosis of each voxel due to the fact that aligning (or registering) MRI data to pathological findings is an unsolved research problem. Qualitative assessment indicates that the technique is more prone to error when the suspicious lesion is contained within grossly enhancing ductal tissue; however, it is capable of delineating the lesion successfully in this context and outperforms the enhancement threshold. An additional limitation of the study is that it only considers individual measures of malignancy but does not investigate combining those measurements as part of a classification system designed to predict malignancy based on a combination of many measurements. An additional limitation of this approach is that its performance will vary based on the distance of the not suspicious ellipse relative to the suspicious ellipse if that distance introduces variation in the type and distribution of non-suspicious normal tissues. The goal is to segment a suspicious lesion away from its surrounding non-suspicious tissue and so the technique naturally benefits from local estimation of the tissue not of clinical interest. It is beneficial for the non-suspicious ellipse to include a variety of non-suspicious tissues in the immediate vicinity of the lesion.
Future work will investigate using supervised learning equations (such as the one we have presented here) towards making a final tissue diagnosis based on accumulated training samples from many lesions (i.e., the normal way a supervised learning algorithm is incorporated into a computer-aided diagnostic system). The proposed method presented in this paper is dependent on user input as it is a semi-automatic segmentation technique. Future work will investigate inter and intraobserver variability by having multiple specialists repeatedly use the tool. Differences in the final computed regions-of-interest will be evaluated. Future work will also investigate methods for automatically acquiring the positive and negative training data that seed the learning algorithm in order to produce a fully automatic system that does not depend on user input.
Conclusions
The proposed method for semi-automatic segmentation of breast MRI lesions has been tested on a challenging screening dataset containing many small lesions. The approach was demonstrated to assist in the extraction of markers for malignancy such as the assessment of a lesion’s margin as well as its tissue’s temporal dynamics. The proposed method was demonstrated to outperform the established enhancement threshold-based segmentation method in this context.
Acknowledgments
The MRI data was acquired using funding from the Canadian Breast Cancer Research Alliance. The authors would also like to thank the Canadian Breast Cancer Foundation and the Canadian Institute for Health Research for their financial support for this research project.
Conflicts of Interest
The authors report no conflict of interest.
References
- 1.Curry SJ. Fulfilling the potential of cancer prevention and early detection. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- 2.Ford S, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner E, et al. Systematic review: using magnetic resonance imaging to screen women at high risk for breast cancer. Annals of Internal Medicine. 2008;148(9):671–679. doi: 10.7326/0003-4819-148-9-200805060-00007. [DOI] [PubMed] [Google Scholar]
- 4.Saslow D, et al. American Cancer Society Guidelines for Breast Screening with MRI as an adjunct to mammography. Cancer J Clin. 2007;57:75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 5.Berg W, et al. Detection of breast cancer with addition of annual screening ultrasound or a single screening MRI to mammography in women with elevated breast cancer risk. Journal of the American Medical Association. 2012;307(13):1394–1404. doi: 10.1001/jama.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren R, et al. A test of performance of breast MRI interpretation in a multicentre screening study. Magn Reson Imaging. 2006;24(7):917–929. doi: 10.1016/j.mri.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Giger M, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Academic Radiology. 2006;13(1):63–72. doi: 10.1016/j.acra.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Giger M, Bick U, Newstead G. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Medical Physics. 2006;33:2878. doi: 10.1118/1.2210568. [DOI] [PubMed] [Google Scholar]
- 9.Nie K, et al. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Academic Radiology. 2008;15(12):1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q, et al., Interactive lesion segmentation on dynamic contrast enhanced breast MRI using a Markov model. Proceedings SPIE Medical Imaging 2006: Image Processing, 6144, 2006, San Diego, USA
- 11.Xiaohua C, Brady M, Lo J, Moore N. Simultaneous segmentation and registration of contrast-enhanced breast MRI. Information Processing in Medical Imaging Lecture Notes in Computer Science. 2005;3565:126–137. doi: 10.1007/11505730_11. [DOI] [PubMed] [Google Scholar]
- 12.Woods B, et al. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. Journal of Magnetic Resonance Imaging. 2007;25(3):495–501. doi: 10.1002/jmri.20837. [DOI] [PubMed] [Google Scholar]
- 13.Greenman RL, et al. Bilateral imaging using separate interleaved 3D volumes and dynamically switched multiple receive coil arrays. Magn Reson Med. 1998;39:108–115. doi: 10.1002/mrm.1910390117. [DOI] [PubMed] [Google Scholar]
- 14.Martel AL, et al. Evaluating an optical-flow-based registration algorithm for contrast-enhanced magnetic resonance imaging of the breast. Phys Med Biol. 2007;52(13):3803–3816. doi: 10.1088/0031-9155/52/13/010. [DOI] [PubMed] [Google Scholar]
- 15.Levman J, et al. Classification of dynamic contrast-enhanced magnetic resonance breast lesions by support vector machines. IEEE Transactions on Medical Imaging. 2008;27(5):688–696. doi: 10.1109/TMI.2008.916959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levman J, et al. A vector machine formulation with application to the computer-aided diagnosis of breast cancer from DCE-MRI screening examinations. Journal of Digital Imaging. 2014;27:145–151. doi: 10.1007/s10278-013-9621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levman J, et al. Effect of the enhancement threshold on the computer-aided detection of breast cancer using MRI. Academic Radiology. 2009;16(9):1064–1069. doi: 10.1016/j.acra.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levman J, Martel AL. A margin sharpness measurement for the diagnosis of breast cancer from magnetic resonance imaging examinations. Academic Radiology. 2011;18(12):1577–1581. doi: 10.1016/j.acra.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Warner E, et al. Surveillance of BRCA1 and BRCA2 mutation carriers with magnetic resonance imaging, ultrasound, mammography, and clinical breast examination. Journal of the American Medical Association. 2004;29(11):1317–1325. doi: 10.1001/jama.292.11.1317. [DOI] [PubMed] [Google Scholar]