Abstract
Prader-Willi Syndrome (PWS) is a neurogenetic disorder caused by the deletion of imprinted genes on the paternally inherited human chromosome 15q11-q13. This locus harbours a long non-protein-coding RNA (U-UBE3A-ATS) that contains six intron-encoded snoRNAs, including the SNORD116 and SNORD115 repetitive clusters. The 3′-region of U-UBE3A-ATS is transcribed in the cis-antisense direction to the ubiquitin-protein ligase E3A (UBE3A) gene. Deletion of the SNORD116 region causes key characteristics of PWS. There are few indications that SNORD115 might regulate serotonin receptor (5HT2C) pre-mRNA processing. Here we performed quantitative real-time expression analyses of RNAs from the PWS locus across 20 human tissues and combined it with deep-sequencing data derived from Cap Analysis of Gene Expression (CAGE-seq) libraries. We found that the expression profiles of SNORD64, SNORD107, SNORD108 and SNORD116 are similar across analyzed tissues and correlate well with SNORD116 embedded U-UBE3A-ATS exons (IPW116). Notable differences in expressions between the aforementioned RNAs and SNORD115 together with the host IPW115 and UBE3A cis-antisense exons were observed. CAGE-seq analysis revealed the presence of potential transcriptional start sites originated from the U-UBE3A-ATS spanning region. Our findings indicate novel aspects for the expression regulation in the PWS locus.
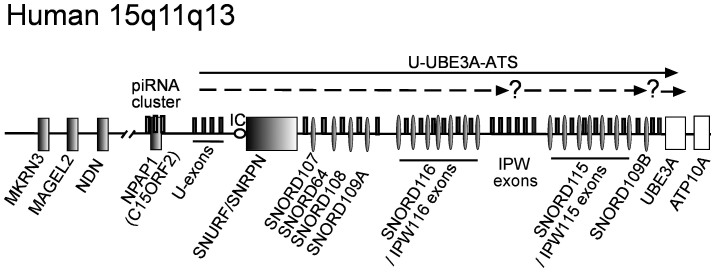
The Prader-Willi (PWS) and Angelman syndromes (AS) are neurogenetic disorders that are caused by inactivation or inherited deletion of paternally and maternally expressed genes on human chromosome 15q11-q13, respectively. AS is characterized by cognitive impairment, absence of speech, seizures, ataxia and mental retardation. Genetically, AS is linked to deletion or inactivating mutations of the ubiquitin E3 ligase encoding gene (UBE3A) on human maternally inherited chromosome 15 (Figure 1). Individuals with PWS suffer initially from hypotonia, neonatal feeding difficulties and failure to thrive. The PWS symptoms of later stages include hyperphagia, which if not controlled leads to obesity, general and continued development delay, small hands and feet, behavioral problems and mild intellectual disability1. PWS disorder results from the loss of function of one or more imprinted, paternally expressed genes on the proximal long arm of chromosome 15 (Figure 1)2,3. The PWS locus consists of several paternally expressed protein-coding genes, a piRNA gene cluster and six different C/D-box snoRNA species4,5. Two of them: SNORD116 (HBII-85) and SNORD115 (HBII-52) are organized in large, tandemly repeated clusters, containing 29 and 48 gene copies respectively (Figure 1). Other snoRNAs are present either as single copy (SNORD64 (HBII-13), SNORD107 (HBII-436) and SNORD108 (HBII-437)) or double copy (SNORD109A/B (HBII-438A/B)) genes3,4,6. The piRNA cluster is located within the human X15ORF2/NPAP1 gene, suggesting that it can be recognized and silenced as an invasive gene by RNA-mediated mechanisms7. Several studies indicate that all snoRNAs are likely to be processed from the introns of a long primary non-protein coding transcript (U-UBE3A-ATS) (Figure 1)3. This large >600 kb long transcript, which reportedly initiates from U-exons upstream from SNURF/SNRPN encoding gene, includes the PWS-imprinting center (PWS-IC) and extends to overlap in antisense orientation with the UBE3A gene region (Figure 1)3,8,9,10. However, some observations point to the possible presence of additional regulatory elements or differential processing of the distal region of the U-UBE3A-ATS transcript (UBE3A-ATS)11. There are reports indicating that SNORD64, SNORD107, SNORD108, SNORD109A, SNORD116 and their hosting exons of U-UBE3A-ATS RNA (including IPW116 exons: previously known as IPW-A1 and IPW-A2) are ubiquitously expressed in human tissues12,13. Whereas, the transcript containing intronic copies of SNORD115 embedded by IPW115 exons (previously known as IPW-G1 and IPW-G2) and the SNORD109B together with the distal part of UBE3A antisense region (UBE3A-ATS) is restricted to neurons (Figure 1)12,13. However, there are also studies identifying SNORD115 in a few other human tissues4,14.
Figure 1. Organization of the human PWS locus.
Schematic representation of human 15q11-q13 region (drawing is not to scale). Protein coding and snoRNA genes are marked as boxes and ovals, respectively. Paternally and maternally expressed genes are shown with dark or white colors, respectively. Dark bars indicate alternatively spliced exons of the paternally expressed U-UBE3-ATS transcript. Known and suggested U-UBE3-AS transcripts are shown as black and dashed arrows, respectively. Question marks depict regions with identified internal TSS-tags (see also Supplementary Figure S7). The PWS imprinting center (PWS-IC) is schematically indicated by a circle.
Based on mouse models, two groups reported that deletion of the SNORD116 genes cluster together with IPW116 exons results in postnatal growth retardation15,16. Subsequently, patients with similar micro-deletions of the SNORD116 genomic region that exhibited key characteristics of Prader-Willi syndrome were described17. Nevertheless, the question of the functional significance of SNORD116 in PWS is still unknown. The contribution of the IPW116 exons as part of the U-UBE3A-ATS long non-protein coding RNA in PWS should also be considered11,15,18. Moreover, chimeric long non-protein coding RNA containing the IPW116 exon flanked by two SNORD116 RNAs (previously known as PWCR119; GenBank: AF241255.1) was shown to sequester the Fox2 splicing factor resulting in alteration of pre-mRNAs splicing pattern20. In addition, the cumulative impact to the full-fledged PWS phenotype of different genes located in the PWS region is a viable possibility21,22,23.
Although the involvement of SNORD115 in PWS syndrome is questionable, as loss of the snoRNA cluster does not lead to PWS phenotype17,24, an antisense element within SNORD115 is complementary to an alternatively spliced exon of the 5HT-2C serotonin receptor pre-mRNA. The snoRNA target region is located within a sequence that is subject to post-transcriptional editing4. Subsequently, it was shown that the SNORD115 guide element might regulate A to I editing. However, this occurs only if the target RNA is transcribed in the nucleolus under control of RNA polymerase I promoter25. In vitro experiments show that SNORD115 could interfere with alternative splicing of the 5HT-2C serotonin receptor pre-mRNA. However, in these experiments the corresponding splice site of the pre-mRNA was mutated26. In addition, the transcription of long cis-antisense non-protein coding UBE3A-ATS RNA (that is predicted to include part of IPW115 exons) on the paternal chromosome has been reported to negatively regulate UBE3A expression in neurons, thereby determining the tissue-specific imprinting of the UBE3A gene in human and mouse brain12,13,27,28.
We have performed a novel comprehensive analysis of the expression profile of long and small non-protein coding RNAs from PWS locus in 20 different human tissues. In addition, an expression profile of the Angelman syndrome gene (UBE3A) across investigated tissues was also determined. Obtained data demonstrate that SNORD64, SNORD107, SNORD108, SNORD116 snoRNAs and IPW116 exons exhibit expression patterns different from SNORD115 and IPW115 exons. Expression profiles of investigated UBE3A cis-antisense exons of UBE3A-ATS transcript did not correlate with snoRNAs or with IPW116 and IPW115 exons of long non-protein coding RNAs (npcRNAs). The UBE3A expression revealed minimal deviation across tissues. We also performed analysis of RNA-seq reads derived from Cap Analysis of Gene Expression (CAGE-seq) libraries of human cell lines and tissues. The transcripts potentially initiated within and between snoRNA clusters were identified. Our findings provide evidence for widespread expression of the investigated non-protein coding RNAs from the PWS locus and point to additional aspects in regulation of SNORD115 and the distal region of the UBE3A-ATS transcript in human.
Results
Assembling a npcRNA ‘Expression Ruler'
Quantitative real-time PCR (qPCR) gene expression analysis is a powerful tool that requires minimal sample material, time and efforts. However, considering the technical and experimental variation in starting materials due to e.g. RNA yield and efficiency of cDNA synthesis, there is a need for suitable reference genes serving as internal controls to reduce possible errors between different samples and runs, hence allowing accurate interpretation of qPCR data29. Common reference genes to normalize mRNA expression levels are protein-coding genes (frequently termed housekeeping genes, HKGs), which are constitutively expressed in a wide variety of tissues/cell types with minimal deviation between different tissue samples and/or under different experimental conditions30,31,32,33. In a previous study on the analysis of non-protein coding RNA abundance, we proposed the use of ubiquitously expressed housekeeping npcRNAs (HKRs) as reference genes. Since the biogenesis of most npcRNAs differs from that of protein-coding mRNAs, the use of HKGs might thus lead to the misinterpretation of qPCR data34. In addition to being ubiquitously and constitutively expressed, the selected reference genes should be present in similar abundance to the RNA target under investigation.
To determine a set of constitutively expressed npcRNAs with a wide dynamic range of expression (reference control), RT-qPCR expression data of eleven selected housekeeping npcRNAs (HKRs) were analyzed in twenty human tissue samples. HKRs are defined as npcRNAs exhibiting ubiquitous expression patterns with low tissue to tissue variation34. The HKRs belong to different structural and functional classes, are located on different chromosomes to avoid co-regulatory effects in their expression patterns and were named according to the snoRNA database35 or as defined by the HUGO gene nomenclature committee (HGNC; Supplementary Table S1).
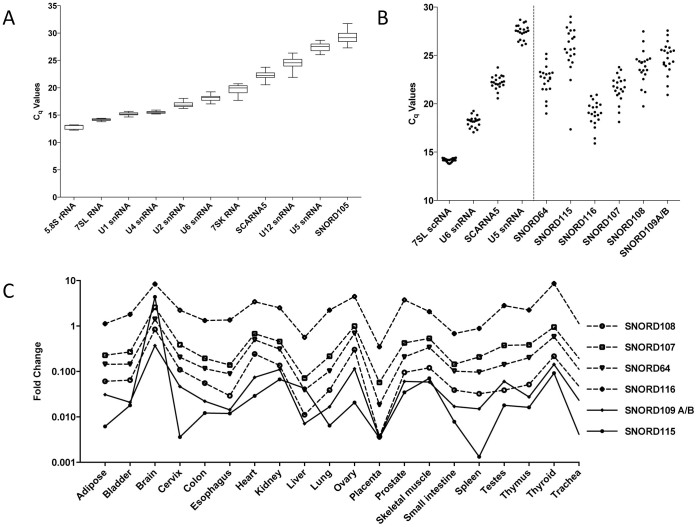
For each HKR, the mean quantification cycle (Cq) value, which represents the number of PCR cycles required to obtain a fluorescence signal above a defined threshold level36, was calculated in 20 human tissues. In general, a Cq difference of 3 in qPCR analysis indicates a 10-fold change in expression level. The mean Cq values were calculated for the test set of snoRNAs alongside a set of HKRs using cDNAs prepared using oligo (dT) 10-12 plus random hexamers. HKRs were grouped into arbitrary categories as reflected by their mean Cq values. The first category comprised: 5.8S rRNA, 7SL scRNA, U1 snRNA and U4 snRNA exhibiting Cq values below 16. For further analysis, 7SL scRNA was included due to its lower tissue-to-tissue expression variation. U2 snRNA, U6 snRNA, 7SK RNA and SCARNA5 (U87 scaRNA) with mean Cq values between 16–25 were grouped into the second category, whereas U12 snRNA, U5 snRNA and SNORD105 (U105 snoRNA) with Cq values above 26 were grouped into the third category (Figure 2A). Notably, RNA structure and/or post-transcriptional modifications could influence reverse transcription (RT) efficiency during cDNA synthesizes and thus significantly affect qPCR-determined true RNA abundance measurements37. We selected at least one HKR from each category (7SL RNA, U6 snRNA and/or SCARNA5, and U5 snRNA) and set up an npcRNA expression ruler that could be applied in the data analysis of novel transcripts.
Figure 2. Assembly of the npcRNA ‘expression ruler' and its application for analysis of snoRNAs from PWS locus.
(A) RT-qPCR mean Cq values for 11 npcRNA HKRs in 20 human tissue cDNA samples. The median Cq values are shown as lines, 25 to 75 Cq percentile as boxes and the range of Cq values from 20 cDNA samples as whiskers. (B) The “Expression Ruler” applied to investigate snoRNAs from PWS-locus. (C) RT-qPCR analysis of the human PWS locus snoRNAs. Normalized expression data showing fold change values obtained in 20 different human tissues by qPCR.
Comparative expression analysis of snoRNA genes from PWS locus
snoRNA genes from the PWS-locus were identified more than a decade ago. However, a detailed analysis of their expression in different human tissues has not been reported4,14. In order to gain further insight into the non-protein coding RNA expression within the PWS-locus, we initially set out to investigate snoRNA genes located in this region (Figure 1). For these analyses, the npcRNA expression ruler concept (see above) was applied. Earlier reports have shown that PWS locus derived snoRNAs are highly expressed in the brain and some of them could be found in a few other human tissues4,11. However, as quantitative data concerning their expression in various tissues were missing, we investigated snoRNAs expression in 20 different human tissues using RT-qPCR (Figure 2B). All primer pairs were designed and evaluated according to a standardized protocol described previously34. In short, primer specificity was assessed by end-point PCR using human brain cDNA as a template and only single amplicons were detected (data not shown). Amplification efficiencies were determined in a qPCR assay using a 10-fold dilution series of human brain cDNA.
Overall, PWS locus snoRNAs exhibit a difference in expression range of more than four orders of magnitude and demonstrate the need of a set of HKRs for accurate data analysis (Figure 2B). In general, SNORD116 expression levels correlate with the expression of U6 snRNA and in some tissues with SCARNA5, indicating its high to medium abundance in the 20 human tissues tested. The SNORD115 snoRNA gene cluster showed a primarily low expression level similar to the U5 snRNA expression in the ruler, except in the brain. Expression of SNORD115 in the brain correlates with U6 snRNA indicating its high expression. Hence, consistent with previous reports, our RT-qPCR data showed that the snoRNAs are differentially expressed, exhibiting their highest levels in human brain. Notably, consistent results were obtained for 3 different panels of total RNA samples using random or gene specific priming during cDNA synthesis (Supplementary Figure S1A-E). Upon further comparison of the expression patterns of SNORD116, SNORD64, SNORD107 and SNORD108 snoRNA, highly similar profiles among investigated human tissues were observed (Figure 2C). These snoRNA genes are located upstream of the SNORD116 gene cluster and were suggested to co-transcribe with the proximal part of the host - U-UBE3A-ATS long npcRNA (Figure 1)3. Hence, in line with previous observations our data indicate the possibility of co-regulatory expression or processing of aforementioned snoRNA genes (Figure 2C). When expression profiles of the SNORD115 gene cluster were compared with other snoRNA genes from the PWS locus, their expression patterns deviated substantially (Figure 2C and 3A). The SNORD109 represents a special case, as there are two copies of the gene located at different positions within the PWS locus (Figure 1). The SNORD109A gene is located proximal to the SNORD116 cluster, whereas SNORD109B is distal to the SNORD115 gene cluster. Since their sequence is identical, it is not possible to distinguish between their expression profiles. The detected combined expression profile of SNORD109A and SNORD109B genes does not correlate to SNORD116 and SNORD115 genes, additionally arguing for differential expression of npcRNA in investigated tissues (Supplementary Figure S2).
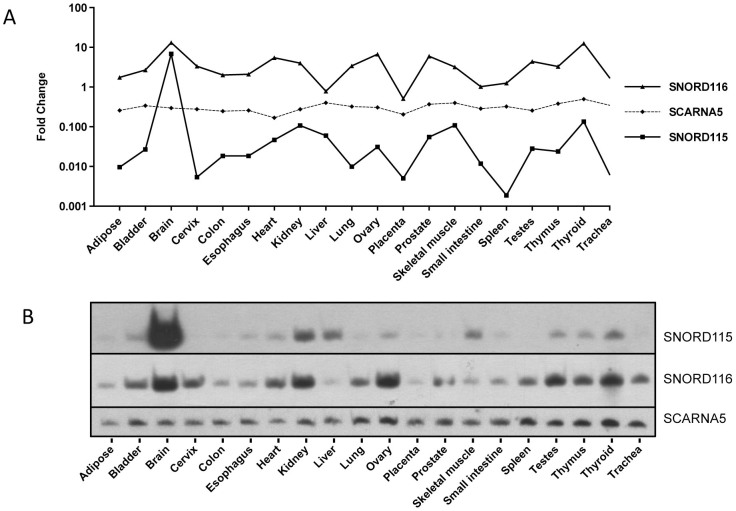
Figure 3. Comparative expression analysis of SNORD115 and SNORD116.
Expression analysis of SNORD115 and SNORD116 among 20 different human tissues performed by: (A) RT-qPCR and (B) northern blot hybridization. (A) RT-qPCR data is represented as fold change after normalization with the geometric mean of “Expression Ruler”. (B) SCARNA5 serve as loading controls in northern blot hybridization experiments.
To further support the RT-qPCR data, we performed northern hybridizations for SNORD116 and SNORD115 snoRNAs, as well as SCARNA5 using specific deoxy-oligonucleotide probes. Expression patterns shown by northern blot analysis correlated well with that of RT-qPCR data (Figure 3A and 3B, Supplementary Figure S3). Notably, in agreement with a recent analysis, we did not detect psnoRNAs (processed snoRNAs) transcripts (Supplementary Figure S3)38. However, a longer RNA of approximately 110 nts in length was detected by northern blot hybridization in a human brain RNA with a SNORD115 specific probe (Supplementary Figure S3). This RNA could potentially represent a longer SNORD115 isoform, analogous to the previously reported L-Snord115 in rat brain39.
The normalized RT-qPCR data is represented as fold change and is interdependent from the geometric mean of the four reference RNAs (7SL scRNA, U6 snRNA, SCARNA5 and U5 snRNA). In our analysis, the normalized HKRs showed variance of around 2-fold change in expression levels between the individual tissues underlining their eligibility as reference genes in RT-qPCR. SNORD116 showed a fold change level above 10 with the highest expression in brain, ovary and thyroid tissue. The lowest expression was observed in liver and placenta, in both RT-qPCR and northern blot analyses (Figure 3A and 3B). In contrast, SNORD115 showed a more than 1000-fold change in expression levels between individual tissues indicating a highly variable expression pattern (Figure 3A and 3B). The highest expression was observed in brain, whereas moderate expression was seen in kidney, liver, skeletal muscle and thyroid. The expression in the remaining tissues was low and according to our analysis, the lowest expression was observed in the spleen (Figure 3B). Interestingly, Castle et al. 2010, reported the expression of SNORD116 and SNORD115 in 11 different human tissues14. In correlation with our results, the authors showed that SNORD116 was present in all tested tissues while SNORD115 was predominantly expressed in brain with lower levels in kidney, skeletal muscle, liver and testis. However, SNORD115 expression in other tissues was barely detected by Castle et al. 201014.
Our next goal was to analyze tissues with the most prominent deviations in expression patterns between the SNORD116 and SNORD115 gene clusters. When snoRNAs expression patterns were compared across 20 different human tissues, striking differences were observed in cervix, colon, kidney, liver, lung, prostate, skeletal muscle, small intestine and spleen (Figure 3A and 3B).
Expression analysis of U-UBE3A-ATS and UBE3A genes
We performed a further assessment of the expression profile of U-UBE3A-ATS long non-protein coding RNA across 20 different human tissues. Human U-UBE3A-ATS RNA transcription is initiated from paternal unmethylated chromosomal region(s) (U-exons), located proximally from the PWS-IC center. This long npcRNA is composed of multiple alternatively spliced exons including repetitive subtypes of IPW116 and IPW115 exons flanking SNORD116 and SNORD115 RNA copies, respectively (Figure 1). Based on exonic sequences and genomic location, U-UBE3A-ATS transcript could be divided into two parts. The first is the proximal region, which harbors intronic SNORD64, SNORD107, SNORD108, SNORD109A genes and SNORD116 cluster. It is composed of different exons including U-exons and repetitive IPW116 subtypes (Figure 1). The second region is UBE3A-ATS transcript, which harbors intronic SNORD109B and the SNORD115 cluster, and extends distally to overlap the UBE3A gene in cis-antisense orientation (Figure 1). There are reports suggesting that the expression of UBE3A-ATS is restricted to the brain, whereas the proximal transcript could be identified in other tissues (for review40). To evaluate the expression of different U-UBE3A-ATS regions, we have designed primers targeting the IPW116, IPW115 and UBE3A cis-antisense exons (Supplementary Figure S4). Overall, different exons of U-UBE3A-ATS transcript(s) exhibit a difference in expression range of more than five orders of magnitude and demonstrate the need of a set of HKRs for accurate data analysis (Supplementary Figure S5). When the expression profile of IPW116 exons was investigated, we detected patterns nearly identical to the SNORD64, SNORD107, SNORD108 and SNORD116 gene clusters across 20 human tissues (Figure 4A). This observation strongly supports the notion that investigated RNAs are derived from the same differentially regulated precursor transcript.
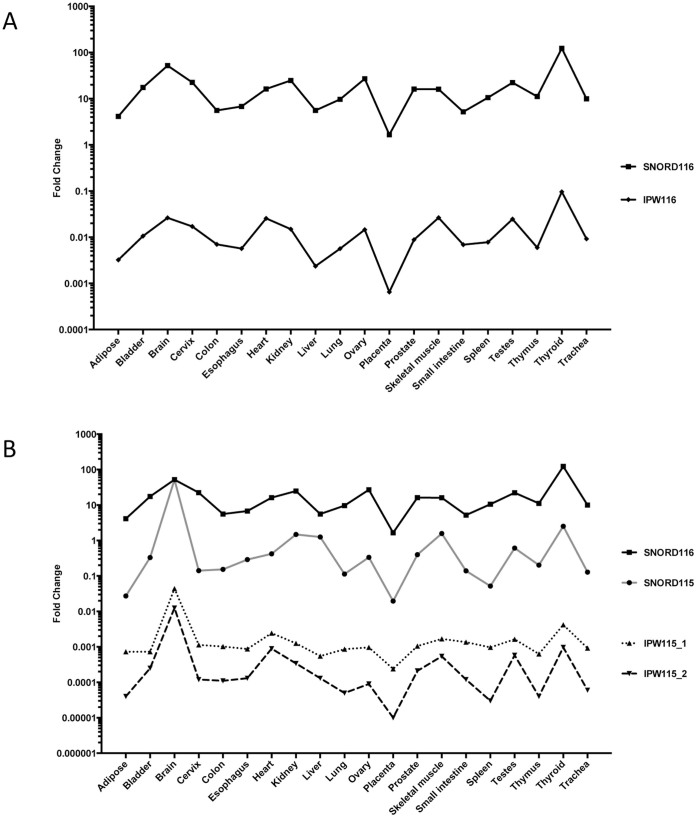
Figure 4. Expression analysis of IPW116 and IPW115 exons.
Normalized expression data of IPW116 and IPW115 exons showing fold change values obtained in 20 different human tissues by qPCR. Expression results indicate that IPW116 exons exhibit an expression pattern similar to SNORD116 (A), whereas IPW115 exons show similarity to SNORD115 expression across analyzed tissues (B).
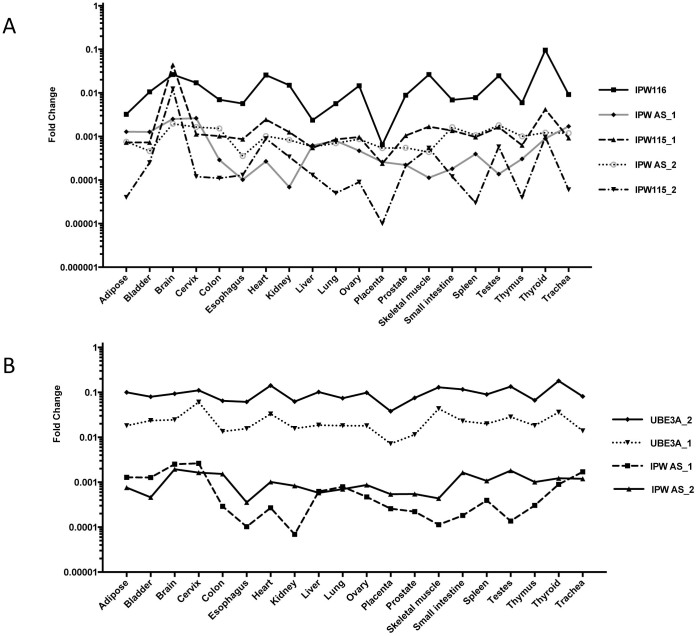
To gain insight into the regulation of the distal end of the U-UBE3-ATS RNA, we first determined the expression profile of IPW115 exons in different human tissues. We detected the presence of IPW115 exons in all investigated tissues, arguing for its ubiquitous nature in humans (Figure 4B). When expression profiles of IPW115 exons and snoRNAs were compared, a closer pattern was observed with SNORD115, indicating that these RNAs are potentially derived from the same precursor transcript (Figure 4B). The observed tissue-specific variations between expression profiles of proximal IPW116 and distal IPW115 exons are in line with differences detected among PWS – snoRNA genes. Hence, our results suggest additional layers of expression regulation, namely transcription initiation within the PWS RNA coding region.
Next, we set out to evaluate tissue-specific expression of the UBE3A cis-antisense exons from UBE3A-ATS long npcRNA. We designed primer combinations for two assays: IPW-AS-1 and IPW-AS-2 targeting different splice exons of UBE3A-ATS RNA. The AS-2 forward primer was located on the SNORD109B 3′-flanking exon, whereas the reverse primer targeted the UBE3A-ATS region partly antisense to the protein-coding exon of UBE3A mRNA (Supplementary Figure S4). The IPW-AS-1 assay aimed at targeting UBE3A-ATS exons that are cis-antisense to the 5′-region of UBE3A gene. Surprisingly, in addition to human brain tissue, both assays could detect UBE3A cis-antisense long non-protein coding transcripts within all analyzed tissues. The expression level of long npcRNA(s) detected by IPW-AS1 and IPW-AS2 assays was comparable, and in some tissues even higher than in IPW115 exons (Figure 5A). However, the IPW115 containing transcript was highly unregulated in human brain. Unexpectedly, we did not detect pronounced differences in the abundance of UBE3A cis-antisense exons in this tissue (Figure 5B). Overall, the expression profiles of cis-antisense exons did not correlate with IPW116 or IPW115 transcripts, indicating the possibility of independent regulation.
Figure 5. Comparative expression analysis of UBE3A cis-antisense exons and the UBE3A mRNA.
(A) Expression profiles of IPW and UBE3A cis-antisense exons in different tissues. (B) Comparative analysis of UBE3A and UBE3A cis-antisense transcripts in 20 different human tissues.
The UBE3A gene is biallelically expressed in most human tissues, but is imprinted and exclusively expressed from the maternally inherited chromosome in brain. Its specific expression appears to be controlled, at least in part, by the AS imprinting center (AS-IC) that is located at approximately 35–40 kb upstream of PWS-IC41,42. There are reports suggesting that epigenetic silencing of UBE3A in human neurons is controlled and regulated by the expression of UBE3A-ATS non-protein coding RNA from paternal chromosome40.
When the expression profile of the UBE3A gene was investigated, we detected a minimal, mostly negligible deviation across 20 human tissues. Its expression variation was lower than that of 7SL RNA and U6 snRNA, which are among the most abundant and commonly used reference RNAs. Based on abundance and low tissue-to-tissue variation, we suggest the use of the UBE3A transcript as an internal reference control for future expression analysis (Supplementary Figure S6). The UBE3A expression profile showed neither positive nor negative co-regulation with cis-antisense or IPW-exons (Figure 5B). Hence, our obtained expression data indicate a more complex regulatory network in the PWS/AS locus and raise a reasonable concern for the proposed neuronal-specific function of the UBE3A cis-antisense transcript(s) in human.
Analysis of CAGE tags
The expression profiles of the investigated PWS locus derived non-protein coding RNAs prompted us to examine the possible presence of additional regulatory elements and transcription start sites within the distal regions of the U-UBE3A-ATS spanning region. For our analysis we studied RNA-seq data derived from the 5′-Cap Analysis of Gene Expression (CAGE) tags from various tissues/cell lines and subcellular compartments43. A complete collection of CAGE-tags representing potential transcription start sites (TSS) predicted based on Hidden Markov Modeling (HMM) was obtained from the ENCODE/RIKEN dataset freely available on the BLAT human genome browser44. All CAGE tags from the human chromosome 15q11-q13 region were restricted to selected coordinates (chr15: 25354000 -25598033) of the UCSC Genome Browser on the human genome, (Feb. 2009 (GRCh37/hg19) Assembly) and were extracted using UNIX script and manually inspected. In total, 745 CAGE-tag contigs supported by the Hidden Markov model were identified (Supplementary Table S2). About two-thirds of the TSS tags were localized to the region between the SNORD116 and SNROD115 gene clusters, suggesting possible independent transcription initiation of the distal region of the U-UBE3A-ATS transcript (Supplementary Figure S7A). The majority of the remaining CAGE-tags were mapped distal to the SNORD115 gene array, but proximal to UBE3A cis-antisense exons of the long npcRNA (Supplementary Figure S7B). This finding further suggests an internal initiation of transcription within the PWS non-protein coding RNA containing locus, and partly explains the difference in expression between the IPW115 and UBE3A cis-antisense exons of the U-UBE3A-ATS transcript. Interestingly, analysis of CAGE data from the PWS-locus revealed a number of TSS tags that map to the U-UBE3A-ATS transcript in cis-antisense orientation (Supplementary Table S3). Most of them were derived from the same regions as sense TSS tags, potentially indicating the presence of euchromatin in this genomic locus. This could potentially shed light on the ubiquitous expression of the U-UBE3A-ATS derived npcRNAs in humans.
Discussion
Expression analysis of RNA molecules from the human transcriptome provides essential information about tissue specificity and regulation, which is important for further functional studies. We set out to analyze the expression of PWS locus derived non-protein coding RNAs in various human tissues. We chose quantitative real-time PCR (qPCR) because of its high sensitivity and very low sample quantity consumption/requirement45,46,47. However, qPCR needs careful assay design and reaction optimization to maximize sensitivity and accuracy48,49. The selection of suitable internal controls for data normalization is pivotal to reduce errors caused by experimental variations such as starting material, RNA extraction, cDNA preparations, etc.31. In addition, the differences between expression levels of the reference gene and the gene of interest could also lead to misinterpretation. Here, we proposed a set of npcRNAs with vastly different expression levels as an ‘Expression Ruler' (Figure 2A). We recommend using a multiple reference gene concept to cover a wide expression range for analysis of novel or yet poorly characterized transcripts.
We applied the Expression Ruler concept, to analyze the expression levels of small and long non-protein coding RNAs from the PWS locus in 20 different human tissues. We clearly revealed related profiles from SNORD64, SNORD107, SNORD108 and SNORD116 snoRNA (Figure 2C), implying that these RNAs are derived from the same transcript. A notable difference in expression was observed between the aforementioned snoRNAs and SNORD115 RNA, suggesting involvement of additional regulatory elements in regulation of the distal region of the U-UBE3A-ATS transcript around the SNORD115 gene clusters (Figure 2C). To further support this notion, we investigated the expression of IPW116 and IPW115 exons of U-UBE3A-ATS transcript(s). The observed differences in expression profiles of exons of the host gene were similar to the intronic snoRNAs. In contrast, when the expression profiles of UBE3A cis-antisense exons were analyzed, we observed neither a correlation with snoRNAs nor with IPW exons. These results argue for an independent transcription regulation of UBE3A cis-antisense exons in human tissues. In agreement with our expression data, detailed analysis of TSS-tags could identify independent transcriptional start sites within the U-UBE3A-ATS spanning region. Collectively, in addition to well-known U-exons, our data suggest the presence of at least two different regions containing transcription start sites. The first is located between the SNORD116 and SNORD115 gene clusters, and the second is upstream from the UBE3A cis-antisense region. Our findings indicate new facets in the regulation of expression of non-protein coding RNAs in the PWS/AS locus and open an opportunity for a better understanding of molecular mechanisms associated with human disorders.
Methods
RNA samples and first-strand cDNA synthesis
Total RNA samples were purchased from FirstChoice® Human Total RNA Survey Panel (Ambion); these included adipose, bladder, brain, cervix, colon, esophagus, heart, kidney, liver, lung, ovary, placenta, prostate, skeletal muscle, small intestine, spleen, testes, thymus, thyroid and trachea tissues. For each tissue, total RNA samples were isolated from respective tissues collected from three different individuals (Supplementary Table S4).
Total RNA samples were DNase treated, certified for purity and integrity as tested on the Agilent Bioanalyzer. Total RNA concentration and purity were verified using a NanoDrop spectrophotometer ND-1000 (Thermo Scientific) by measuring absorbance at OD260/280. Furthermore, the presence of DNA contamination was assessed by PCR as described earlier34.
First strand cDNA synthesis was performed by one of two methods: 1. SuperScript® II reverse transcriptase using oligo(dT) and random hexamer primers (Invitrogen), or 2. reverse transcription using transcriptor reverse transcriptase (Roche) and pool of gene specific primers.
1. In brief, 5 µg of total RNA was incubated with 0.5 µl of oligo(dT)12-18 (500 µg/µl), 1 µl of random hexamer primers (3 µg/µl) 1 µl of dNTP mix (25 mM mix) and 7.5 µl of DEPC-treated water (Ambion) for 5 min at 65°C. The reaction was cooled on ice for 2–3 min and briefly centrifugated. Then, 5 µl of first strand synthesis buffer (5 X, containing 250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2), 2 µl of 0.1 M DTT, 2.5 µl of ribolock RNAase inhibitor (40 U/µl, Fermentas) and 1 µl of (200 U/µl) of SuperScript® II RTase were added and incubated for 90 min at 42°C. Reverse transcription reaction was terminated by heat inactivation for 15 min at 75°C and stored at −20°C. Final cDNA was diluted 1:20 before use in RT-qPCR.
2. Total RNA (0.5 µg) was incubated with 2 µl of 1 µM reverse primer, 1 µl of dNTP mix (25 mM mix) and 9.5 µl of DEPC-treated water for 10 min at 65°C. The reaction was cooled on ice for 2–3 min and briefly centrifuged. Then, 4 µl of first strand synthesis buffer (5 X, containing 250 mM Tris-HCl (pH 8.5), 150 mM KCl, 40 mM MgCl2), 0.5 µl of ribolock RNAase inhibitor (40 U/µl, Fermentas) and 0.5 µl of (20 U/µl) of Transcriptor reverse transcriptase were added and incubated for 60 min at 55°C. Reverse transcription reaction was terminated by heat inactivation for 10 min at 85°C. Reaction volume was increased to 50 µl using nuclease-free water stored at −20°C. cDNA was diluted 1:10 prior to use in a real-time PCR reaction.
Reverse-transcription quantitative real-time PCR
All details on the primers used for RT-qPCR analysis in this study are given in Supplementary Table S1. Primer pairs were taken from a previous study or designed and analyzed accordingly34. All cDNAs prepared with method 1 were used in the RT-qPCR reactions that were performed in triplicate along with non-template-controls in 384-well microtitre plates on an ABI Prism 7900 HT sequence detector system (Applied Biosystems). Amplification was performed in a final reaction volume of 10 µl, starting with a 2 min activation step at 50°C, 10 min template denaturation step at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. SYBR Green assay also included a melt curve analysis step at the end of the cycling protocol, with continuous fluorescence measurement from 60–95°C. All reactions contained 2 µl of cDNA (20 ng), 5 µl of 2 X SYBR Green Master Mix (Applied Biosystems) and 1 µl containing 10 µM of each primer and 2 µl of DEPC treated water (Ambion). Cq values were determined for all runs applying SDS software v. 2.1 (Applied Biosystems) using automatic baseline settings and a threshold value of 0.2. Results were exported to GraphPad Prism for further analysis.
cDNA samples prepared using protocol 2 were used in real-time PCR assays using the Roche Light Cycler 480 SYBR Green master mix. In a 10 µl reaction, 2 µl of diluted cDNA was added to 5 µl of 2X SYBR green master mix (Roche), 2 µl nuclease-free water and 1 µl of 10 µM primer mix. Real-time PCR amplification reaction included 5 minutes of activation/initial denaturation step followed by 40 cycles of 20 sec at 95°C, 40 sec at 60°C and 1 min extension at 72°C. The reaction included single acquisition of fluorescent signal at 60°C for each cycle and continuous acquisition from 50°C to 97°C at the end of the 40 cycles for melt-curve analysis. All the data produced using automatic baseline settings and a threshold value of 0.2 were transferred to Excel files for subsequent analysis.
Data analysis was carried out using a geometric mean of at least 3 selected housekeeping RNAs used in the study and the fold change is represented as 2ΔCq (Supplementary Table S5) Results were exported to a GraphPad Prism for further analysis. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines was followed50. The MIQE checklist is presented as Supplementary Table S6.
Northern blot analysis
Three µg of each total RNA were separated on 8% (w/v) denaturing polyacrylamide gel (PAAG; 7 M urea, 1 X TBE buffer) at 200 V for 90 min. Total RNA samples from PAAG were transferred onto positively charged nylon membranes (BrightStar Plus, Ambion or Hybond-N+, Amersham Biosciences) using a Trans-blot semi-dry blotting apparatus (BioRad) at 400 mA for 45 min in 0.5 X TBE buffer (90 mM Tris, 64.6 mM boric acid, 2.5 mM EDTA, pH 8.3). After immobilizing RNA using a cross linker (STRATAGENE) at 1200 J/cm2, nylon membranes were pre-hybridized for 1–3 hrs in Ultrahyb Oligo hybridization buffer (Ambion) at 42°C.
To determine npcRNA candidates, hybridizations were performed with RT-qPCR reverse primers or reverse complement oligos to the RT-qPCR forward primers. Hybridization probes were prepared by end labeling with γ-[32P]-ATP using T4 polynucleotide kinase (Fermentas). Hybridization was carried out at 42°C in Ultrahyb Oligo hybridization buffer for 12–16 hrs. Blots were washed twice at room temperature in 2 X SSC buffer (20 mM sodium phosphate, 0.3 M NaCl, 2 mM EDTA, pH 7.4) containing 0.5% SDS for 2 X 30 min (washing repeated if the blot showed high counts). Membranes were exposed to MS-film (Kodak) and developed after 1–2 hrs or, if necessary, further exposed for longer time periods at −80°C.
Analysis of CAGE reads for promoter marks
CAGE clusters were accessed via the UCSC server (www.genome.ucsc.edu/cgi-bin/hgBlat?command=start), combined and intersected with the PWS regions. Simple gawk scripts collect all transcriptional start site (TSS) annotated clusters that localize to the human chromosome 15 between 25354000 and 25598033 resides (Feb. 2009 (GRCh37/hg19) Assembly), respectively. Clusters that are expressed on the sense strand are considered to be a TSS - candidate. Gawk scripts again compute the separation in antisense and sense transcripts.
Author Contributions
C.R.G., Z.K. and T.S.R. conceived and designed the study. C.R.G. performed the experimental work and analyzed qPCR data. C.R. did the computational work. C.R.G. wrote the first draft of the paper and all authors provided input and approved the final manuscript
Supplementary Material
Supplementary Information
Acknowledgments
The authors thank Juergen Brosius for critically reading of an earlier version of the manuscript and helpful discussions. This work was supported by the National Genome Research Network [0313358C and 01GS0808]; the Deutsche Forschungs gemeinschaft [BR 754/4-1] and the Max Planck Society for the Advancement of Science.
References
- Butler M. G. Prader-Willi Syndrome: Obesity due to Genomic Imprinting. Curr Genomics 12, 204–215, 10.2174/138920211795677877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. & Wevrick R. Identification of novel imprinted transcripts in the Prader-Willi syndrome and Angelman syndrome deletion region: further evidence for regional imprinting control. Am J Hum Genet 66, 848–858, 10.1086/302817 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runte M. et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10, 2687–2700 (2001). [DOI] [PubMed] [Google Scholar]
- Cavaille J. et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A 97, 14311–14316 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Walker C. L. & Wevrick R. Prader-Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr Patterns 3, 599–609 (2003). [DOI] [PubMed] [Google Scholar]
- Nahkuri S., Taft R. J., Korbie D. J. & Mattick J. S. Molecular evolution of the HBII-52 snoRNA cluster. J Mol Biol 381, 810–815 (2008). [DOI] [PubMed] [Google Scholar]
- Royo H. & Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol cell 100, 149–166, 10.1042/BC20070126 (2008). [DOI] [PubMed] [Google Scholar]
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet 154C, 365–376, 10.1002/ajmg.c.30273 (2010). [DOI] [PubMed] [Google Scholar]
- Chamberlain S. J. RNAs of the human chromosome 15q11-q13 imprinted region. Wiley Interdiscip Rev RNA 4, 155–166, 10.1002/wrna.1150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot M., Cavaille J. & Feil R. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics 7, 1341–1348, 10.4161/epi.22884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P., Royo H., Marty V., Bortolin-Cavaille M. L. & Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J Cell Sci 123, 70–83, 10.1242/jcs.054957 (2010). [DOI] [PubMed] [Google Scholar]
- Chamberlain S. J. et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci U S A 107, 17668–17673, 10.1073/pnas.1004487107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor K. et al. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum Mol Genet 23, 2364–2373, 10.1093/hmg/ddt628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle J. C. et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PloS one 5, e11779, p10.1371/journal.pone.0011779 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin B. V. et al. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet 3, e235 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F. et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PloS one 3, e1709, 10.1371/journal.pone.0001709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T. et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet 40, 719-721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W. T. et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet 22, 4318–4328, 10.1093/hmg/ddt281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T., Schweizer J., Rees C. A. & Francke U. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which Is highly expressed in brain. Am J Hum Genet 67, 1067–1082, 10.1086/303106 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q. F. et al. Long noncoding RNAs with snoRNA ends. Mol cell 48, 219–230, 10.1016/j.molcel.2012.07.033 (2012). [DOI] [PubMed] [Google Scholar]
- Kanber D. et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet 17, 582–590, 10.1038/ejhg.2008.232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzik M. et al. The C15orf2 gene in the Prader-Willi syndrome region is subject to genomic imprinting and positive selection. Neurogenetics 11, 153–161, 10.1007/s10048-009-0231-z (2010). [DOI] [PubMed] [Google Scholar]
- Neumann L. C. et al. The imprinted NPAP1/C15orf2 gene in the Prader-Willi syndrome region encodes a nuclear pore complex associated protein. Hum Mol Genet 21, 4038–4048, 10.1093/hmg/dds228 (2012). [DOI] [PubMed] [Google Scholar]
- Runte M., Varon R., Horn D., Horsthemke B. & Buiting K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum Genet 116, 228–230, 10.1007/s00439-004-1219-2 (2005). [DOI] [PubMed] [Google Scholar]
- Vitali P. et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol 169, 745–753, 10.1083/jcb.200411129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S. & Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232, 1118265 [pii] 10.1126/science.1118265 (2006). [DOI] [PubMed] [Google Scholar]
- Meng L., Person R. E. & Beaudet A. L. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet 21, 3001–3012, 10.1093/hmg/dds130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L. et al. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet 9, e1004039, 10.1371/journal.pgen.1004039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichopad A., Dilger M., Schwarz G. & Pfaffl M. W. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res 31, e122 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonic A. et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313, 856–862 (2004). [DOI] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L. & Orntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64, 5245–5250 (2004). [DOI] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S. & Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6, 279–284 (2005). [DOI] [PubMed] [Google Scholar]
- Galiveti C. R., Rozhdestvensky T. S., Brosius J., Lehrach H. & Konthur Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA 16, 450–461, 10.1261/rna.1755810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L. & Weber M. J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34, D158–162 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker N. J. Tech.Sight. A technique whose time has come. Science 296, 557–559 (2002). [DOI] [PubMed] [Google Scholar]
- Raabe C. A., Tang T. H., Brosius J. & Rozhdestvensky T. S. Biases in small RNA deep sequencing data. Nucleic Acids Res 42, 1414–1426, 10.1093/nar/gkt1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin-Cavaille M. L. & Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res 40, 6800–6807, 10.1093/nar/gks321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo D., Raabe C. A., Reinhardt R., Brosius J. & Rozhdestvensky T. S. Alternative processing as evolutionary mechanism for the origin of novel nonprotein coding RNAs. Genome biol evol 5, 2061–2071, 10.1093/gbe/evt155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S. J. & Lalande M. Angelman syndrome, a genomic imprinting disorder of the brain. J. Neurosci 30, 9958–9963, 10.1523/JNEUROSCI.1728-10.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich B. et al. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNRPN gene. Nat Genet 14, 163–170, 10.1038/ng1096-163 (1996). [DOI] [PubMed] [Google Scholar]
- Buiting K. et al. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet 72, 571–577, 10.1086/367926 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R. et al. CAGE: cap analysis of gene expression. Nature methods 3, 211–222, 10.1038/nmeth0306-211 (2006). [DOI] [PubMed] [Google Scholar]
- Valen E. et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res 19, 255–265, 10.1101/gr.084541.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Nolan T. & Pfaffl M. W. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol 34, 597–601 (2005). [DOI] [PubMed] [Google Scholar]
- Bustin S. A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29, 23–39 (2002). [DOI] [PubMed] [Google Scholar]
- Huttenhofer A. & Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res 34, 635–646 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya M. et al. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn 5, 209–219 (2005). [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Jiang J., Liu Q. & Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32, e43 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622, 10.1373/clinchem.2008.112797 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information