Abstract
To improve data quality and save cost, clinical trials are nowadays performed using electronic data capture systems (EDCS) providing electronic case report forms (eCRF) instead of paper-based CRFs. However, such EDCS are insufficiently integrated into the medical workflow and lack in interfacing with other study-related systems. In addition, most EDCS are unable to handle image and biosignal data, although electrocardiography (EGC, as example for one-dimensional (1D) data), ultrasound (2D data), or magnetic resonance imaging (3D data) have been established as surrogate endpoints in clinical trials. In this paper, an integrated workflow based on OpenClinica, one of the world’s largest EDCS, is presented. Our approach consists of three components for (i) sharing of study metadata, (ii) integration of large volume data into eCRFs, and (iii) automatic image and biosignal analysis. In all components, metadata is transferred between systems using web services and JavaScript, and binary large objects (BLOBs) are sent via the secure file transfer protocol and hypertext transfer protocol. We applied the close-looped workflow in a multicenter study, where long term (7 days/24 h) Holter ECG monitoring is acquired on subjects with diabetes. Study metadata is automatically transferred into OpenClinica, the 4 GB BLOBs are seamlessly integrated into the eCRF, automatically processed, and the results of signal analysis are written back into the eCRF immediately.
Keywords: Clinical trial, Image analysis, Workflow, Electronic data capture, Web services, OpenClinica
Introduction
In controlled clinical trials, patient’s data is nowadays collected using electronic data capture systems (EDCS), which provide electronic case report forms (eCRF) instead of paper-based CRFs [1]. Automatic evaluation of data by range checks or more complex expressions prevents errors immediately during data acquisition. This avoids elaborative query processing, improves quality of data and saves costs. However, EDCS are often designed as standalone software and suffer from insufficient integration of shared data with surrounding study-related systems. In addition, integration and automatic processing of image and biosignal files are not supported at all. Particularly in multicenter clinical trials, available tools inadequately support collaborations in imaging and image analysis [2] although imaging is important in many clinical trials, e.g., in studies for development of neuropharamacological drugs [3]. For this, novel data workflow concepts are required for clinical trials and for personalized medicine, which make efficient use of high-throughput data such as gene expressions [4].
The open source software OpenClinica has been established as one of the world’s leading EDCS and clinical data management systems (CDMS) [5, 6]. The web application offers mighty functionality for collection, management, and storage of subject data in multicenter clinical trials. However, (i) OpenClinica is designed as standalone system and is not interfacing with other clinical trial-related software, neither the clinical trial management systems (CTMS) nor components of hospital information systems, such as picture archiving and communication systems (PACS); (ii) integration of large image and biosignal data obtained from computed tomography (CT), magnetic resonance imaging (MRI), ultrasound (US), and electrocardiography (ECG) into the eCRF as well as management of such binary large objects (BLOBs) is not supported, and (iii) automatic post-processing of BLOBs data for quantitative analysis, measurements, and classification tasks is not possible.
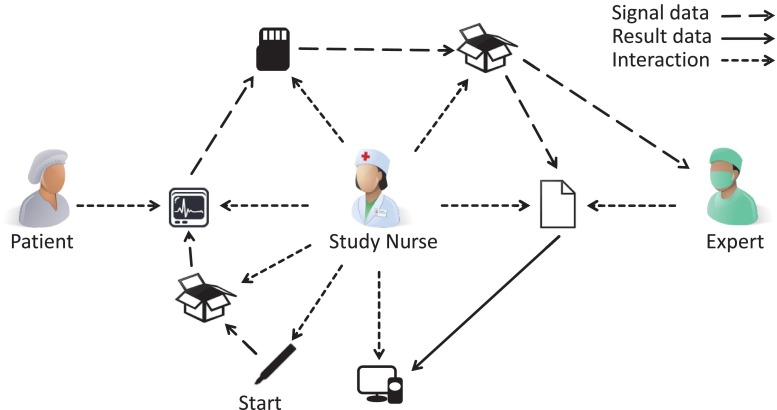
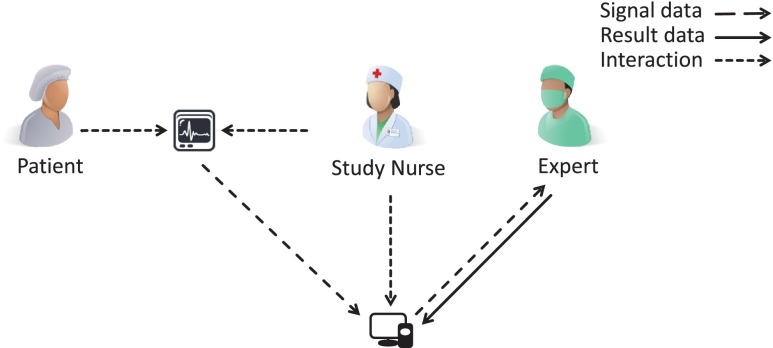
Hence, clinical trials today are characterized by a discontinuous process, which aggravate the workflow of physicians and study nurses. Manual interaction is needed for initialization, image or biosignal data integration and analysis leading to evitable staff effort, error-proneness, latency, as well as data privacy and quality issues. This clearly becomes obvious by tracking a typical multisite trial (Fig. 1). Manual workflow interaction is currently required for (i) preparation and labeling of material for data acquisition (e.g., patient identifier labels on memory cards); (ii) bundling and shipping of material to imaging site via mail; (iii) recording of biosignal or image data on patient’s body; (iv) copying captured data from the recording device to a transferrable memory card or compact disc; (v) shipping of that data storage to an expert for analysis; (vi) expert’s reporting of analysis results, usually send by facsimile, and (vii) manually entering analysis results into the EDCS. Furthermore, these sources of potential failure correspond to the major finding of de Carvalho et al. when analyzing poor data quality due to inappropriate workflow in clinical trial sites [7]: (i) multiplicity of data repositories, (ii) lack of standardized process for data registration at source documents, and (iii) scarcity of decision support systems at the point of research intervention.
Fig. 1.
Conventional EDC workflow in clinical trials with biosignal data acquisition and analysis
Recently, the lung tissue research consortium (LTRC) has established an infrastructure for data and tool sharing in lung disease research [2]: LTRC is a multicenter project, which is sponsored by the National Institutes of Health (NIH) and the NIH Heart, Lung and Blood Institute. LTRC improves disease management by integration of data and tissue repositories. In addition, the LTRC project provides standardized methods for sample collection, de-identification, transport, analysis, result reporting, and preparation for redistribution. Beside of tissue data for lung fluid specimens, CT chest data, and laboratory studies, the LTRC supports application and development of computer-assisted analysis, such as algorithms for automatic texture analysis, vascular and bronchial attenuation pattern recognition, and calculation of lung parenchymal volumes. For this, a research infrastructure has been created, which offers data, image, and tool sharing through web, using digital imaging and communications in medicine (DICOM) and electronic mail. However, LTRC is a rather specific approach designed exclusively to the demands in lung research and bio banking.
The cancer biomedical informatics grid (caBIG) of the National Cancer Institute (NCI) provides a computational network connecting scientists with institutions and offering data sharing and analytical tools [8]. It aims at development of software (e.g., CTMS, pathology tools, tissue banks) for management and analysis of BLOB data to facilitate collaborations across the cancer research. Moreover, caBIG provides its own OpenClinica add-on.1 The caBIG’s OpenClinica package includes web services interfacing OpenClinica with caBIG’s clinical connector. The interface automatically creates study, sites, and subjects in OpenClinica accessing caBIG’s lab data. In addition, caBIG provides a plug-in for single sign-on (SSO) to OpenClinica.2 Although founding principles will be continuously developed in the National Cancer Informatics Program (NCIP), in 2012, caBIG has been retired.3
ImagEDC4 software of Novartis, Switzerland, is an EDCS supporting DICOM data transfer in multicenter clinical trials [9]. It provides functions for de-identification, tagging with study-related identifiers, and cleaning of image data. Additionally, it supports submission of images to caBIG compatible grid services. All record workflow events are captured by a tracking service. ImagEDC’s data transfer service is based on service-orientated architecture (SOA), which is also provided by web services. It is, however, a commercial product with limited access in investigator initiated trials (IIT) due to its high costs.
The retrieve form for data capture (RFD) by the Integrating the Healthcare Enterprise (IHE) initiative [10] is a specification for gathering data in an external application. With RFD, an active application is able to receive a form from a source application and to allow data modifications in their forms, while the user stays in the current system. After finishing, the form is sent back to the source application. Application of RFD compliant systems allows direct eCRF entry on each site, however, any transfer of BLOB data still is impossible.
In summary, existing IT approaches provide methods partly supporting the medical workflow in research and clinical trials. Some systems offer functionality for data sharing and others support BLOB integration and management. In addition, caBIG and LTRC supply collections of data analysis algorithms and ImagEDC offers methods for addressing caBIG services. However, all approaches rather provide tools or data repositories than seamless integration of EDC within the clinical trial workflow. Only caBIG’s OpenClinica package and RFD aim at workflow optimization providing data interfaces. Furthermore, caBIG and LTRC are focused on categorical scientific fields, such as cancer or lung tissue research, disregarding requirements of clinical trials in general.
In this paper, we present a simple concept for seamless workflow integration of EDCS for controlled clinical trials reducing error-proneness and latency. We address the enhancement of data sharing and inclusion of BLOB data into eCRFs. Additionally, we embed automatic analysis of image and biosignal data, which is increasingly used as image-based surrogates or imaging biomarkers. We show the functionality of our work by means of a multicenter clinical trial, in which patient-related medical observations are correlated with ECG recordings, indeed BLOBs of about 4 GB of data.
Material and Methods
Before designing and implementing workflow enhancement in clinical trials, we define general requirements, limitations, and architectural prerequisites. We also describe the test bed used to demonstrate the benefits of the proposed architecture.
Requirements Analysis
Based on the emphasized drawbacks of existing systems and our particular needs, we describe core features required in any clinical trial workflow.
In general (G), a system should
Provide new functionality for data integration, distribution, and automatic analysis.
Extend the ECDS adaptively without modifying its implementation.
Integrate in eCRF design process.
Decompose into modular components, each being flexible and configurable.
Interface with any EDCS.
In consequences, the system architecture is structured in certain modules.
The interface (I) module should
Share data with other systems that already host needed information.
Support data, context, and functional integration.
Offer a mechanism for automatically login.
The data (D) module should
Integrate BLOBs of theoretically unlimited volume, only restricted by free disk space.
Ensure stable and complete data transfer.
Improve usability by multi-selection and auto-compression of files.
Visualize the transfer progress.
Annotate transferred files with context information (e.g., study, subject, and visit ID).
The analysis (A) module should
Integrate java-based algorithms for data analysis.
Extract input parameters automatically, such as the file ID from the eCRF.
Write results back into the eCRF supporting any data type (e.g., numerical values, alphanumerical text, signal and image data).
Specify input and output parameters of black-box modules.
Trigger post-actions.
Existing Components
OpenClinica (Community Edition, Version 3.1.4) is an open source EDCS and CDMS. The web application supports design of studies with user-defined eCRFs and is used for data acquisition in multisite clinical trials. OpenClinica follows industry standards and is approved by regulatory authorities such as the Food and Drug Administration (FDA). The web application is developed with JavaServer Pages and uses a PostgreSQL database. OpenClinica is structured in several packages, e.g., core functionality (OC Core) and web services (OC WS).
To manage and control clinical trials at Uniklinik RWTH Aachen, Aachen, Germany, the study management tool (SMT) has been developed as proprietary CTMS. Meta information such as the name and type of the study, involved study sites, and the study personnel including their roles and access levels are stored. The software has been developed with the Google Web Toolkit (GWT) with Hibernate5 and Gilead6 interfacing a MySQL database. The web application allows entry and modification of data and generates data visualization as animated Flash charts or reports using Jaspers Reports.7
Interface Module
Since SMT as well as OpenClinica process data of clinical trials, similar data types are used, such as study, user, and roles objects. A user of the SMT can easily transfer metadata of a study and its dependencies to OpenClinica [11]. When triggering this action, the selected study and study-related users and sites are created automatically in OpenClinica based on the information already captured in the SMT. A mapping scheme between user roles, such as statisticians in the SMT to data specialist in OpenClinica, joins different view and right concepts of both applications. In addition, transferred users can login to OpenClinica directly from the SMT. For this, a randomized token that is used as OpenClinica password is generated automatically and securely stored in SMT.
The interface module and its implementation can be seen from two points of view: OpenClinica and SMT. OpenClinica already provides web service endpoints with various methods such as for exchange of study data. Additional endpoints have been implemented for user data and corresponding logic (Table 1). The SMT implements web service client functionality to address OpenClinica’s endpoints and its services. The auto-login is implemented by a HTTP request object including the OpenClinica user name and the corresponding password token. These credentials are then sent to the OpenClinica login page with JavaScript.
Table 1.
OpenClinica web service endpoints and provided methods used by the interface module
| Endpoint | Method | Additional |
|---|---|---|
| Study | listAll | |
| getMetadata | ||
| addUserToStudy | X | |
| create | X | |
| User | listAll | X |
| create | X |
Data Module
So far, upload of large data files into OpenClinica is not supported. The data module is designed to overcome those drawbacks [12]. According to our requirements, it is embedded into the eCRF and substitutes the native OpenClinica file upload component. The user is allowed to upload data of volume only limited by the server’s hard disk space. It has been demonstrated at SIIM Annual meeting 2013, and named OC Big.
Following security policies, modern browsers restrict file transfer to a data volume limit of 4 GB. Larger files are automatically decomposed into multiple parts, so called chunks, and such chunks are combined again on the receptor’s site. Optionally, the files are automatically compressed for transfer. In general, all uploaded files are stored on the OpenClinica server and linked to the context (e.g., study, event, and subject). Usability is increased by transfer progress visualization and drag-and-drop support. Uploaded files are integrated automatically in the eCRF. Important settings of the module, such as chunk size and supported BLOB formats, are located in a central configuration file.
Analysis Module
The analysis module has been partly presented in [13]. It is a generic extension of OpenClinica allowing connection with black-box signal and image processing. The outsourced process is triggered after uploading the data from the eCRF. After process completion, the results are imported automatically into the eCRF. The black-box processing is generic designed and can be substituted by any java-based algorithm following specifications for parameter and result transfer.
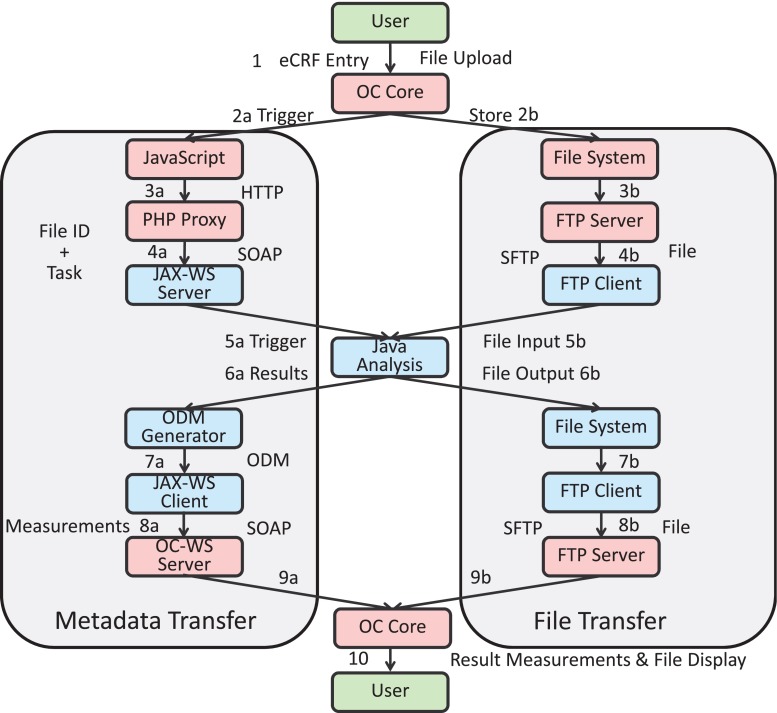
Figure 2 depicts the transfer of metadata and data files on its left and right hand sides, respectively. The workflow of the analysis module starts in step 1 with button click invoking two processes: In step 2a, JavaScript code is triggered to build a HTTP request with metadata. This request is sent to a PHP proxy on the same server, transforming it into a web service message (step 3a). The web service call is then forwarded to the GWT web service server component.
Fig. 2.
Workflow of the analysis module with metadata and file transfer side
The redirection of the metadata information via PHP proxy is necessary because of the same origin policy of modern web browsers [14, 15]. Nowadays, browser software is restricted to security policies to avoid execution of malicious code in the user’s browser on client side. As one of these security mechanisms, the same origin policy prohibits cross domain calls from client side programming languages such as JavaScript. Following the same origin policy, modern browsers do not allow request or modifications of elements without the same origin, namely same hostname, protocol, and port. In general, various workarounds to avoid cross domain calls exists [16, 17]. Here, we use a PHP proxy.
Using the PHP proxy workaround, a web service message is transferred to the WS endpoint on the GWT application server (step 4a). Meanwhile, the uploaded data is stored on the file system by OC Core in step 2b. From here, it is transferred by an SFTP server (step 3b) on the OpenClinica server to a SFTP client on the GWT application server (step 4b). In step 5a, a Java signal file analysis algorithm is triggered by the JAX-WS component, which fetches the file from file transfer side (step 5b). The algorithm for data processing is provided as Java code and packed in a jar file. A so called Worker API is implemented in the GWT application, allowing control of the black-box code by the GWT application and specification of parameters and result values, which are returned by the algorithm in step 6a.
Resulting files are stored on the GWT application server’s file system (step 6b). In the next step, the results are processed by an object data module (ODM) generator, which embeds the measurements in the Clinical Data Interchange Standards Consortium (CDISC)8 ODM format, and forwards the ODM file to the JAX-WS client (step 7a). The ODM format is based on the extensible markup language (XML) and is used for exchange of clinical trial-related data in information systems [18]. OC WS offers a web service endpoint for ODM. Hence, a well-formed XML file with OpenClinica object identifiers (OIDs) for eCRF field elements and corresponding values is sent to the import-method of OC WS (step 8a). As before, the result file is transferred back to the OpenClinica file system via SFTP (steps 7b and 8b). After metadata import, the result measurements and files are embedded into the eCRF of OC Core (steps 9a and 9b) and displayed to the user (step 10).
Evaluation
To assess the impact in clinical trials and to demonstrate seamlessness of the proposed workflow, a certain clinical trial was selected where (i) multiple centers are involved, (ii) OpenClinica is used as EDCS, (iii) data of surrounding systems is available for integration, (iv) ECG long-term recordings of 4 GB are captured, and (v) the data files are analyzed automatically by java-based algorithms.
The selected study aims at identification of surrogate markers for sudden cardiac death in patients with diabetes mellitus and end-stage renal disease (ESRD) and their modulation by beta-blocker treatment. The study is supported by the European Foundation for the Study of Diabetes (EFSD), performed according to ethical requirement and the good clinical practice (GCP) guideline, and registered at ClinicalTrials.gov (NCT02001480).
Based on this exemplary trial, we aim at quantitatively measuring the impact of our approach defining metrics that address failings of existing systems such as latency and additional staff efforts. Staff efforts are quantified counting the number of items/characters which are saved from double entry. Here, we make use of our SMT database hosting about 825 trials at the Uniklinik RWTH Aachen. Latency is measured with respect to Fig. 1, counting estimates for workflow steps that have been avoided.
Results
The resulting system architecture and workflow for the EFSD study is presented based on the interface, data, and analysis module.
System Architecture
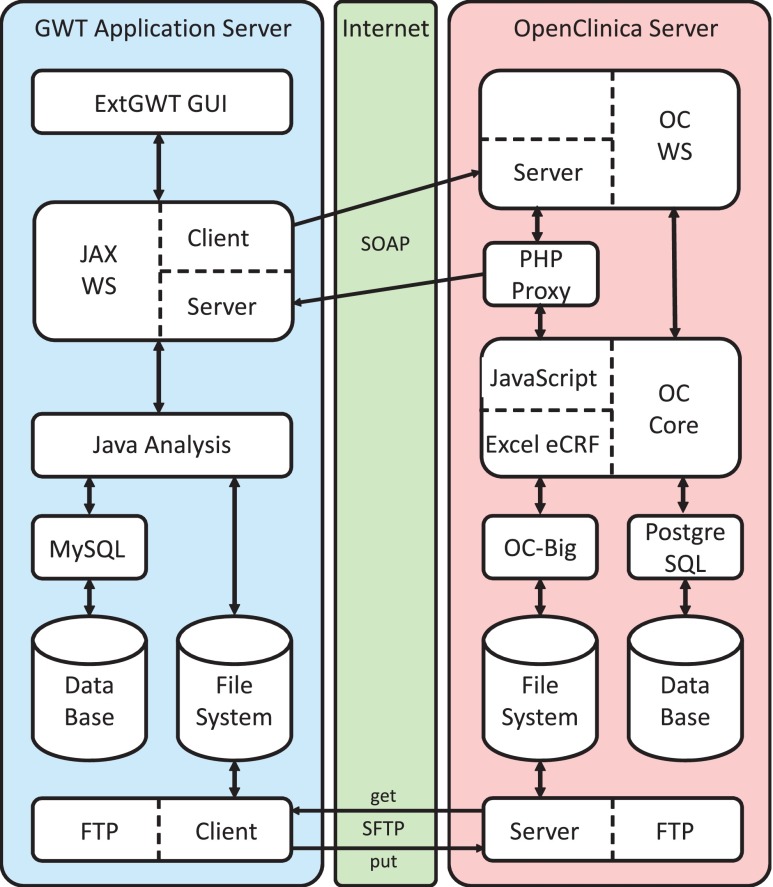
The system architecture consists of the GWT application server representing the SMT (Fig. 3, left side), an OpenClinica server (Fig. 3, right side), and the internet (Fig. 3, middle) as communication channel between both parts.
Fig. 3.
Interfacing and architecture of SMT and OpenClinica
On the GWT side, the architecture consist of the following: (i) an ExtGWT graphical user interface (GUI); (ii) JAX-WS client and server module to trigger, receive, and process web service messages; (iii) a Java analysis component in terms of an embedded jar file providing black-box image and signal processing; (iv) a MySQL database; (v) the server’s file system; and (vi) an SFTP client.
The OpenClinica site is composed of the following: (i) OC WS package, providing a web service server for querying OpenClinica; (ii) a PHP proxy server to work around cross domain calls; (iii) OC Core, which is enriched by JavaScript as web service client for triggering web service messages; (iv) OC Big for upload of large data files to the file system and integration of transferred files into the eCRF; (v) a PostgreSQL database; and (vi) an SFTP server.
ECD Workflow
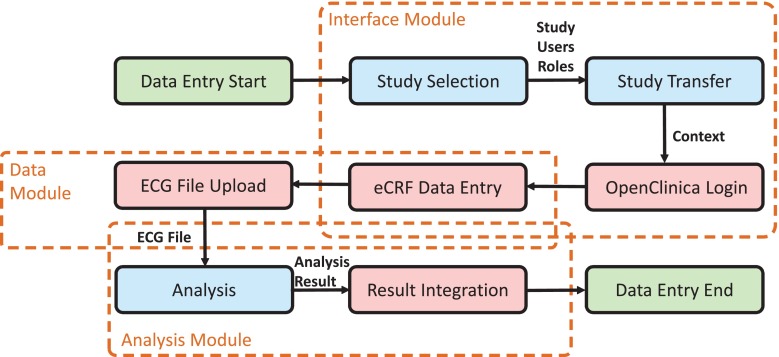
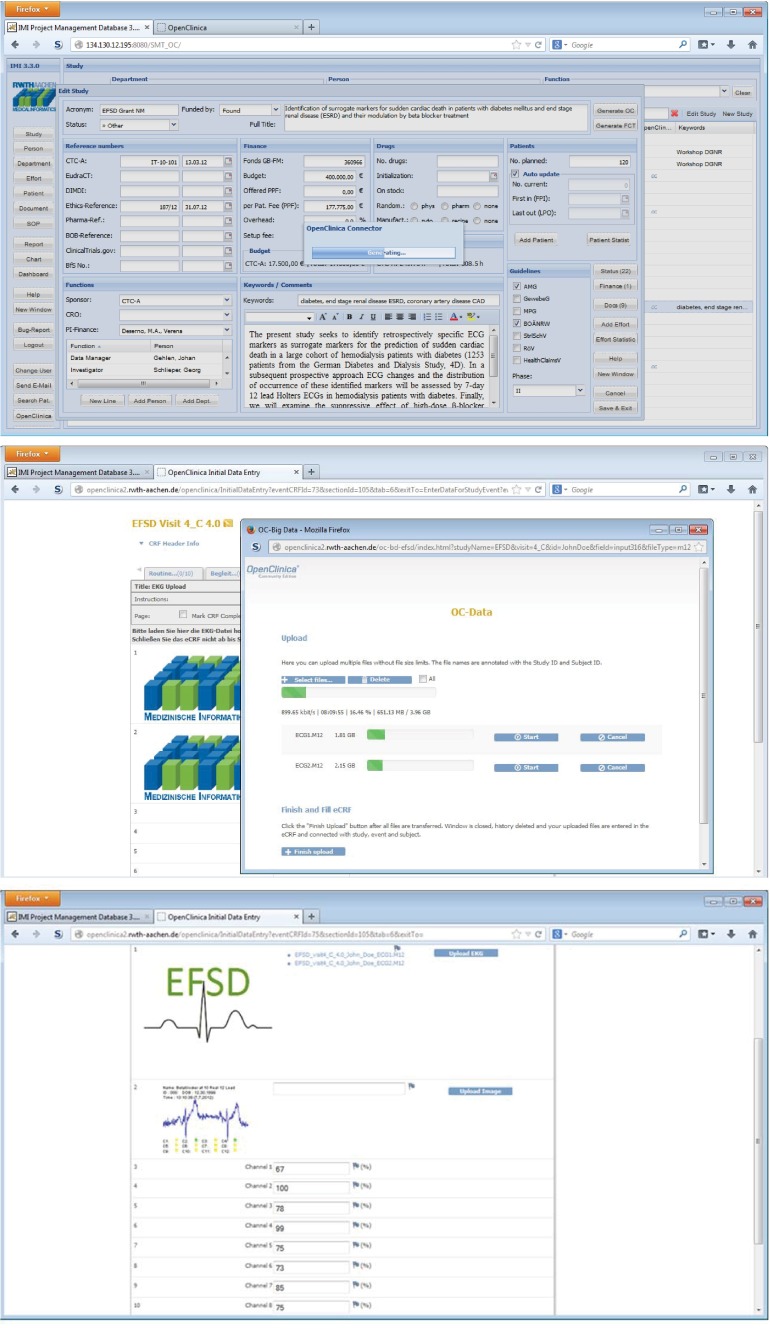
The ECD workflow for the EFSD study was implemented according to our proposed scheme using the interface module, data module, and analysis module (Fig. 4). The data entry process is triggered by the user, e.g., a study nurse, starting in the SMT with an overview of all trials. After selection of the EFSD study, the user initiates the transfer of objects, including study metadata, users, and role objects into OpenClinica using the interface module. After successful transfer, a study-related OpenClinica login button appears in SMT, which logs the user automatically into OpenClinica (Fig. 5, top). Now, OpenClinica’s subject matrix is shown and the user can open an eCRF and enter data. After filling the eCRF with patient’s data, the user uploads ECG recordings (Fig. 5, middle). The ECG signal files contain patient’s activity data for 7 days and 24 h, resulting in data volumes about 4 GB per day. Using the data module OC Big, a robust transfer of these files is ensured. Filenames are annotated with context information by appending study, event, and subject name. Resulting links for download are integrated into the eCRF. After data transfer, the ECG files are sent to the analysis module. Here, a quality check is performed calling a black-box algorithm. The resulting quality measurements are inserted for all channels in designated fields (Fig. 5, bottom).
Fig. 4.
Integrated data entry workflow
Fig. 5.
Results. Metadata is shared between SMT and EDCS using the interface module (top); ECG recordings are integrated in OpenClinica using the data module OC Big (middle); and results of quality checks are entered into the eCRF using the analysis module (bottom)
Requirement Analysis
Regarding our concept in general, all of the defined requirements are fulfilled. The system offers functionality for data sharing, large image and signal file integration, and analysis (Req. G1) without modifying existing OpenClinica code (Req. G2). All OpenClinica-related modifications are based on OC WS by adding new endpoints. Hence, compatibility between our extended version of the OC WS package and new releases of OC Core is ensured. System integration is done by embedding JavaScript code snippets into the eCRF (Req. G3). Furthermore, our system is structured in specific modules, which can be individually activated (Req. G4). In addition, our concept is transferable to all EDCS, which provide interfaces for data exchange (Req. G5). Although it is not guaranteed that other EDCS provide web service functionality, mechanism for data exchange can be transferred to other technologies.
The interface module shares data with surrounding study-related systems (Req. I1). SSO is offered from SMT to OpenClinica (Req. I3). The concept of storing a hidden randomized token as OpenClinica password is advantageous, since the user neither has to share passwords with other systems nor needs to remember several passwords. However, context and functional integration are not provided, since OpenClinica automatically selects the last active study of each user after login. This only can be changed by critical modifications of the OpenClinica database or general changes in OC Core, which could be a better solution but may cause problems with new releases (Req. G2). Hence, requirement I2 is not fulfilled yet.
Integration of large volume data into OpenClinica’s eCRFs is provided by the data module. Since data files are chunked into multiple parts before transfer, data volume is only restricted by server’s free hard disc space and not by browser limits (Req. D1). Chunking also ensures a stable transfer, since transfers errors only affect the current part (Req. D2). The data module includes several features improving usability, for instance, multiselection of files, drag-and-drop, and queue transfer. Specific features can be configured providing easy adaption for various applications. Moreover, compression functionality is supplied as optional feature (Req. D3). During data transfer detailed progress information are visualized (Req. D4) and files are automatically annotated with eCRF context information (Req. D5) ensuring unambiguous association of de-identified data with the study subject.
Integration of black-box java-based data processing is given by the analysis module (Req. A1). The file name is forwarded automatically from the data module. In general, such parameters are extracted from the eCRF after upload (Req. A2) and result measurements are automatically written back to the eCRF (Req. A3). Results are directly integrated into the eCRF and not affected by OpenClinica session timeouts, although eCRF modifications are still protected. In addition, the Worker API integrates and controls black-box algorithms without any knowledge on implementation details (Req. A4) and allows triggering of post-actions (Req. A5).
Impact on ECD Workflow
We measure the efforts of the study data manager with and without the application of our system. Without the interface module, a study in the EDCS is initialized entering manually the study name, all study sites and all user names and account data. For instance in OpenClinica, a study is described by 10 fields (Unique Protocol ID, Brief Title, Official Title, Secondary IDs, Principle Investigator, Brief Summary, Detailed Description, Sponsor, and Collaborators), in which data has to be entered manually. With respect to the Official Title field only and based on all 825 clinical trials monitored in the SMT in average and the ESFD study in particular, savings are counted to 158.33 and 177 characters, respectively. Furthermore 2.70 and 5 sites as well as 5.08 and 14 persons are linked to a study, respectively. Again, persons and sites require several fields to be filled. Using our system, all that information is transferred automatically.
Once the study is initialized, the EDC workflow is performed for any patient enrolled to the trial. Using our approach, the EFSD study workflow is majorly simplified (Fig. 6): (i) ECG is captured on the patient using any memory device; (ii) data is integrated into the EDCS and linked to the patient; (iii) data is analyzed by an expert, and (iv) results are stored back into the EDCS by the expert. In the EFSD study, the human expert is replaced by fully automatic computer analysis. In comparison to the conventional workflow (Fig. 1), both shipping actions introducing greatest latency are avoided. In the entire process for the ESFD study, electronic data transfer times for approx. 4 GB of ECG recording is the only latency remaining, which depends on the internet connection of the local site.
Fig. 6.
Workflow of EDC in the EFSD study with support by an EDCS interfacing data and analysis functionality
Discussion
Almost 15 years ago, Pavlovic, Kern & Miklavcic have proven the diverse advantages of electronic over paper-based CRFs in controlled clinical trials [1]. Electronic data capture has fairly been established nowadays, but mostly, EDCS are driven stand-alone, not being interconnected to other eHealth applications. In particular, handling of image and biosignal data still is supported insufficiently. This also holds for open-source clinical trial data management, which, in line with Fegan & Lang [19], is seen as the future of workflow management. Many others have already said that there is no “one-size-fits-all” solution for web-based documentation of clinical trials [20], and that open-source allows easily combining several components to fulfill the demands, as proposed in our approach.
In consequence, a system has been built, which reduces error-prone manual working steps for study personal in the EDC workflow, resulting in time (and cost) savings and quality improvements. We estimate 30 to 60 min working time for initialization of a study, the study sites and the persons and roles, which is saved completely. This estimate is based on data in a private study management database. In ClinicalTrials.gov, a study title contains 89.72 characters (as averaged on all 163,376 studies listed on 20th of March, 2014), which is somewhat below our findings. This may be due to language and translations issues and is not considered as general inconsistency.
Introduction of the data and analysis module obviously reduces necessary working steps for the study nurse in multisite trials, since patient’s data and processing result is directly exchanged using the EDCS and time-costly mailing is avoided. Hence, staff effort is reduced, while patient’s data privacy is improved, since no temporal storage device is needed anymore. This may shorten the entire cycle from weeks or days (Fig. 1) down to hours or just seconds (Fig. 6).
Despite the remaining problems with existing EDCS, new challenges are already identified for the near future of web-based clinical trials, i.e., (i) internationally of trial subjects in BRIC9 and VISTA10 multinational studies [7], (ii) self-filled CRFs by study subjects [21], and (iii) image-based surrogate endpoints from signals (1D images) to moving volumes (4D images) [22].
In order to make EDCS more efficient, Franklin, Guidry & Brinkley performed a 2-year qualitative evaluation [6]. They found that the importance of ease of use and training materials outweighed the number of features and functionality. Nonetheless, importing or exporting of data is seen as one out of five major requirements to EDCS functionality. Improved data exchange is addressed with all of our three proposed modules: the interface module connects EDCS with CTMS, the data module connects EDCS with data sources, such as PACS, and the analysis module allows plug-and-play of any Java-based data processing.
In conclusion, our work provides technical ideas for workflow improvement when using EDCS in clinical trials. Our concept has been implemented by means of OpenClinica, but general ideas can easily be transferred to other EDCS. In particular, our implementation works directly for web-based EDCS supporting web services. According to [23], PACS and DICOM support will be integrated in near future. For this, patient’s privacy data can then be directly removed from the DICOM data, since correct mapping to patient’s eCRF is ensured by the EDCS.
Acknowledgments
This research was partly supported by European Foundation for the Study of Diabetes (EFSD 74550-94555).
Footnotes
Brazil, Russia, India, and China
Vietnam, Indonesia, South Africa, Turkey, and Argentina
References
- 1.Pavlović I, Kern T, Miklavcic D. Comparison of paper-based and electronic data collection process in clinical trials: costs simulation study. Contemp Clin Trials. 2009;30(4):300–316. doi: 10.1016/j.cct.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Langer S, Bartholmai B. Imaging informatics: challenges in multi-site imaging trials. J Digit Imaging. 2011;24(1):151–159. doi: 10.1007/s10278-010-9282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uppoor RS, Mummaneni P, Cooper E, Pien HH, Sorensen AG, Collins J, et al. The use of imaging in the early development of neuropharmacological drugs: a survey of approved NDAs. Clin Pharmacol Ther. 2007;84(1):69–74. doi: 10.1038/sj.clpt.6100422. [DOI] [PubMed] [Google Scholar]
- 4.Baker SG, Sargent DJ. Designing a randomized clinical trial to evaluate personalized medicine: a new approach based on risk prediction. J Natl Cancer Inst. 2010;102(23):1756–1759. doi: 10.1093/jnci/djq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leroux H, McBride S, Gibson S. On selecting a clinical trial management system for large scale, multicenter, multi-modal clinical research study. Stud Health Technol Inform. 2011;168:89–95. [PubMed] [Google Scholar]
- 6.Franklin JD, Guidry A, Brinkley JF. A partnership approach for electronic data capture in small-scale clinical trials. J Biomed Inform. 2011;44:103–108. doi: 10.1016/j.jbi.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Carvalho EC, Batilana AP, Claudino W, Reis LF, Schmerling RA, Shah J, Pietrobon R. Workflow in clinical trial sites & its association with near miss events for data quality: ethnographic, workflow & systems simulation. PLoS One. 2012;7(6):e39671. doi: 10.1371/journal.pone.0039671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenstermacher D, Street C, McSherry T, Nayak V, Overby C, Feldman M. The cancer biomedical informatics grid (caBIG) Conf Proc IEEE Eng Med Biol Soc. 2005;1:743–746. doi: 10.1109/IEMBS.2005.1616521. [DOI] [PubMed] [Google Scholar]
- 9.El-Ghatta SB, Cladé T, Snyder JC. Integrating clinical trial imaging data resources using service-oriented architecture and grid computing. Neuroinformatics. 2010;8(4):251–259. doi: 10.1007/s12021-010-9072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Fadly A, Rance B, Lucas N, Mead C, Chatellier G, Lastic P, et al. Integrating clinical research with the Healthcare Enterprise: from the RE-USE project to the EHR4CR platform. J Biomed Inform. 2011;44:S94. doi: 10.1016/j.jbi.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Deserno TM, Samsel C, Haak D, Spitzer K: Data, function, and context integration of OpenClinica using web services. Proc GMDS 2012 (in German)
- 12.Haak D, Gehlen J, Sripad P, Marx N, Deserno TM: Extension of OpenClinica for context-related integration of large data volume. Proc GMDS 2013 (in German)
- 13.Deserno TM, Haak D, Samsel C, Gehlen J, Kabino K: Integration image management and analysis into OpenClinica using web services. Proc SPIE 2013; 8674: 0F1-10
- 14.Hallaraker O, Vigna G: Detecting malicious JavaScript code in Mozilla. ICECCS Proc. 2005; 85–94
- 15.Flanagan D: JavaScript: The Definitive Guide. O’Reilly Media, Inc, 2011
- 16.Ullmann C: Calling Cross Domain Web Services in AJAX. Technical Report 2006. Available from: http://www.simple-talk.com/dotnet/asp.net/calling-cross-domain-web-services-in-ajax/
- 17.Brinzarea-Iamandi B, Darie C: AJAX and PHP: Building Modern Web Applications: Build User-Friendly Web 2.0 Applications with JavaScript and PHP. Packt Pub; 2009
- 18.Breil B, Kenneweg J, Fritz F, Bruland P, Doods D, Trinczek B, et al. Multilingual medical data models in ODM format: a novel form-based approach to semantic interoperability between routine healthcare and clinical research. Appl Clin Inform. 2012;3:276–289. doi: 10.4338/ACI-2012-03-RA-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fegan GW, Lang TA. Could an open-source clinical trial data-management system be what we have all been looking for? PLoS Med. 2008;5(3):e6. doi: 10.1371/journal.pmed.0050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reboussin D, Espeland MA. The science of web-based clinical trial management. Clin Trials. 2005;2(1):1–2. doi: 10.1191/1740774505cn059ed. [DOI] [PubMed] [Google Scholar]
- 21.Musick BS, Robb SL, Burns DS, Stegenga K, Yan M, McCorkle KJ, Haase JE. Development and use of a web-based data management system for a randomized clinical trial of adolescents and young adults. Comput Inform Nurs. 2011;29(6):337–343. doi: 10.1097/NCN.0b013e3181fcbc95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai J, Chang Z, Wang Z, Paul Segars W, Yin FF. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: a feasibility study. Med Phys. 2011;38(12):6384–6394. doi: 10.1118/1.3658737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aryanto KY, Broekema A, Oudkerk M, van Ooijen PM. Implementation of an anonymisation tool for clinical trials using a clinical trial processor integrated with an existing trial patient data information system. Eur Radiol. 2012;22(1):144–151. doi: 10.1007/s00330-011-2235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]