Abstract
Genome sequencing efforts have revealed a strikingly large number of uncharacterized genes, including poorly or uncharacterized metabolic enzymes, metabolites, and metabolic networks that operate in normal physiology, and also those enzymes and pathways that may be rewired under pathological conditions. Though deciphering the functions of the uncharacterized metabolic genome is a challenging prospect, it also presents an opportunity for identifying novel metabolic nodes that may be important in disease therapy. In this review, we will discuss the chemoproteomic and metabolomic platforms employed in identifying, characterizing, and targeting nodal metabolic pathways important in physiology and disease, describing an integrated workflow for functional mapping of metabolic enzymes.
Introduction
One of the most provocative findings to come out of the Human Genome Project was the discovery of a large number of genes encoding proteins with unknown function, including many uncharacterized enzymes that participate in the metabolism of small-molecule metabolites (Venter et al., 2001). These data revealed that our knowledge of cellular metabolism was far less complete than we thought, and opened up the possibility for a yet undiscovered landscape of metabolites and metabolic pathways. Indeed, even our understanding of well-characterized enzymes and their metabolic functions in normal physiology remains largely incomplete, especially in the pathological states where these pathways may be rewired or possess unique or novel functions. We are now faced with the grand challenge of deciphering these uncharacterized metabolic networks and disentangling the normal and disease roles of previously described metabolic pathways. This undiscovered metabolic space presents an exciting opportunity for discoveries in basic biology and opens up the potential for targeting unique or novel metabolic drivers of diseases related to dysregulated metabolism, such as obesity, diabetes, atherosclerosis, cancer, infection, and inflammatory diseases. Recent work has also demonstrated the regulatory importance of metabolite flux through a given pathway and the diverse roles of small biomolecules beyond classical metabolism, including signaling and epigenetic, transcriptional, and post-translational regulation of critical cell function.
In this review we will describe how innovative metabolic mapping techniques have been used to successfully identify, characterize, and pharmacologically target nodal metabolic pathways important in mammalian physiology and disease. Specifically, we will discuss chemoproteomic and metabolomic approaches that have been useful in globally assessing enzyme activities, developing chemical tools to interrogate enzyme function, and mapping the metabolic pathways and metabolite-driven regulation controlled by these enzymes.
Chemoproteomic approaches to assess the functional state of enzymes in complex biological systems
One of the key challenges of studying enzyme function has been the ability to assay for explicit metabolic enzyme activities of specific proteins in complex biological systems, especially for enzymes with no known substrate or function. Developing a method for global assessment of enzyme functionality remains difficult as: 1) enzymes can be regulated by post-translational events in vivo, which are poorly detected by standard gene and protein expression profiling; 2) a substantial proportion of the proteome remains functionally uncharacterized, preventing the use of substrate-activity assays; and 3) the physicochemical properties of many enzymes complicate their analysis in biological samples (e.g. low abundance, difficulty in enrichment).
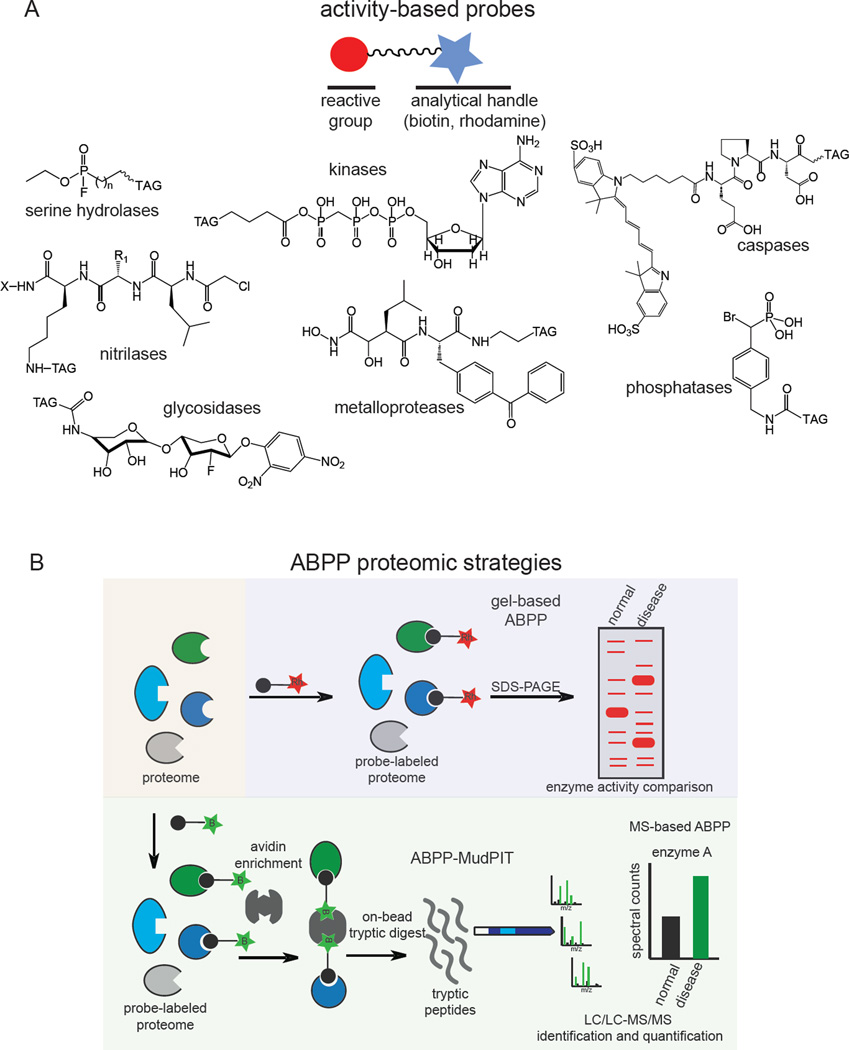
One powerful method developed to address these challenges is activity-based protein profiling (ABPP), a chemoproteomic platform that employs activity-based probes (ABPs) that measure the functional state of enzymes en masse in complex biological samples (Evans and Cravatt, 2006; Moellering and Cravatt, 2012; Nomura et al., 2010a). An ABP consists of a chemical group that covalently reacts with the active sites of enzymes across a particular enzyme class based on chemical reactivity within a conserved catalytic architecture, and an analytical handle that facilitates a simultaneous read out enzyme activities (Figure 1A). This analytical handle can be a fluorophore for visualizing enzyme activities, or a biotin handle for enrichment, identification, and quantification of activities by mass spectrometry-based proteomics (Figure 1B). To date, there exist ABPs for more than a dozen enzyme classes, including hydrolases, proteases, kinases, phosphatases, glycosidases, caspases, oxygenases, oxidoreductases, and nitrilases (Adam et al., 2001; Barglow and Cravatt, 2006; Kato et al., 2005; Kidd et al., 2001a; Liu et al., 1999; Patricelli et al., 2007; Saghatelian et al., 2004a; Walls et al., 2009; Weerapana et al., 2008; Williams et al., 2006; Xiao et al., 2013).
Figure 1.
Activity-based protein profiling (ABPP). A) Examples of activity-based probes. B) gel-based ABPP and ABPP-MudPIT platforms for fluorescent and mass-spectrometry-based analysis of enzyme activities. “Rh” denotes rhodamine and “B” denotes biotin.
ABPP overcomes many of the traditional challenges facing enzyme activity assessment in complex samples. First, these probes selectively and simultaneously label all the active, but not inactive enzymes in a class, revealing changes in enzyme activity distinct from alterations in protein or transcript expression level (Jessani et al., 2005; Kidd et al., 2001a). Second, ABPs enable enzyme activity assessment of uncharacterized enzymes, since these probes react with active sites based on class-wide conserved chemical reactivity (Bachovchin et al., 2010a; Chiang et al., 2006; Weerapana et al., 2008). Third, ABPs allow enrichment of specific classes of enzymes based on shared functional properties, facilitating characterization of enzymes that may be in low abundance or are embedded in a membrane (Bachovchin et al., 2010a; Weerapana et al., 2010).
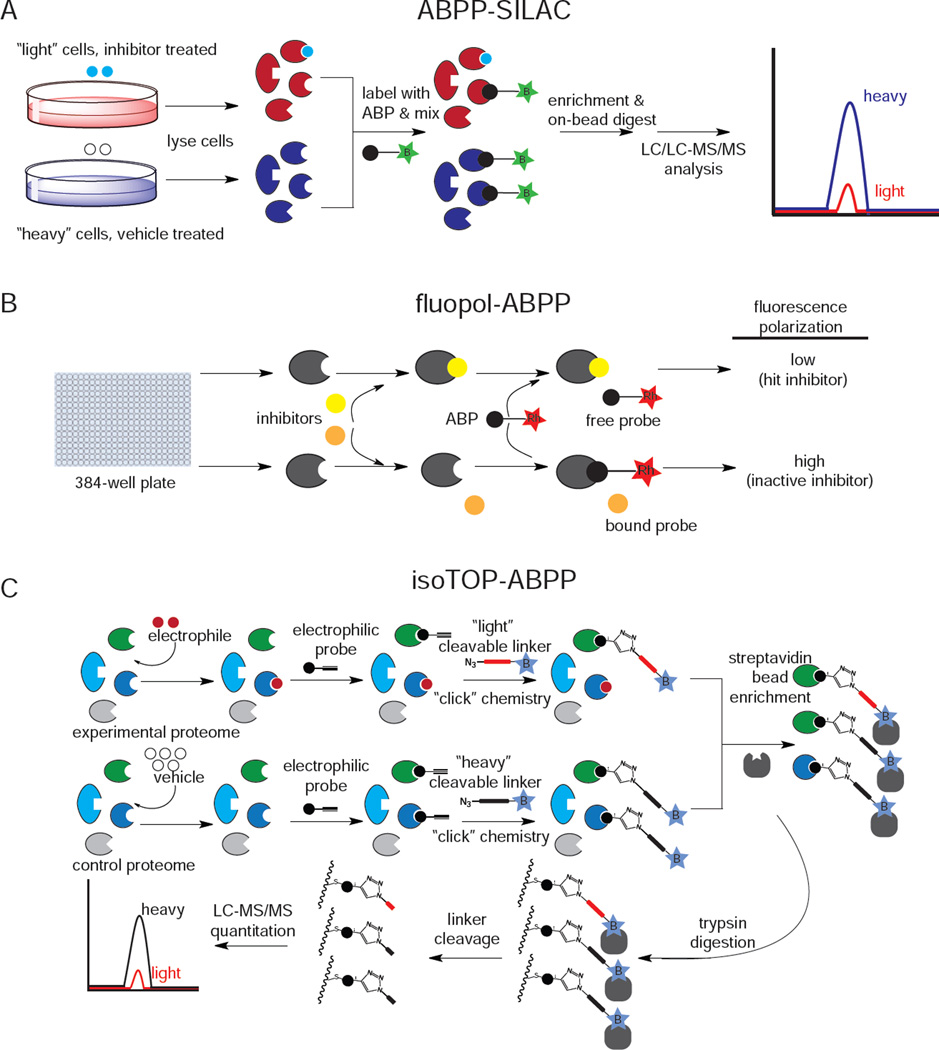
Since the initial development by Cravatt and Bogyo, ABPP platforms have incorporated chemical and analytical methods that enable detecting enzyme activities in cells or in vivo, mapping sites of probe labeling in the proteome, and quantitative assessment of enzyme activities. One of the most significant advances in ABPP platforms has been the complementation with bioorthogonal “click chemistry” methods (Speers et al., 2003). Bioorthogonal ABPs bearing an alkyne handle (instead of rhodamine or biotin) can be treated in vitro, in situ, or even in vivo to label active enzymes, facilitating target identification by subsequently appending an analytical handle (e.g. rhodamine-azide or biotin-azide) in vitro through copper-catalyzed “click chemistry” (Speers et al., 2003). Another development has coupled ABPP with stable isotopic labeling of cells (ABPP-SILAC) (Figure 2A) for quantitative proteomic analysis of enzyme activities (Adibekian et al., 2011). To improve the throughput of gel-based or mass spectrometry-based ABPP, Bachovchin and colleagues adapted a method for high-throughput screening using fluorescence polarization (fluopol-ABPP) (Figure 2B), primarily used for inhibitor screening (Bachovchin et al., 2009) . Recently, Weerapana and colleagues developed an ABPP platform called tandem orthogonal proteolysis ABPP (TOP-ABPP) (Weerapana et al., 2008).to identify hyper-reactivity and functionality of specific amino acids within the proteome. In TOP-ABPP, alkyne-bearing ABPs or reactivity-based probes (RBPs) are “clicked” to azide-bearing tags that contain chemically- or TEV-protease cleavable linkers and a biotin handle, allowing enrichment of probe-bound proteins and subsequent release of probe-bound peptides upon tandem digestions with trypsin and TEV protease. This TOP-ABPP can be adapted for quantitative proteomics by incorporating an isotopically “heavy” labeled valine into the linker and measuring the ratio of heavy to light labeling on a protein or at a particular residue, a platform called isotopic TOP-ABPP (isoTOP-ABPP) (Figure 2C) (Weerapana et al., 2010). These ABPP platforms have been successfully used to identify and characterize enzyme activities in various human diseases, including cancer, obesity, neurodegenerative diseases, and microbial infection (Blais et al., 2010; Dominguez et al., 2014; Nomura et al., 2010b, 2011a; Sadler et al., 2012; Singaravelu et al., 2010).
Figure 2.
Competitive ABPP platforms. A) ABPP-SILAC; B) fluopol-ABPP; c) isotope-ABPP
We review here several representative examples of how ABPP has been used to discover metabolic drivers of disease. ABPP has been widely used to study the serine hydrolase superfamily of enzymes using the serine hydrolase-directed ABPs fluorophosphonate (FP)-rhodamine and FP-biotin that covalently phosphorylate the active-site serine of nearly all of the >200 serine hydrolase enzymes (Bachovchin et al., 2010b; Kidd et al., 2001b; Liu et al., 1999). The serine hydrolase family is one of the largest metabolic enzyme classes in the mammalian genome, though many of the family members are poorly or incompletely characterized (Long and Cravatt, 2011). This class encompasses many types of enzymes, including hydrolases, esterases, lipases, proteases, thioesterases, and peptidases (Long and Cravatt, 2011). Through mining serine hydrolase activities with ABPP, several key metabolic or proteolytic drivers and biomarkers of cancer have been identified, including KIAA1363, monoacylglycerol lipase (MAGL), and retinoblastoma-binding protein 9 (RBBP9) (Chiang et al., 2006; Jessani et al., 2002; Nomura et al., 2010b; Shields et al., 2010). Serine hydrolase profiling was also used to discover that mutations in the poorly characterized alpha/beta-hydrolase domain-containing 12 (ABHD12) in patients with a neurodegenerative disease known as PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataracts) encoded a functionally inactive ABHD12. Despite the lack of functional information regarding ABHD12, Blankman and colleagues used ABPP to reveal inactive ABHD12 in PHARC tissues and followed with metabolomics techniques (discussed later in this review) to describe ABHD12 metabolic activity and link that loss of function to PHARC (Blankman et al., 2013). Serine hydrolase ABPs have also been used to identify important enzyme activities in bacterial and viral infections such as carboxylesterase 1 (CES1) as an upregulated enzyme activity in hepatitis C virus (HCV)-infected hepatoma cells critical in maintaining viral replication (Blais et al., 2010).
Many other ABPs have been generated and validated in complex proteomes, including but not limited to: 1) 2-oxoglutarate-dependent oxygenase probes that employ a hydroxyquinoline template coupled to a photoactivatable crosslinking group and biotin handle (Rotili et al., 2011), 2) S-adenosylmethionine (SAM)-dependent methyltransferase probes that consist of S-adenosylhomocysteine analogs with amino linkers attached to scaffolds containing photocrosslinkers and a biotin handle (Dalhoff et al., 2010), 3) a suite of bioorthogonal cytochrome P450 ABPs against a wide cross-section of human P450s (Wright and Cravatt, 2007; Wright et al., 2009), and 4) pargyline and deprenyl-based bioorthogonal ABPs for monoamine oxidase (Krysiak et al., 2012).
Bogyo and colleagues have been on the forefront of using ABPP in visualizing enzyme activities in cancer, generating ABPs to track cysteine protease activity in cancer cell progression and proteosomal substrate specificity (Greenbaum et al., 2000; Nazif and Bogyo, 2001). They have used quenched near-infrared fluorescent ABP (qNIRF-ABP) to image cysteine protease activities in tumor xenografts in vivo in mice (Blum et al., 2007) and have also developed a highly selective aza-peptidyl asparadinyl epoxide qNIRF-ABP probe for legumain, a lysosomal cysteine protease upregulated in multiple human cancers, to visualize tumors. Recently, they generated a caspase-directed ABP to visualize and quantify dexamethasone-induced apoptosis in the thymus and Apomab-induced apoptosis in tumor xenografts in vivo in mice (Edgington et al., 2009).
ABPP platforms have also been extended to map the endogenous reactivity of the proteome with reactive electrophile-based reactivity-based probes (RBPs). Carroll and colleagues developed bioorthogonal dimedone and sulfenome RBP probes that selectively react with sulfenic acid cysteine modifications in the proteome and used these probes to identify redox regulated pathways such as the cysteine sulfenic acid-modified Gpx3 regulation of Yap1 in yeast, involved in regulating epidermal growth factor receptor tyrosine kinase activity (Leonard et al., 2009; Paulsen and Carroll, 2009; Paulsen et al., 2012; Reddie et al., 2008; Seo and Carroll, 2011). Weerapana et al. used isoTOP-ABPP to perform a massive quantitative proteomic profiling effort to comprehensively profile hyper-reactive cysteines in complex proteomes, in which cells were first labeled with the cysteine-reactive iodoacetamide-alkyne bioorthogonal probe, after which proteomes were subjected to click chemistry with an isotopically-labeled azide-linked cleavable biotin linker for enrichment and release of cysteine-labeled peptides for subsequent analysis by quantitative proteomics (Weerapana et al., 2010). The authors uncovered a wide range of hyper-reactive cysteines in the proteome that were enriched in functional cysteines involved in a wide range of activities, including nucleophilic and reductive catalysis and sites of oxidative modification for both characterized and uncharacterized proteins across many different protein classes, including some metabolic enzymes (Weerapana et al., 2010). As demonstrated, isoTOP-ABPP is a broad and quantitative approach capable of assessing the specific sites involved in catalytic and regulatory function of large numbers of proteins and metabolic enzymes.
As described above, ABPP has proven to be a powerful technology in probing enzyme activities across a wide range of enzyme classes and across many physiological and cellular contexts, though like any approach, this technology is not without its limitations. While many ABPs and RBPs have been developed by the chemical biology community, there are still many metabolic enzyme classes that cannot be assayed by ABPP-based methods. Furthermore, while higher throughput ABPP methods have been developed to screen large numbers of compounds against one enzyme, the broad profiling of large numbers of enzyme activities is still medium to low-throughput using gel-based and proteomic-based methods. These limitations notwithstanding, as described here, ABPP is a versatile platform can be used not only for target identification of enzymes important in various diseases, but also for characterization of unknown enzymes, for visualizing enzyme activities, and even uncovering hyper-reactivity and function across the proteome.
Chemoprotomics for Developing Selective Small-Molecule Inhibitors for Metabolic Enzymes
In addition to target discovery, and imaging applications, ABPP can also be used in a competitive mode to screen for enzyme inhibitors (Moellering and Cravatt, 2012). Inhibitor screening by competitive ABPP exhibits several advantages over conventional substrate assays. First, enzymes can be tested in native proteomes without the need for recombinant expression or purification (Adibekian et al., 2011; Chang et al., 2011; Chiang et al., 2006; Long et al., 2009), and second, inhibitors can be developed for uncharacterized enzymes without prior knowledge of endogenous substrates (Adibekian et al., 2011; Bachovchin et al., 2010b; Chang et al., 2011; Chiang et al., 2006; Li et al., 2007). Because inhibitors are tested against many enzymes in parallel, inhibitor potency can be simultaneously assessed with selectivity, guiding medicinal chemistry efforts to develop highly specific and effective enzyme inhibitors (Chang et al., 2011; Long et al., 2009; Bachovchin et al., 2009, 2011; Adibekian et al., 2011). Thus, competitive ABPP provides a universal assay for inhibitor discovery applicable to any enzyme regardless of existing knowledge of its function as long as there is a cognate ABP or RBP for the enzyme. Competitive ABPP has emerged as a powerful approach for developing potent and selective small-molecule inhibitors for both characterized and uncharacterized enzymes, which are then used to inform the functions of metabolic enzymes in complex systems. As described previously with standalone ABPP approaches, competitive ABPP can be employed in a low-throughput/high resolution mass spectrometry-based proteomics format with biotin-tagged activity-based probes using a Multidimensional Protein Identification Technology (ABPP-MudPIT), a medium-throughput gel-based format with fluorescent activity-based probes (gel-based ABPP), or a high-throughput screening format using fluorescence polarization with ABPs against large compound libraries (fluopol-ABPP) (Adibekian et al., 2011; Bachovchin et al., 2009, 2010b; Chang et al., 2011).
Competitive ABPP platforms have been remarkably successful in generating small-molecule inhibitors of serine hydrolases using the fluorophosphonate ABP. Chang et al. developed the compound JW480, a highly selective, irreversible, in vivo efficacious, and orally bioavailable inhibitor of the previously uncharacterized enzyme KIAA1363 that selectively inhibited KIAA1363 activity in various tissues and in tumor xenografts, and impaired cancer cell migration and in vivo tumor growth (Chang et al., 2011). Bachovchin et al. generated a library of >140 serine hydrolase inhibitors based on the carbamate scaffold and tested all of these inhibitors against a library of >70 recombinantly expressed serine hydrolases in a “library versus library” screening effort, and successfully identified lead inhibitors for >40 % of enzymes tested (Bachovchin et al., 2010b). Competitive ABPP platforms have also been used to generate selective inhibitors for the serine hydrolases MAGL and fatty acid amide hydrolase (FAAH), which degrade the endocannabinoid signaling lipids 2-arachidonoylglycerol (2-AG) and anandamide, respectively. MAGL inhibitors found through a competitive ABPP screen of a structurally diverse carbamate library and subsequent optimization led to the development of the carbamate JZL184 as the first potent, selective, and in vivo active MAGL inhibitor (Long et al., 2009). As described later in this review, JZL184 has been used extensively to characterize the biochemical functions of MAGL using metabolomics approaches, in the process revealing this enzyme as a therapeutic target for cancer, inflammation and inflammatory diseases, neurodegenerative diseases, anxiety, and pain. Ahn et al. have also used ABPP to generate the highly selective and in vivo efficacious biaryl ether piperidine urea FAAH inhibitor PF-3845. The authors also generated a bioorthogonal analog of PF-3845 bearing an alkyne handle to show that PF3845-yne only inhibited FAAH in vivo (Ahn et al., 2009).
Adibekian et al. used competitive ABPP-SILAC platforms to show that the 1,2,3-triazole-urea scaffold is ideal for generating irreversible serine hydrolase inhibitors. The authors generated a library of triazole urea inhibitors and optimized the highly selective inhibitors AA74-1, AA39-2, and AA44-2 for acyl peptide hydrolase, platelet activating factor acetylhydrolase 2, and uncharacterized hydrolase ABHD11, respectively (Adibekian et al., 2011). Using this scaffold, Hsu et al. have generated the triazole urea inhibitors KT109 and KT172 for the 2-AG-synthesizing enzyme diacylglycerol lipase (DAGL) and confirmed selectivity of these inhibitors in situ by ABPP-SILAC and in vivo by ABPP-MudPIT (Hsu et al., 2012).
Screening large inhibitor libraries via high-throughput fluopol-ABPP has led to identification of several small molecule inhibitors of metabolic enzymes. These include selective inhibitors of anti-cancer targets protein methyl esterase 1 (PME1), glutathione transferase-omega (GSTO), and RBBP9, as well as an inhibitor for the anti-inflammatory target protein arginine deaminase 4 (Bachovchin et al., 2009, 2011; Knuckley et al., 2010; Tsuboi et al., 2011).
As demonstrated above, competitive ABPP platforms are very useful in developing inhibitors for metabolic enzymes. The ability to assess inhibitor selectivity and target occupancy of inhibitors in cells or even in vivo has been a particularly useful feature of this approach, towards providing highly specific chemical tools for further biological discovery, leads for clinical development, and biomarkers for inhibitor efficacy (Moellering and Cravatt, 2012).
Metabolomics to Annotate the Functions of Uncharacterized Metabolic Enzymes
Chemoproteomic strategies like ABPP have greatly facilitated efforts to assess metabolic enzyme activities and developing chemical tools to disrupt these activities in complex biological systems. However, these strategies still need to be integrated with functional metabolomic approaches to decipher the metabolites that are regulated by these enzymes and how these metabolites and their associated metabolic pathways are involved in (patho)physiology. The metabolome is generally considered a collection of small-molecule metabolites that include nutrients and their biosynthetic intermediates to provide biomass (nucleic acids and DNA/RNA, amino acids and proteins, fatty acids and membrane lipids) and energy for cell growth and function. However, this view is rapidly expanding to encompass diverse metabolite constituents that serve as intracellular and extracellular signaling molecules influencing physiological processes such as neurotransmission (e.g. acetylcholine, glutamate, 2-AG, anandamide) (Fisher and Wonnacott, 2012; Hassel and Dingledine, 2012; Kohnz and Nomura, 2014), inflammation (e.g. sphingosine-1-phosphate, prostaglandins) (Wymann and Schneiter, 2008), and cancer (e.g. eicosanoids and lysophosphatidic acid) (Mills and Moolenaar, 2003; Wang and Dubois, 2010), as well as endogenous nuclear hormone receptor ligands that influence transcriptional regulation (Evans and Mangelsdorf, 2014), and metabolites that confer post-translational and epigenetic regulation onto the proteome and genome (e.g. UDP-GlcNAc and glycosylation, acetyl-coA and acetylation) (Wellen and Thompson, 2012). The metabolome is the functional output of enzymes that generate, degrade, or convert biomolecules. Thus functional metabolomic strategies, uncovering metabolite changes that occur upon disruption of a specific metabolic enzyme, are essential in deciphering biochemical functions and (patho)physiological roles of enzymes in complex living systems.
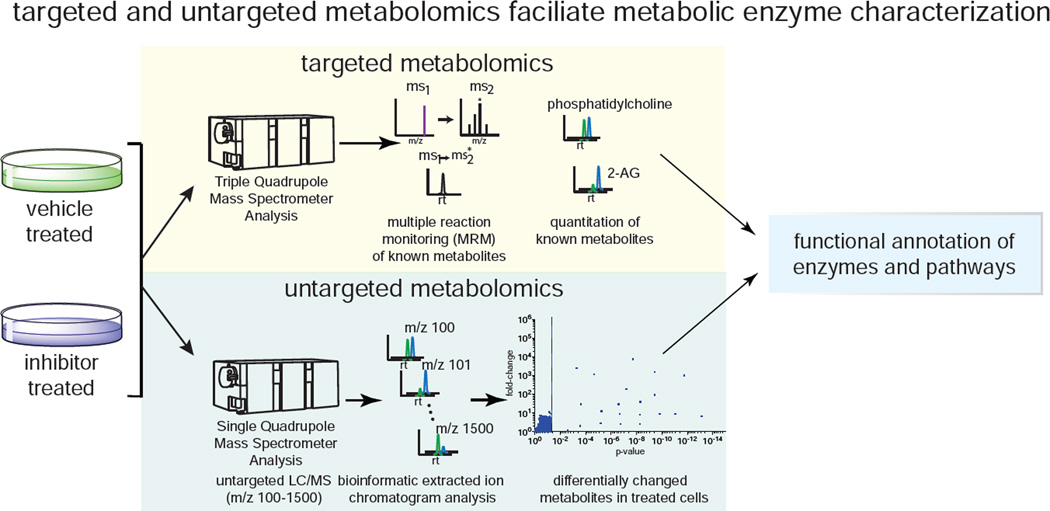
Compared to the genome and the proteome, the metabolome presents unique challenges for global analysis due to the significant physicochemical diversity in metabolite size, molecular weight, hydrophobicity, chemical stability, charge, volatility, abundance, and ionization inherent in biological samples. Several types of technologies and methodologies have been used in metabolomic profiling to attempt near global detection and analysis. NMR, gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-MS (LC-MS) are the most common techniques used for metabolomic profiling (Patti et al., 2012), and metabolomics analysis is often performed using “targeted” or “untargeted” methods that capture complementary information (Figure 3). Using targeted metabolomic methods, a series of known metabolites are quantitatively measured, in which a mass spectrometer targets a list of known metabolites by their mass-to-charge ratio (m/z) and/or the transition of the “parent” m/z of metabolites to their respective ms2 fragment “daughter” ions (known as multiple reaction monitoring). Targeted metabolomics enables sensitive, simultaneous quantification of hundreds of known metabolites based on availability of standards, and is very useful in studying defined metabolic pathways and quantifying specific, very low abundance metabolites (Patti et al., 2012). However, targeted metabolomic detection is restricted to quantifying known metabolites for which there are existing standards. Untargeted metabolomics can prove especially useful for uncovering the function(s) of uncharacterized enzymes, when deciphering unique and novel roles of previously characterized enzymes, or identifying new metabolites. Untargeted metabolomic analyses in which the mass spectrometer is set to scan a wide m/z range and collect all mass spectra are used as a complementary approach to targeted analyses to improve metabolome coverage (Patti et al., 2012; Saghatelian and Cravatt, 2005; Saghatelian et al., 2004b; Vinayavekhin et al., 2010). This large amount of collected mass spectral data can then be analyzed by bioinformatic platforms such as XCMS or MAVEN to identify, integrate, and compare all detectable ions to identify those ions that are changed between comparison groups (Clasquin et al., 2012; Patti et al., 2012; Smith et al., 2006; Tautenhahn et al., 2012). This data can then be used to identify potentially novel metabolites altered in abundance between groups using metabolomic databases like METLIN, HMDB, and Lipid Maps and traditional analytical chemistry methods (Fahy et al., 2007; Nikolskiy et al., 2013; Smith et al., 2005; Wishart et al., 2013).
Figure 3.
Targeted and untargeted metabolomic profiling platforms. Targeted metabolomic approaches oftentimes are performed by multiple reaction monitoring-based LC-MS/MS methods. Untargeted metabolomic approaches are performed through collected all mass spectra and using bioinformatics tools (e.g. XCMS) to identify, integrate, and compare ions between comparison groups.
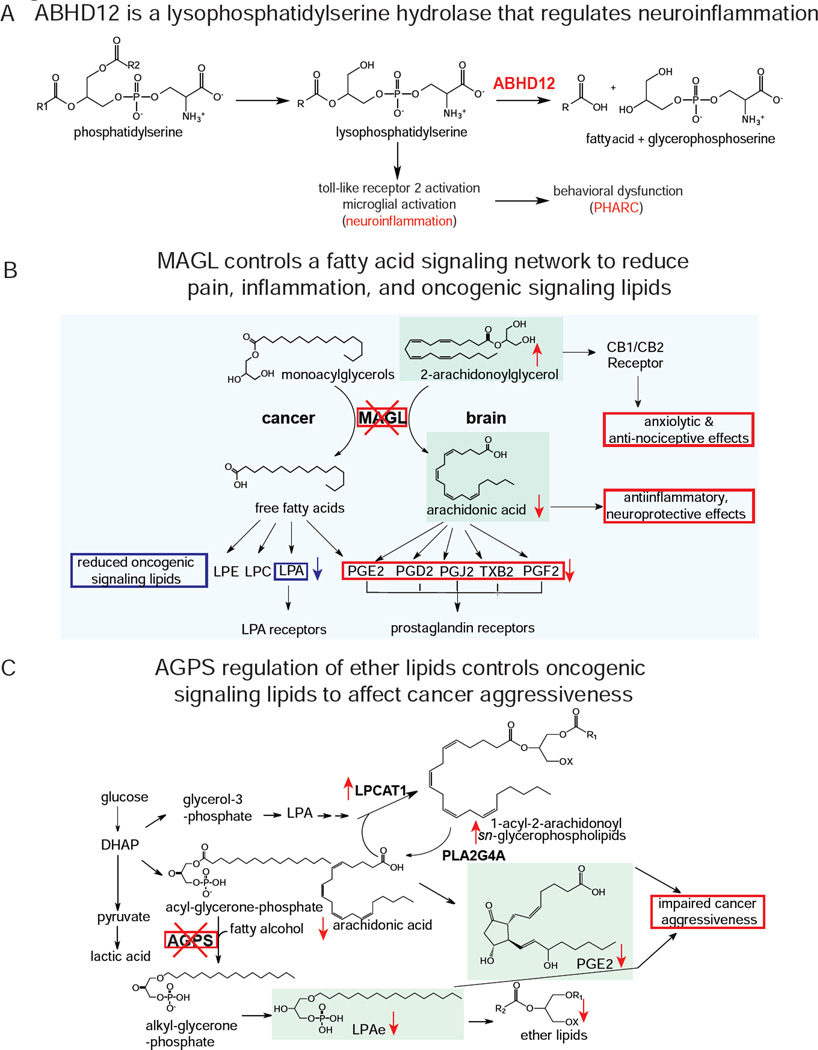
Both targeted and untargeted functional metabolomic approaches have recently revealed the functions of previously uncharacterized enzymes. Blankman and colleagues used untargeted metabolomics to uncover the function of the previously uncharacterized enzyme α/β-hydrolase domain-containing 12 (ABHD12), a serine hydrolase mutationally inactivated in patients with PHARC, as a lysophosphatidylserine (LPS) hydrolase (Figure 4A) (Blankman et al., 2013). Blankman et al. showed that ABHD12-deficient mice show elevated levels of the toll-like receptor agonist LPS, leading to neuroinflammation and motor and auditory defects reminiscent of PHARC (Blankman et al., 2013).
Figure 4.
Examples of metabolic pathways elucidated by metabolomic profiling. A) ABHD12 was characterized as a LPS hydrolase. ABHD12 deficiency leads to an accumulation in LPS, activates toll-like receptor 2, induces neuroinflammation, and causes a neurodegenerative disease known as PHARC. B) MAGL was shown to play critical roles of controlling endocannabinoid and eicosanoid signaling in the brain and fatty acid and fatty acid-derived oncogenic signaling lipids in cancer. MAGL blockade in brain leads to elevations in 2-AG endocannabinoid signaling and anxiolytic and anti-nociceptive effects, while also lowering the primary arachidonic acid precursor pool for pro-inflammatory eicosanoid production, leading to reduced inflammation, and neuroprotection against neurodegenerative diseases. In cancer, MAGL blockade leads to reduced fatty acids and fatty acid derived signaling lipids such as prostaglandins and lysophosphatidic acid, which impairs cancer pathogenicity. C) In cancer, AGPS was shown to not only control ether lipid synthesis, but also fatty acid metabolism that balances structural lipids with signaling lipids that fuel cancer. AGPS knockdown in cancer cells leads to reductions in the tumor-promoting lipid lysophosphatidic acid-ether, leading to a diversion of arachidonic acid away from oncogenic prostaglandins and towards structural lipids, leading to a net impairement in cancer aggressiveness.
Inhibitors developed through competitive ABPP platforms have also been used to interrogate the functions of uncharacterized enzymes, which then led to insights into their pathophysiological roles. Using untargeted metabolomics and the selective KIAA1363 inhibitor AS115 developed by competitive ABPP efforts, Chiang et al. discovered that KIAA1363 is a 2-acetyl-monoalkylglycerol (2-acetyl MAGE) hydrolase that generates monoalkylglycerol ether (MAGE) leading to the generation of the oncogenic signaling lipid alkyl-lysophosphatidic acid, which in turn fueled cancer cell pathogenicity and tumor growth (Chiang et al., 2006).
Both targeted and untargeted metabolomics have been used in conjunction with genetic manipulation to reveal the functions of bacterial metabolic enzymes. Baran et al. used the combined metabolomic platforms to characterize and validate genes related to specific metabolite utilization in bacteria by profiling libraries of mutant strains in Escherichia coli and Shewanella oneidensis MR-1. Through this approach, the authors identified genes with known functions as well as novel transport proteins and enzymes required for utilization of tested metabolites. Specifically, they uncovered a predicted ABC transporter encoded by genes SO1043 and SO1044 required for citrulline utilization and a predicted histidase encoded by the gene SO3057 required for utilization of ergothioneine by S. oneidensis (Baran et al., 2013).
Metabolomics to Reveal Unique and Novel Roles for Previously Characterized Enzymes
While many “characterized” metabolic enzymes have putative biochemical functions, often these enzyme functions have only been determined in vitro or may be described solely based on sequence homology or by association with an enzyme family, and may not have been validated in in vivo systems. Furthermore, enzymes may play alternate or additional roles or be linked to different metabolic pathways depending on the tissue or cell type, or in dysregulated and rewired disease states. Functional metabolomics has proven critical in mapping rewired, retasked, or novel functions of enzymes in tissue- or cell-specific or disease-specific contexts.
De Carvalho et al. utilized an untargeted metabolomic approach to describe the Mycobacterium tuberculosis enzyme Rv1248c, which was characterized at the time as a thiamine diphosphate-dependent α–ketoglutarate decarboxylase. Using metabolomic approaches, the authors found that Rv1248c was misannotated and that its actual function was to catalyze the conjugation of α–ketoglutarate and glyoxylate to yield 2-hydroxy-3-oxoadipate, which decomposes to 5-hydroxylevulinate, possibly involved in glyoxylate detoxification, glutamine metabolism, or heme biosynthesis (de Carvalho et al., 2010).
Targeted and untargeted metabolomics were also used to show the tissue-specific and disease-specific roles of MAGL in coordinating multiple lipid signaling pathways that underlie inflammation, pain, mood, and cancer (Figure 4B). Competitive ABPP was used to generate JZL184, the first selective MAGL inhibitor, subsequently used to show that MAGL blockade caused large elevations in the levels of the endocannabinoid 2-AG in mouse brain, leading to cannabinoid receptor type 1 (CB1)-dependent antinociceptive, anxiolytic, and anti-inflammatory effects (Kinsey et al., 2009, 2010, 2011; Long et al., 2009; Sciolino et al., 2011). Using both targeted and untargeted metabolomic approaches, subsequent studies showed that MAGL blockade in specific tissues such as brain, liver, and lung also lowered arachidonic acid and arachidonic acid-derived pro-inflammatory eicosanoids such as prostaglandins and thromboxanes. This resulted in neuroprotective and hepatoprotective effects in degenerative and inflammatory diseases through suppressing inflammation, thereby linking anti-inflammatory endocannabinoid signaling to pro-inflammatory eicosanoid signaling through MAGL (Long et al., 2009; Nomura et al., 2008a, 2008b, 2011a; Schlosburg et al., 2010; Chen et al., 2012; Nomura et al., 2011b; Piro et al., 2012).
Metabolomics has also been fundamental in understanding how cancer cells alter their metabolism to fuel their pathogenic properties. In a very unique discovery of a neopmorphic function for a mutated enzyme, Dang et al. used untargeted metabolomic profiling to show that a mutant form of the tricarboxylic acid cycle enzyme isocitrate dehydrogenase 1 (IDH1), IDH1 R132H, found in multiple types of cancers, generated a novel oncometabolite 2-hydroxyglutarate (2-HG) (Dang et al., 2009), which in turn caused epigenetic changes that fuels cancer progression (Xu et al., 2011).
Nomura et al. used untargeted metabolomic platforms to find that MAGL plays a distinct role in regulating fatty acid release for the generation of fatty acid-derived lysophospholipids and eicosanoids that drive aggressive features in cancer (Nomura et al., 2010c, 2011c). In another example, Benjamin et al. used targeted and untargeted metabolomic approaches to show that inactivating the ether lipid-generating enzyme alkylglycerone phosphate synthase (AGPS) in aggressive cancer cells dramatically reduced structural and oncogenic signaling ether lipid levels. AGPS inactivation also diverted the flux of arachidonic acid away from other tumor-promoting signaling lipids such as prostaglandins, and towards structural acylglycerophospholipids, leading to impaired cancer pathogenicity and tumorigenesis (Benjamin et al., 2013) (Figure 4C).
Looking further at cancer metabolism, Locasale and Possemato independently showed phosphoglycerate dehydrogenase (PHGDH) is a critical metabolic node in cancer cells, diverting glucose metabolism into serine and glycine metabolism (Locasale et al., 2011; Possemato et al., 2011). Locasale et al. utilized heteronuclear single quantum coherence spectroscopy NMR and isotopic tracing using targeted LC/MS-based metabolomics of 13C-glucose labeled cells to show significant 13C incorporation into 3-phosphoserine and serine pathways through PHGDH (Locasale et al., 2011). Using functional metabolomics, Locasale et al. found that inactivating PHGDH in melanoma cancer cells lowered phosphoserine levels and caused an accumulation in glycolytic intermediates (Locasale et al., 2011). In breast cancer cells, Possemato et al. used functional metabolomics to show that nearly half of α-ketoglutarate was derived from the serine pathway and that PHGDH inactivation in breast cancer cells reduced the levels of multiple TCA cycle metabolites, leading to impaired cancer pathogenicity (Possemato et al., 2011).
Metabolomics has also been useful in defining metabolic drivers of viral infection. Grady et al. used an siRNA screen to show that argininosuccinate synthase 1 (ASS1) knockdown increased virus yield and subsequently used metabolomic profiling to show that ASS1 inactivation resulted in a metabolic signature that closely resembled HSV-1 infection, in which levels of aspartate, carbamoyl-aspartate, one of the first committed metabolites on the pathway to nucleotide synthesis, and nucleotides and their precursors were reduced (Grady et al., 2013).
Collectively, targeted and untargeted metabolomic platforms can be used to functionally characterize not only the substrate/product relationships of metabolic enzymes, but also the metabolic networks that these enzymes control in (patho)physiological settings. These techniques have proven useful in discovering the mechanisms through which these enzymes control disease progression and in elucidating the therapeutic potential of manipulating specific metabolic pathways.
Post-Translational and Epigenetic Regulation of the Proteome and Genome by Metabolic Pathways
While small-molecule metabolites have long been known to confer post-translational and epigenetic modifications onto the proteome and genome, these types of regulation have been considered to be primarily regulated by the enzymes directly involved in adding or removing these modifications. These metabolites, their modifications, and their respective enzymes include acetyl-CoA and acetylation/deacetylation by acetyltransferases and deacetylases, S-adenosylmethionine (SAM) and methylation/demethylation by methyltransferases and demethylases, and ATP and phosphorylation/dephosphorylation by kinases and phosphatases (Prabakaran et al., 2012). However, recent studies have shown that the metabolic enzymes and metabolic fluxes that generate these cofactors may play an important role in regulating the levels of post-translational and epigenetic modifications (Wellen and Thompson, 2012). This realization brings forth the exciting prospect of controlling metabolic, signaling, and transcriptional networks simultaneously through directly manipulating metabolic pathways. Here, we provide some recent examples of metabolite-driven protein and gene regulation in controlling pathophysiological processes.
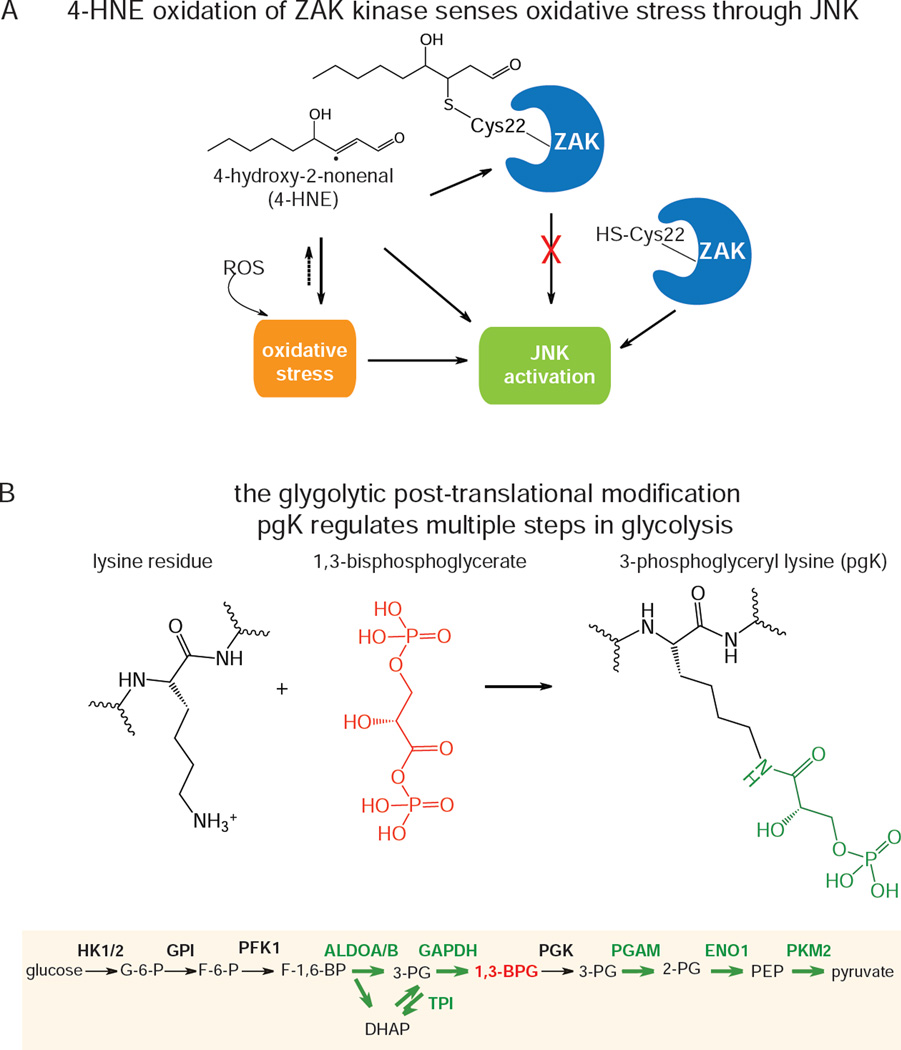
Chemoproteomic platforms can also be used to identify metabolite-driven post-translational modifications in the proteome. Wang et al. used a competitive isoTOP-ABPP platforms to globally map the targets of 4-hydroxy-2-nonenal (HNE), a common lipid product of lipid peroxidation, whereby HNE was competed against iodoacetamide-alkyne labeling in cells. The authors showed that a unique cysteine in ZAK kinase was the most sensitive target to HNE, and suggested that ZAK is a special node in MAPK signaling that is sensitive to oxidative stress (Wang et al., 2014) (Figure 5A).
Figure 5.
Examples of post-translational modifications regulated by specific metabolites. a) isoTOP-ABPP was used to map distinct hyper-reactive cysteines that were particularly sensitive to modification by the endogenously produced reactive electrophile 4-hydro2-nonenal. B) A novel post-translational regulation by a glycolytic product 1,3-bisphosphoglycerate that modifies lysines on glycolytic enzymes and inhibits their activity.
Moellering et al. recently discovered a novel post-translational modification, 3-phosphoglyceryl-lysine (pgK), generated by nonenzymatic covalent lysine modifications through the glycolytic intermediate 1,3-bisphosphoglycerate (1,3-BPG), the product of glyceraldehyde 3-phosphate dehydrogenase (Figure 5B). The authors showed that this modification accumulated on several glycolytic enzymes in cells exposed to high glucose, leading to inhibition of activity and redirection of glycolytic intermediates to biosynthetic pathways that support cancer cell pathogenicity (Moellering and Cravatt, 2013).
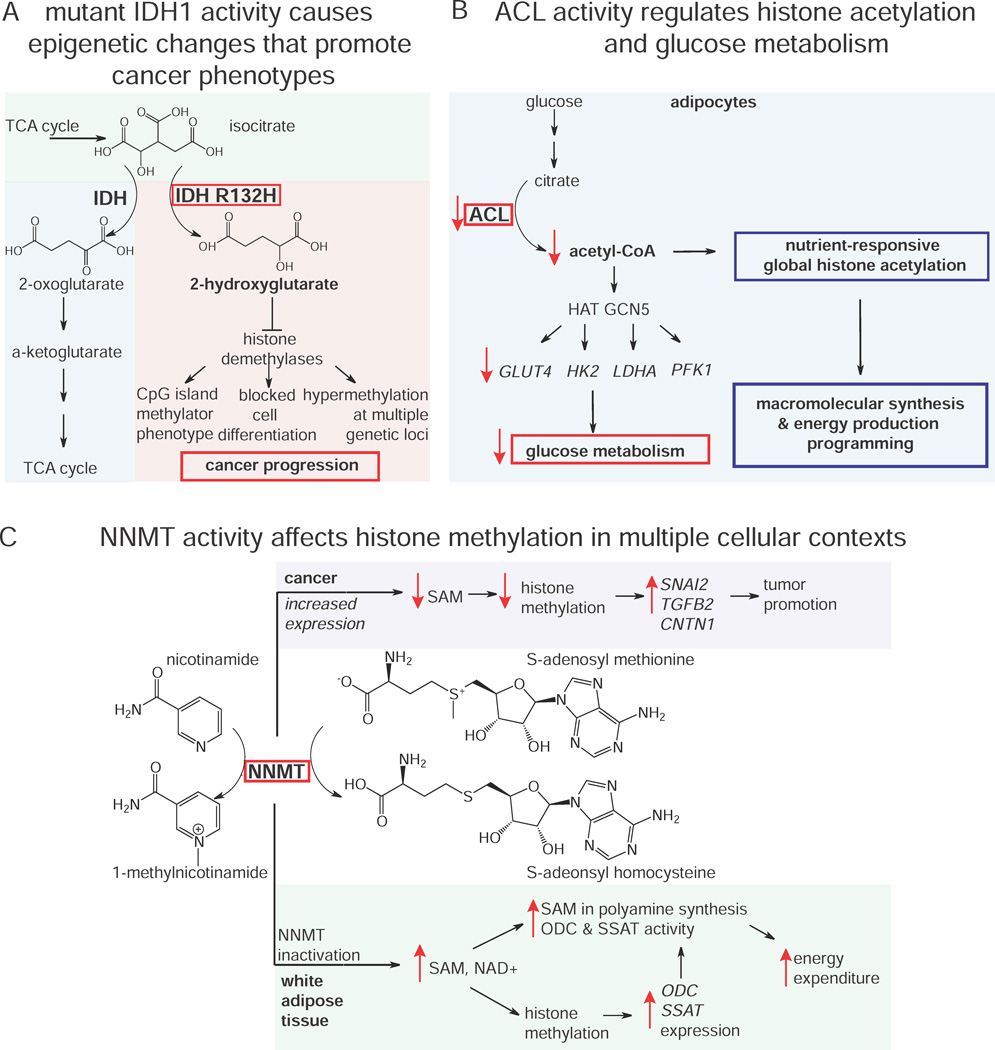
We described in the previous section how mutant IDH1 in cancer cells generate the oncometabolite 2-HG that drives cancer pathogenicity. Studies have shown that IDH1 mutations are correlated with hypermethylation at specific loci known as the CpG island methylator phenotype in glioma and that mutant IDH1 causes hypermethylation at many genetic loci (Figure 6A) (Turcan et al., 2012). Work by Xu et al. and Lu et al. have since shown that 2-HG acts as a competitive inhibitor of multiple α-KG-dependent dioxygenases, and that IDH1 mutations can impair histone demethylation resulting in a block to cell differentiation (Lu et al., 2012; Xu et al., 2011). A mutation in IDH1 generates the novel oncometabolite 2-HG, which then confers large scale epigenetic alterations altering the expression of large numbers of genes, thus linking central carbon metabolism to gene regulation and cancer pathogenicity.
Figure 6.
Metabolic control of epigenetic features. A) Mutant IDH1 R132H generates 2-HG, inhibiting histone demethylases to fuel cancer progression through multiple mechanisms. 2) ACL activity controls acetyl-CoA levels to confer nutrient-responsive histone acetylation and gene expression, altering glucose metabolism and cellular programming of macromolecular synthesis and energy production.C) Increased expression of NNMT in cancer decreases histone methylation, increasing expression of tumor promoting genes. In white adipose tissue, inactivation of NNMT increases polyamine synthesis and histone methylation, elevating gene expression and activity of enzymes critical for high energy expenditure.
Recent studies have also highlighted the importance of acetyl-CoA levels in histone acetylation and transcriptional regulation. Tu et al. used two-dimensional GC/MS-based metabolomic profiling to show changes in acetyl-CoA levels periodic with yeast cell cycle phase, suggesting that acetyl-coA levels may control temporal regulation of cell cycle processes (Tu et al., 2007). Subsequently, Cai et al. showed that an increase in acetyl CoA levels led directly to the Gcn5p/SAGA-catalyzed acetylation of histones at genes important for growth, thus promoting growth transcriptional programming in yeast and serving as a metabolic rheostat to initiate cell growth through acetylation of specific histones (Cai and Tu, 2011). Similarly, Wellen et al. showed in mammalian cells that ATP-citrate lyase (ACL) influences cell growth and differentiation through controlling acetyl-CoA levels, driving nutrient-responsive histone acetylation and selective gene expression prompting growth factor-induced increases in nutrient metabolism and reprogramming of intracellular metabolism to utilize glucose for ATP production and macromolecular synthesis (Figure 6B) (Wellen et al., 2009).
In another example, Ulanovskaya showed that N-methyltransferase (NNMT) can also influence the histone methylation and epigenetic regulation that drives cancer aggressiveness through methylating nicotinamide to generate N-methylnicotinamide (Figure 6C). This leads to reduced SAM levels, thus diverting SAM methylation away from histone methylation. The authors showed that NNMT overexpression leads to upregulation of many tumor-promoting gene products including SNAI2, TGFB2, and CNTN1, via the altered epigenetic landscape of cancer cells (Ulanovskaya et al., 2013). Interestingly, Kraus et al. showed that NNMT also alters epigenetic and metabolic landscapes in white adipose tissue (WAT) (Kraus et al., 2014). Through these mechanisms, the metabolic enzyme NNMT regulates metabolism and epigenetic regulation to drive both cancer and obesity.
Future Challenges
The resurgence of interest in metabolism has spurred a rapid pace of advancements in the decade following the completion of the human genome project, providing insight into fundamental biochemistry as well as revealing mechanistic details of diseases with metabolic bases such as cancer, infection, and obesity and diabetes. Development of innovative chemoproteomic and metabolomic platforms has enabled the characterization and description of enzyme function in complex living systems, the metabolic pathways that these enzymes regulate, and even metabolic pathway-driven post-translational and epigenetic regulation of the proteome and genome. While these technologies have revealed previously uncharacterized aspects of metabolism and clarified existing ones, the majority of the metabolic map still remains obscured. Higher throughput technologies for metabolomics and improved genetic or pharmacological manipulation of enzyme activities in complex systems are required to decode the function of the currently concealed metabolic genome.
Furthermore, major challenges still exist in elucidating enzyme function. While we have provided successful examples of characterizing enzymes in complex living systems, there are at least an equal number of examples from personal experience where inactivating or overexpressing an enzyme causes no detectable metabolomic changes upon metabolomic profiling. There are many reasons that may account for this apparent metabolomic intransigence including technical and methodological limitations such as 1) extraction procedures and LC/MS chromatography conditions that confer metabolite instability; 2) the metabolites regulated by the enzyme are too low abundance or are otherwise undetectable by traditional metabolomic methods (e.g. does not ionize well, not volatile, or does not have a derivatization method that improves detection); and 3) the metabolite change may be localized to a specific cell-type in a tissue or a specific intracellular compartment and the metabolite changes are masked by the remainder of the extracted metabolome. Elucidating enzyme function is also complicated by: 4) mischaracterized enzymes that may be described as acting on a specific class of small-molecule metabolites but may actually be operating on alternative metabolite classes or even protein, peptide, or genomic substrates that may not be amenable to analysis by metabolomic platforms; and 5) enzymes that may be acting on a yet unknown post-translational protein modification.
The information quality challenge of mischaracterized enzymes is likely a larger problem than currently appreciated, as many enzymes are named based on sequence homology to other proteins, but may not share similar substrate specificity. Metabolomic approaches are not amenable for substrate profiling of larger substrates such as protein, peptides, and post-translational modifications. Nonetheless, innovative protease and peptidase substrate profiling methods have been developed to functionally define metabolic enzymes that may have protein and peptide substrates, including subtiligase-mediated degradomic strategies, protein topography and migration analysis platform, and peptidomic profiling methods (Dix et al., 2008, 2012; Kim et al., 2012; Mahrus et al., 2008; Nolte et al., 2009; Tinoco et al., 2010). While there are innovative proteomic methods for mapping well-characterized post-translational modifications such as phosphorylation and acetylation by phosphoproteomic or acetylomic methods (Choudhary and Mann, 2010), unfortunately, there are currently few to no methods for characterizing the functions of enzymes that act on a yet unknown post-translational modifications, since there are currently no proteomic strategies for globally identifying novel or unknown protein modifications across the proteome.
What is quite clear is that achieving the goal of large scale functional characterization and description of metabolic enzymes in complex physiological and disease systems will require integrating multidimensional metabolic mapping technologies. This will likely include the chemoproteomic and functional metabolomic approaches described here in addition to newly developed chemical strategies that will expand our access into protein function, our ability to generate pharmacological tools, and our capacity to accelerate throughput of these analyses. These advances, coupled with increased resolution and depth of analytical platforms will support our drive to interrogate the unexplored aspects of the metabolome, proteome, and peptidome. The complex interplay between enzyme function, metabolomic landscape, post-translational and epigenetic regulation, and metabolite-protein-signaling networks described in this review presents a great challenge for scientists, but also offers exciting opportunities to understand complex biology and treat disease by understanding fundamental metabolic function.
Highlights.
Chemoproteomic approaches can be used to assess the functional state of enzymes
Chemoproteomic approaches can be used to develop inhibitors for enzymes
Metabolomics can be used to assign biochemical function of enzymes
Metabolic pathways can control post-translational and epigenetic regulation
Significance.
In the postgenomic era, we are faced with the challenge of attributing biochemical functions to a vast array of uncharacterized enzymes and assigning these functions to metabolic networks and (patho)physiological functions. Recent efforts have uncovered exciting examples of intersections between metabolites and metabolic pathways, as well as demonstrating control over signaling pathways and post-translational and epigenetic regulation, suggesting many more unidentified nexus of coordination between the metabolome, proteome, and genome. Meeting this challenge requires integrating multiple complementary types of modern metabolic mapping technologies. In this review, we will describe how innovative metabolic mapping techniques have been used to successfully identify, characterize, and pharmacologically target nodal metabolic pathways important in mammalian physiology and disease.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R21CA170317, R01CA172667, P42ES004705), the Searle Scholar Foundation, and the Center for Environment Research on Toxics
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam GC, Cravatt BF, Sorensen EJ. Profiling the specific reactivity of the proteome with non-directed activity-based probes. Chem. Biol. 2001;8:81–95. doi: 10.1016/s1074-5521(00)90060-7. [DOI] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Wang C, Hsu K-L, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat. Chem. Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem. Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat. Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U. S. A. 2010a;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U. S. A. 2010b;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schürer SC, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran R, Bowen BP, Price MN, Arkin AP, Deutschbauer AM, Northen TR. Metabolic footprinting of mutant libraries to map metabolite utilization to genotype. ACS Chem. Biol. 2013;8:189–199. doi: 10.1021/cb300477w. [DOI] [PubMed] [Google Scholar]
- Barglow KT, Cravatt BF. Substrate mimicry in an activity-based probe that targets the nitrilase family of enzymes. Angew. Chem. Int. Ed Engl. 2006;45:7408–7411. doi: 10.1002/anie.200603187. [DOI] [PubMed] [Google Scholar]
- Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais DR, Brûlotte M, Qian Y, Bélanger S, Yao SQ, Pezacki JP. Activity-based proteome profiling of hepatoma cells during hepatitis C virus replication using protease substrate probes. J. Proteome Res. 2010;9:912–923. doi: 10.1021/pr900788a. [DOI] [PubMed] [Google Scholar]
- Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1500–1505. doi: 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat. Chem. Biol. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- Cai L, Tu BP. Acetyl-CoA drives the transcriptional growth program in yeast. Cell Cycle Georget. Tex. 2011;10:3045–3046. doi: 10.4161/cc.10.18.17000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho LPS, Zhao H, Dickinson CE, Arango NM, Lima CD, Fischer SM, Ouerfelli O, Nathan C, Rhee KY. Activity-based metabolomic profiling of enzymatic function: identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase. Chem. Biol. 2010;17:323–332. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem. Biol. 2011;18:476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhang J, Wu Y, Wang D, Feng G, Tang Y-P, Teng Z, Chen C. Monoacylglycerol lipase is a therapeutic target for Alzheimer’s disease. Cell Rep. 2012;2:1329–1339. doi: 10.1016/j.celrep.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, Rabinowitz JD. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2012. LC-MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhoff C, Hüben M, Lenz T, Poot P, Nordhoff E, Köster H, Weinhold E. Synthesis of S-adenosyl-L-homocysteine capture compounds for selective photoinduced isolation of methyltransferases. Chembiochem Eur. J. Chem. Biol. 2010;11:256–265. doi: 10.1002/cbic.200900349. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Wang C, Okerberg E, Patricelli MP, Cravatt BF. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell. 2012;150:426–440. doi: 10.1016/j.cell.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E, Galmozzi A, Chang JW, Hsu K-L, Pawlak J, Li W, Godio C, Thomas J, Partida D, Niessen S, et al. Integrated phenotypic and activity-based profiling links Ces3 to obesity and diabetes. Nat. Chem. Biol. 2014;10:113–121. doi: 10.1038/nchembio.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat. Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SK, Wonnacott S. Chapter 13 - Acetylcholine. In: Brady ST, Siegel GJ, Albers RW, Price DL, editors. Basic Neurochemistry. Eighth Edition. New York: Academic Press; 2012. pp. 258–282. [Google Scholar]
- Grady SL, Purdy JG, Rabinowitz JD, Shenk T. Argininosuccinate synthetase 1 depletion produces a metabolic state conducive to herpes simplex virus 1 infection. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E5006–5015. doi: 10.1073/pnas.1321305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- Hassel B, Dingledine R. Chapter 17 - Glutamate and Glutamate Receptors. In: Brady ST, Siegel GJ, Albers RW, Price DL, editors. Basic Neurochemistry. Eighth Edition. New York: Academic Press; 2012. pp. 342–366. [Google Scholar]
- Hickey SF, Hammond MC. Structure-guided design of fluorescent S-adenosylmethionine analogs for a high-throughput screen to target SAM-I riboswitch RNAs. Chem. Biol. 2014;21:345–356. doi: 10.1016/j.chembiol.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K-L, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. DAGLβ inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat. Chem. Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh D-Y, Yates JR, Jeffrey SS, 3rd, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat. Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KAH, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry (Mosc.) 2001a;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry (Mosc.) 2001b;40:4005–4015. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- Kim Y-G, Lone AM, Nolte WM, Saghatelian A. Peptidomics approach to elucidate the proteolytic regulation of bioactive peptides. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8523–8527. doi: 10.1073/pnas.1203195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey SG, Long JZ, O’Neal ST, Abdullah RA, Poklis JL, Boger DL, Cravatt BF, Lichtman AH. Blockade of endocannabinoid-degrading enzymes attenuates neuropathic pain. J. Pharmacol. Exp. Ther. 2009;330:902–910. doi: 10.1124/jpet.109.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey SG, Long JZ, Cravatt BF, Lichtman AH. Fatty acid amide hydrolase and monoacylglycerol lipase inhibitors produce anti-allodynic effects in mice through distinct cannabinoid receptor mechanisms. J. Pain Off. J. Am. Pain Soc. 2010;11:1420–1428. doi: 10.1016/j.jpain.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey SG, O’Neal ST, Long JZ, Cravatt BF, Lichtman AH. Inhibition of endocannabinoid catabolic enzymes elicits anxiolytic-like effects in the marble burying assay. Pharmacol. Biochem. Behav. 2011;98:21–27. doi: 10.1016/j.pbb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Jones JE, Bachovchin DA, Slack J, Causey CP, Brown SJ, Rosen H, Cravatt BF, Thompson PR. A fluopol-ABPP HTS assay to identify PAD inhibitors. Chem. Commun. Camb. Engl. 2010;46:7175–7177. doi: 10.1039/c0cc02634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnz RA, Nomura DK. Chemical approaches to therapeutically target the metabolism and signaling of the endocannabinoid 2-AG and eicosanoids. Chem. Soc. Rev. 2014 doi: 10.1039/c4cs00047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, Pirinen E, Pulinilkunnil TC, Gong F, Wang Y, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysiak JM, Kreuzer J, Macheroux P, Hermetter A, Sieber SA, Breinbauer R. Activity-based probes for studying the activity of flavin-dependent oxidases and for the protein target profiling of monoamine oxidase inhibitors. Angew. Chem. Int. Ed Engl. 2012;51:7035–7040. doi: 10.1002/anie.201201955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J. Am. Chem. Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavón FJ, Serrano AM, Selley DE, Parsons LH, et al. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat. Chem. Biol. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cravatt BF. How chemoproteomics can enable drug discovery and development. Chem. Biol. 2012;19:11–22. doi: 10.1016/j.chembiol.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazif T, Bogyo M. Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2967–2972. doi: 10.1073/pnas.061028898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskiy I, Mahieu NG, Chen Y-J, Tautenhahn R, Patti GJ. An untargeted metabolomic workflow to improve structural characterization of metabolites. Anal. Chem. 2013;85:7713–7719. doi: 10.1021/ac400751j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte WM, Tagore DM, Lane WS, Saghatelian A. Peptidomics of prolyl endopeptidase in the central nervous system. Biochemistry (Mosc.) 2009;48:11971–11981. doi: 10.1021/bi901637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat. Chem. Biol. 2008a;4:373–378. doi: 10.1038/nchembio.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Hudak CSS, Ward AM, Burston JJ, Issa RS, Fisher KJ, Abood ME, Wiley JL, Lichtman AH, Casida JE. Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg. Med. Chem. Lett. 2008b;18:5875–5878. doi: 10.1016/j.bmcl.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 2010a;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S-W, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010b;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S-W, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010c;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MCG, Ward AM, Hahn YK, Lichtman AH, Conti B, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011a;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MCG, Ward AM, Hahn YK, Lichtman AH, Conti B, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011b;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem. Biol. 2011c;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S. Imaging approach for monitoring cellular metabolites and ions using genetically encoded biosensors. Curr. Opin. Biotechnol. 2010;21:45–54. doi: 10.1016/j.copbio.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry (Mosc.) 2007;46:350–358. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. Chemical dissection of an essential redox switch in yeast. Chem. Biol. 2009;16:217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro JR, Benjamin DI, Duerr JM, Pi Y, Gonzales C, Wood KM, Schwartz JW, Nomura DK, Samad TA. A dysregulated endocannabinoid-eicosanoid network supports pathogenesis in a mouse model of Alzheimer’s disease. Cell Rep. 2012;1:617–623. doi: 10.1016/j.celrep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo H-K, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddie KG, Seo YH, Muse Iii WB, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol. Biosyst. 2008;4:521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotili D, Altun M, Kawamura A, Wolf A, Fischer R, Leung IKH, Mackeen MM, Tian Y-M, Ratcliffe PJ, Mai A, et al. A photoreactive small-molecule probe for 2-oxoglutarate oxygenases. Chem. Biol. 2011;18:642–654. doi: 10.1016/j.chembiol.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler NC, Angel TE, Lewis MP, Pederson LM, Chauvigné-Hines LM, Wiedner SD, Zink EM, Smith RD, Wright AT. Activity-based protein profiling reveals mitochondrial oxidative enzyme impairment and restoration in diet-induced obese mice. PloS One. 2012;7:e47996. doi: 10.1371/journal.pone.0047996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Cravatt BF. Discovery metabolite profiling--forging functional connections between the proteome and metabolome. Life Sci. 2005;77:1759–1766. doi: 10.1016/j.lfs.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. U. S. A. 2004a;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry (Mosc.) 2004b;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, et al. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat. Neurosci. 2010;13:1113–1119. doi: 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciolino NR, Zhou W, Hohmann AG. Enhancement of endocannabinoid signaling with JZL184, an inhibitor of the 2-arachidonoylglycerol hydrolyzing enzyme monoacylglycerol lipase, produces anxiolytic effects under conditions of high environmental aversiveness in rats. Pharmacol. Res. Off. J. Ital. Pharmacol. Soc. 2011;64:226–234. doi: 10.1016/j.phrs.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew. Chem. Int. Ed Engl. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- Shields DJ, Niessen S, Murphy EA, Mielgo A, Desgrosellier JS, Lau SKM, Barnes LA, Lesperance J, Bouvet M, Tarin D, et al. RBBP9: a tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2189–2194. doi: 10.1073/pnas.0911646107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaravelu R, Blais DR, McKay CS, Pezacki JP. Activity-based protein profiling of the hepatitis C virus replication in Huh-7 hepatoma cells using a non-directed active site probe. Proteome Sci. 2010;8:5. doi: 10.1186/1477-5956-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco AD, Tagore DM, Saghatelian A. Expanding the dipeptidyl peptidase 4-regulated peptidome via an optimized peptidomics platform. J. Am. Chem. Soc. 2010;132:3819–3830. doi: 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Bachovchin DA, Speers AE, Spicer TP, Fernandez-Vega V, Hodder P, Rosen H, Cravatt BF. Potent and selective inhibitors of glutathione S-transferase omega 1 that impair cancer drug resistance. J. Am. Chem. Soc. 2011;133:16605–16616. doi: 10.1021/ja2066972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem. Biol. 2010;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- Walls C, Zhou B, Zhang Z-Y. Activity-based protein profiling of protein tyrosine phosphatases. Methods Mol. Biol. Clifton NJ. 2009;519:417–429. doi: 10.1007/978-1-59745-281-6_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nat. Rev. Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Weerapana E, Blewett MM, Cravatt BF. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods. 2014;11:79–85. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 2008;4:405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SJ, Hekmat O, Withers SG. Synthesis and testing of mechanism-based protein-profiling probes for retaining endo-glycosidases. Chembiochem Eur. J. Chem. Biol. 2006;7:116–124. doi: 10.1002/cbic.200500279. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AT, Cravatt BF. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 2007;14:1043–1051. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AT, Song JD, Cravatt BF. A suite of activity-based probes for human cytochrome P450 enzymes. J. Am. Chem. Soc. 2009;131:10692–10700. doi: 10.1021/ja9037609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Xiao J, Broz P, Puri AW, Deu E, Morell M, Monack DM, Bogyo M. A coupled protein and probe engineering approach for selective inhibition and activity-based probe labeling of the caspases. J. Am. Chem. Soc. 2013;135:9130–9138. doi: 10.1021/ja403521u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, Xiao M-T, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]