Abstract
Adrenomedullin (ADM), a secretory peptide with multiple functions in physiological to pathological conditions, is upregulated in several human cancers, including brain, breast, colon, prostate, and lung cancer. However, the molecular mechanisms underlying the regulation of ADM expression in cancerous cells are not fully understood. Here, we report that oncostatin M (OSM), a cytokine belonging to the interleukin-6 family, induces ADM expression in astroglioma cells through induction of signal transducer and activator of transcription-3 (STAT-3) phosphorylation, nuclear translocation, and subsequent DNA binding to the ADM promoter. STAT-3 knockdown decreased OSM-mediated expression of ADM, indicating that ADM expression is regulated by STAT-3 in astroglioma cells. Lastly, scratch wound healing assay showed that astroglioma cell migration was significantly enhanced by ADM peptides. These data suggest that aberrant activation of STAT-3, which is observed in malignant brain tumors, may function as one of the key regulators for ADM expression and glioma invasion.
Malignant gliomas are the most common subtype of primary brain tumors. They are characterized by cellular pleomorphism, microvascular proliferation, areas of necrosis, and extensive invasion into the surrounding brain tissues, which leads to poor prognosis for patients1,2. Because of the extraordinary ability to invade the surrounding healthy brain tissue, complete elimination of malignant gliomas by surgical resection is almost impossible3. Thus, the identification of molecular mechanisms involved in invasion is an important objective in glioma research, to develop an effective therapeutic modality for this particular tumor.
In glioma, a large number of microglia/macrophages are found within the tumor mass, and they are known to be involved in the tumor microenvironment which favors glioma growth and invasion, through releasing several microglia/macrophages-derived molecules4. Oncostatin M (OSM), one of interleukin-6 (IL-6) family cytokines, is secreted by activated macrophages and microglia5,6. Increased OSM expression has been reported in a variety of cancers, including malignant glioma7. OSM mainly activates signal transducer and activator of transcription (STAT)-3, which is involved in glioma development and progression8,9,10,11. Constitutive activation and phosphorylation of STAT-3 is frequently detected in glioma, and this activation is believed to promote tumor formation and progression via transcriptional activation of downstream genes12,13.
Adrenomedullin (ADM), a 52-amino acid ring-structure peptide originally isolated from a human pheochromocytoma, is expressed in human cancer cell lines, including brain, breast, colon, prostate, and lung cancer cells14. In physiologic conditions, ADM performs important roles as a vasodilator, bronchodilator, regulator of hormone secretion, neurotransmitter, antimicrobial agent, and controller of renal functions15. In brain tumors, the extent of ADM mRNA expression is related to the tumor type and grade16. However, the stimuli involved in the increased expression of ADM and the molecular mechanisms regulating ADM expression in brain tumors are not fully understood.
In the present study, we showed that OSM induces ADM upregulation through the activation of STAT-3 and that ADM contributes to increased invasion activity in human astroglioma cell lines. Our data support the notion that ADM expression level is influenced by OSM, which is secreted by activated microglia/macrophages, providing the first evidence that ADM-mediated glioma invasion can be facilitated by the inflammatory tumor microenvironment.
Results
OSM induces ADM expression in astroglioma cells
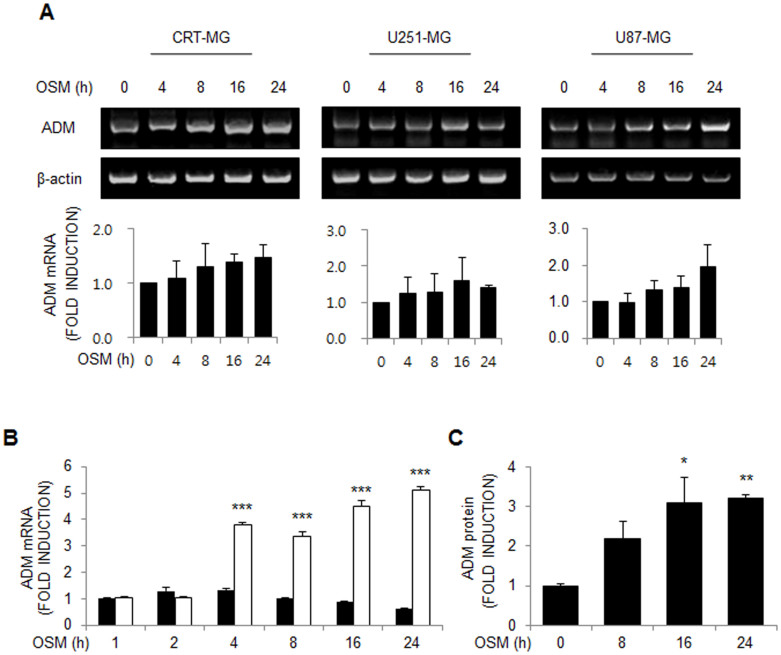
To test whether ADM expression level is affected by OSM, astroglioma cell lines were incubated in the presence of human oncostatin M (hOSM; 10 ng/mL) for various periods. Total RNA was extracted and then subjected to reverse transcription-polymerase chain reaction. As shown in Figures 1A and 1B, ADM mRNA expression levels in the OSM-treated astroglioma cells were enhanced in a time-dependent manner compared to that in the untreated cells. ADM secretion was also upregulated by OSM treatment in a time-dependent manner in CRT-MG cell culture supernatant (Fig. 1C).
Figure 1. OSM induces ADM expression in astroglioma cells.
(A) CRT-MG, U251-MG and U87-MG astroglioma cells were either untreated controls (time zero) or treated with hOSM (10 ng/mL) as indicated, and RNA were extracted and analyzed by RT-PCR for ADM mRNA. Data are presented as the fold induction compared with each untreated control cells. Data shown are representative of three independent experiments. (B) qRT-PCR analysis was performed for CRT-MG cells incubated in the absence or presence of hOSM (10 ng/mL) for 0–24 h. ***P < 0.001 vs. each untreated control cells. (C) ADM protein from the culture supernatants was analyzed by ELISA. Data are presented as the fold induction in relative ADM protein levels compared with the untreated cells at the time point 0 h. Data shown are representative of at least three experiments. *P < 0.05, **P < 0.01 vs. untreated control. OSM, oncostatin M.
OSM induces STAT-3 activation and migration in astroglioma cells
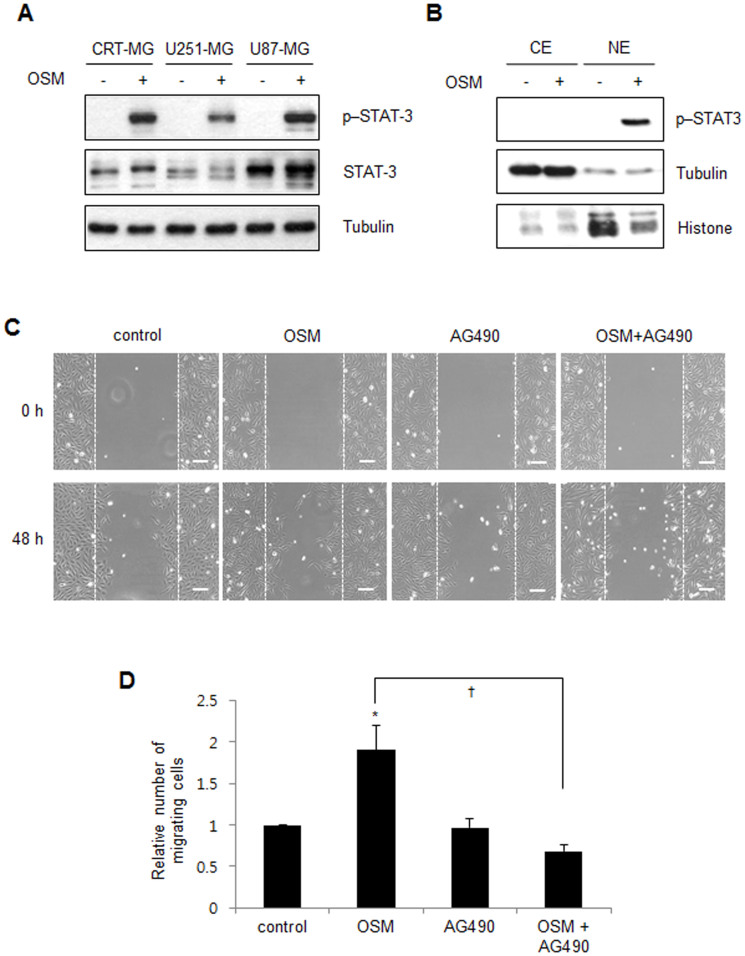
OSM is known to predominantly activate the STAT-3 signaling pathway17,18. To examine whether OSM induces STAT-3 phosphorylation in human astroglioma cells, CRT-MG, U251-MG, and U87-MG cells were incubated in the absence or presence of hOSM (10 ng/mL) for 30 min and then analyzed by immunoblotting. STAT-3 phosphorylation at residue Tyr705 was significantly enhanced by OSM treatment in all cell lines (Fig. 2A). Next, to examine whether OSM induced the translocation of phosphorylated STAT-3 to the nucleus, CRT-MG cells were incubated in the absence or presence of hOSM for 30 min and cytosolic and nuclear extracts were prepared. A strong STAT-3 phosphorylation signal, induced by OSM, was observed mainly in the nuclear fraction, but barely detected in the cytoplasmic fraction (Fig. 2B). Recent investigations revealed a role for OSM in tumor invasion mainly through STAT-319,20. To further examine whether OSM enhances migration of astroglioma cells and OSM-induced STAT-3 activation is involved in this process, scratch wound healing assays were performed. OSM treatment induced enhanced CRT-MG cell migration. However, co-treatment with AG490, one of the JAK/STAT pathway inhibitors, inhibited OSM-induced cell migration (Figs. 2C and 2D). These data indicate that OSM induces STAT-3 activation, which, in turn, mediates astroglioma cell migration.
Figure 2. OSM induces STAT-3 activation and migration in astroglioma cells.
(A) Whole cell lysates from the CRT-MG, U251-MG and U87-MG cells treated with OSM (10 ng/mL) for 30 min were analyzed by immunoblotting against total STAT-3 and p-Y705-STAT-3. Tubulin was used as the loading control. Cropped blots are used. Full-length blots are shown in supplementary Figure 1. (B) Nuclear and cytoplasmic extracts from CRT-MG cells treated with hOSM (10 ng/ml) for 30 min and analyzed by immunoblotting against total STAT-3 and p-Y705-STAT-3. Tubulin and Histone were used as loading and purity controls of each cellular fraction. Data shown are representative of at least three experiments. P, phospho; CE, cytoplasmic extracts; NE, nuclear extracts. Full-length blots are shown in supplementary Figure 1. (C) Representative micrographs of the scratch wound healing assay in CRT-MG cells in the absence or presence of either OSM or AG490 at 48 h, as indicated. Bar = 100 μm. (D) Confluent CRT-MG cells were assayed for migration into the wound at 48 h after scratching, in the absence or presence of OSM and AG490, as indicated. Migrating cells were measured by counting the cell numbers. The results are expressed as the mean ± SD values (compared with untreated controls at 48 h time point); normalized values are shown. The data shown are representative of three independent experiments. (*P < 0.05 vs. untreated control; †P < 0.05 between OSM-treated cells and OSM/AG490-treated cells.).
STAT-3 binds to the ADM promoter in the presence of OSM
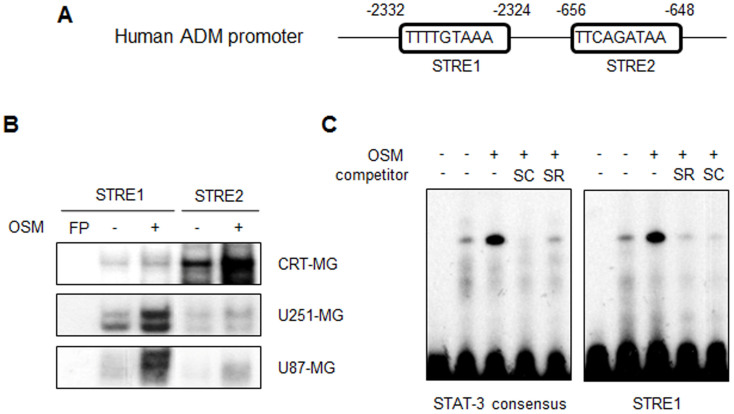
The promoter region of human ADM contains two putative STAT-3 response elements (STRE1 and STRE2; TTN5AA) (Fig. 3A). To test whether activated STAT-3 binds to the ADM promoter, three human astroglioma cell lines were treated with hOSM for 30 min. The nuclear extracts (NEs) from each cell line were then subjected to an electrophoretic mobility shift assay (EMSA) (Fig. 3B). In CRT-MG cells, DNA binding of STAT-3 to STRE2 in human ADM promoter was strongly enhanced by OSM treatment, while STAT-3 binding to STRE1 was slightly increased. In U251-MG and U87-MG cells, STAT-3 binding to STRE1 was strongly enhanced by OSM and a modest increase in STRE2 binding was observed. The same NEs obtained from CRT-MG cells were then exposed to the STAT-3 consensus oligonucleotide (Fig. 3C). STAT-3 binding to the ADM promoter STRE1 and STAT-3 consensus oligonucleotide was enhanced by OSM treatment. Competition with an excess of unlabeled STRE1 or STAT-3 oligonucleotide abrogated the complex formation.
Figure 3. STAT-3 binds to the ADM promoter in the presence of OSM.
(A) The human ADM promoter. The putative STAT-3 response elements are indicated as STRE1 and STRE2. (B) The NEs from CRT-MG, U251-MG and U87-MG astroglioma cells treated with hOSM (10 ng/mL) for 30 min were incubated in the presence of a radiolabeled DNA probe for the human ADM promoter and subjected to an electrophoretic mobility shift assay. (C) NEs from CRT-MG cells were analyzed with a radiolabeled DNA probe of either STAT-3 consensus oligonucleotide or human ADM promoter (STRE1). The competition assay was performed by adding a 100-fold molar excess of cold STRE1 and the STAT-3 probe. Data shown is representative of at the least three experiments. STRE, STAT-3 response elements; FP, free probe; SC, STAT-3 consensus; SR, STRE1. Lane 1: FP; lane 2: CRT-MG nuclear extract; lane 3: OSM-treated CRT-MG nuclear extract; lanes 4–5: OSM-treated CRT-MG nuclear extract with competing unlabeled probe as indicated.
STAT-3 knockdown reduces OSM-induced ADM expression
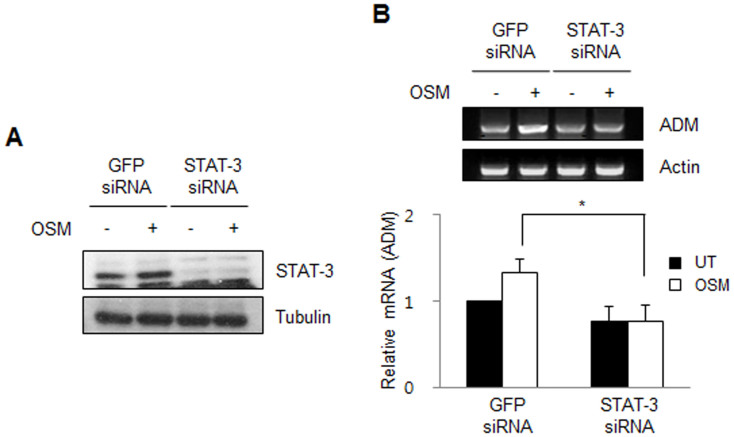
To determine whether the enhanced ADM expression level in the presence of OSM is mediated by STAT-3, siRNA-mediated downregulation of STAT-3 was performed. CRT-MG cells were transiently transfected with a STAT-3 siRNA or a green fluorescence protein (GFP) siRNA (negative control) and incubated with or without hOSM. As shown in Figure 4B, ADM mRNA expression level in the STAT-3 siRNA-transfected cells was decreased compared to that in the GFP siRNA-transfected cells. The reduction in the expression of STAT-3 protein in the STAT-3 siRNA-transfected cells was confirmed by immunoblotting (Fig. 4A). These results demonstrate that STAT-3 knockdown reduces OSM-mediated transcription of the ADM gene, indicating that ADM expression is regulated by STAT-3 in astroglioma cells.
Figure 4. STAT-3 knockdown reduces OSM-induced ADM expression in astroglioma cell lines.
(A) CRT-MG cells were transiently transfected with STAT-3 siRNA or GFP siRNA, as negative control. Two days after transfection, cells were incubated in the absence or presence of hOSM (10 ng/mL) for 24 h. Effective siRNA-mediated suppression of STAT-3 protein expression was verified for each assay by immunoblotting. Cropped blots are used. Full-length blots are shown in supplementary Figure 1. (B) ADM mRNA expression was analyzed by RT-PCR and normalized by actin. The relative expression of ADM was calculated as the normalized amount divided by the normalized amount of CRT-MG cells which were transiently transfected with GFP siRNA in the absence of OSM, and the expression level of parental cells was arbitrarily set at 1. Data shown are representative of three independent experiments. *P < 0.05 between STAT-3 siRNA-transfected cells and GFP siRNA-transfected cells in the presence of OSM.
ADM enhances astroglioma cell invasion
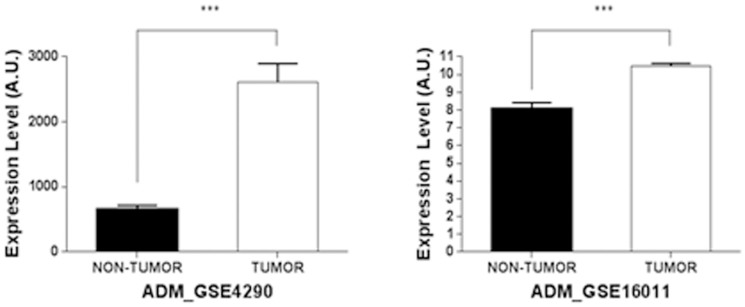
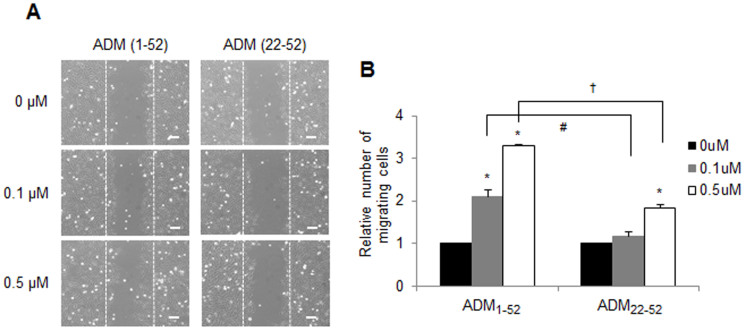
A previous study demonstrated that the extent of ADM mRNA expression is related to the brain tumor type and grade16. In two independent data sets, ADM mRNA was found to be consistently overexpressed in brain tumor samples when compared to non-cancerous samples (Fig. 5, brain samples from patients with epilepsy in GSE4290 and normal adult brain samples in GSE16011). To explore the effect of enhanced ADM on astroglioma cells, CRT-MG cells were incubated in the absence or presence of full-length ADM peptide (ADM1-52) or the truncated ADM peptide (ADM22-52) for 48 h in a medium containing 1% FBS, and wound-healing assay was performed. As shown in Figure 6, the number of cells migrating to the wound region significantly increased in the ADM1-52-treated cells, in a dose-dependent manner, compared to the untreated cells. Compared to the ADM1-52-treated cells, ADM22-52 treatment of CRT-MG cells displayed a modest enhancement of cell migration.
Figure 5. ADM mRNA expression is increased in human astroglioma.
Relative ADM mRNA expression levels were compared in non-tumorous and tumorous brain samples from two independent microarray data-sets in the GEO database (GSE4290 and GSE16011). Patient astrocytoma samples represent the brain tumor group. Epilepsy (GSE4290) or normal adult brain (GSE16011) samples were used as the non-brain tumor group. ***P < 0.001 between non-tumor and tumor group.
Figure 6. ADM enhances the invasion activity of human astroglioma cells.
(A) Representative micrographs of the scratch wound healing assay in CRT-MG cells in the absence or presence of either ADM1-52 or ADM22-52 at 48 h. Bar = 100 μm. (B) Confluent CRT-MG cells were assayed for migration into the wound at 48 h after scratching, in the absence or presence of either ADM1-52 or ADM22-52, as indicated. Migrating cells were measured by counting the cell numbers that advanced into the cell-free space. The results are expressed as the mean ± SD values (compared with untreated controls at 48 h time point); normalized values are shown. The data shown are representative of three independent experiments. The error bars represent SD. (*P < 0.05 vs. each untreated control; #P < 0.05 and †P < 0.05 between ADM1-52-treated cells and ADM22-52-treated cells.).
Discussion
In this study, we demonstrated, for the first time, that OSM enhances ADM expression levels in astroglioma cells. OSM induced STAT-3 phosphorylation and nuclear translocation, leading to enhanced DNA binding of STAT-3 to the ADM promoter, increased ADM mRNA expression, and increased secretion of ADM. Moreover, we showed that increased ADM expression contributes to astroglioma cell migration.
Several reports have demonstrated that STAT-3 or ADM is upregulated in a variety of cancers and contribute to tumor progression14,16,21,22. Hsieh et al. reported that ADM level correlates with elevation of STAT-3 phosphorylation in breast cancer, suggesting that ADM is one of the potential downstream genes regulated by STAT-323. However, the molecular mechanisms involved in ADM upregulation and the direct relationship between STAT-3 and ADM are yet to be fully understood. Our results provide initial evidence that ADM is induced by OSM, a multifunctional cytokine, through a STAT-3-dependent mechanism.
STAT-3 is a cytoplasmic transcription factor that is activated in response to a variety of cytokines, chemokines, and growth factors. The IL-6 family of cytokines, including OSM, preferentially activates STAT-3, leading to dimerization, nuclear translocation, and binding to interferon-γ-activated site-like DNA elements24. Since STAT-3 is a convergence point for several signaling pathways that promote growth, proliferation, and metastasis of cancer cells and since STAT-3 aberrant activation is found in several human cancers, including gliomas, STAT-3 is thought to be a promising target for cancer therapy21. STAT-3 is known to induce the expression of several genes such as survivin, vascular endothelial growth factor, c-Myc, and cyclin D1 that regulate apoptosis and proliferation in cancer12. In addition to these genes, our present results provide additional confirmation that ADM is one of the downstream targets of STAT-3, which contributes to tumor invasion. Previous reports showed that OSM promotes tumor invasion in cervical cancer and osteosarcoma, mainly through STAT-319,20. Consistent with these findings, our study demonstrated that OSM enhanced migration of CRT-MG cells, which was abrogated by co-treatment with AG490 (Fig. 2D).
It has been reported that increased ADM expression levels are found in glioblastoma, and that ADM functions as a potent inducer of glioblastoma cell growth16. In addition, ADM is known to affect cell proliferation and angiogenesis in cancer15,22,25. ADM22-52 is a truncated peptide, derived from the full-length ADM. In contrast to the full-length ADM (ADM1-52) peptide, the truncated ADM (ADM22-52) peptide is reported to act as an agonist or an antagonist, depending on the cell type26. In our system, ADM seemed to be an inducer of tumor invasion and migration. Although there was a difference in the extent of migration, both ADM1-52 and ADM22-52 enhanced astroglioma cell migration (Fig. 6B).
Cancer cells synthesize and release many secretory molecules such as growth factors and inflammatory chemokines27. These factors promote and aggravate tumor progression, neoangiogenesis, and metastasis by remodeling both tumor cells and their microenvironment. Consistent with these findings, our results demonstrate that astroglioma cells express and secrete ADM in response to OSM, indicating that astroglioma cells provide a substantial source of ADM. Inflammatory cytokines, such as TNF-α, IL-1β, and oxidative stress, are known to stimulate ADM synthesis and secretion28,29, although ADM constitutive secretion was reported in keratinocytes30. With regard to oxidative stress, hypoxia is also known to be a potent inducer of ADM in various cancers31,32. Once a tumor develops, numerous inflammatory cells infiltrate into the tumor region and the tumor cells are exposed to these inflammatory cells and mediators as well as to hypoxic conditions. Thus, it would be interesting to compare ADM expression patterns in response to inflammatory conditions and/or hypoxic conditions and to identify the major transcription factors that regulate ADM expression in certain cellular contexts.
Methods
Cells
The CRT-MG, U87-MG and U251-MG human astroglioma cell lines were maintained at 37°C and 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with L-glutamine, 100 U/ml penicillin, 10 μg/ml streptomycin, and 10% FBS.
Reagents and antibodies
Recombinant human OSM (hOSM) was purchased from Stem cell technologies Inc (Grapevine, TX, USA). Antibody to p-Y705-STAT-3 was purchased from Cell Signaling Technology (Beverly, MA, USA). Antibody to STAT-3 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and anti-tubulin antibody was purchased from Sigma-Aldrich, Co. (St. Louis, MO, USA). Human ADM peptide 1-52 and ADM fragment 22-52 were purchased from Sigma-Aldrich, Co. (St. Louis, MO, USA).
RNA isolation, reverse transcription polymerase chain reaction (RT-PCR)
Total cellular RNA was isolated from confluent monolayers of cells using RNA extract kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. cDNA was synthesized from 5 μg of total RNA with reverse transcription and PCR was performed using 3 μl cDNA with a thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The primer sequences were as follows: ADM, forward 5′-AAGAAGTGGAATAAGTGGGCT-3′, reverse 5′-TGTGAACTGGTAGATCTGGT-3′.
Quantitative real-time PCR (qRT-PCR)
cDNA was prepared as describe above and qRT-PCR was performed on an ABI StepOnePlus™ Real-Time PCR machine (Applied Biosystems, Foster City, CA) using a Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Generation of a single gene-specific PCR product was confirmed by melting curve analysis. Relative expression was calculated using the comparative Ct method as described previously33. All PCR reaction samples were prepared in triplicate for each gene. The relative mRNA expression levels were determined by. The primer sequences were as follows: ADM, forward 5′-TGCCCAGACCCTTATTCGG-3′, reverse 5′-AGTTGTTCATGCTCTGGCGG-3′.
Enzyme-linked immunosorbent assay (ELISA)
Cells (5 × 106) were plated and incubated with or without hOSM (10 ng/ml) for 0–24 h. At the end of the incubation, medium was collected and concentrated using centricon YM-3 filter devices (Millipore Co., IL, USA). ADM protein levels were measured by competitive ELISA method using human adrenomedullin EIA kit (Phoenix Pharmaceuticals, CA, USA). Prepared sample, peptide standard, rehydrated primary antibody and rehydrated biotinylated peptide incubated in 96-well plates for 2 h at room temperature. After four washes, plates were incubated with SA-HRP for 1 h at room temperature. After washing, plates were incubated with TMB substrate solution for 1 h at room temperature. Absorbance was measured spectrophotometrically at 450 nm using a microplate reader. The concentration of ADM in each sample was determined by reference to a standard curve generated using known amounts of ADM.
Nuclear and cytoplasmic fractionation
Cells (2 × 106) were plated and incubated with medium or hOSM (10 ng/ml) for 30 min and then collected. Nuclear and cytoplasmic fractionations were purified using the NE-PER kit (Rockford, IL, USA). Cell pellet was resuspended in 200 μl CER I for 30 min on ice and then 11 μl of ice-cold CER II was added to and incubated for 1 min on ice. After centrifugation (12,000 × g) at 4°C for 5 min, the supernant (cytoplasmic extract) was transferred to clean pre-chilled tube. The pellet was resuspended in 100 μl of ice-cold NER for 40 min on ice with continued vortexing for 15 sec every 10 min. The tube was centrifuged (12,000 × g) at 4°C for 20 min and the supernant (nuclear extract) was transferring to clean pre-chilled tube. 15 μg of nuclear extracts and 40 μg of cytoplasmic extracts were analyzed by immunoblotting.
Immunoblotting
Cells were plated and incubated with or without hOSM (10 ng/ml) for 30 min, followed by lysis in cold radioimmunoprecipitation assay (RIPA) buffer with protease inhibitors (1% Nonidet-P40, 0.5% sodium deoxycholate, 10 mM disodium hydrogen phosphate, 150 mM sodium chloride, 1 mM EDTA, 0.1% SDS, 1 mM sodium orthovanadate, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 1 mM PMSF) for 30 min on ice. Lysates were centrifuged (12,000 × g) at 4°C for 30 min and 20 μg of total protein was subjected to 10% SDS/PAGE, and then blots were probed with STAT-3, p-Y705-STAT-3 and and tubulin antibodies. After incubation with secondary antibody, blots were developed using ECL chemiluminescence system (Amersham, Buckinghamshire, UK).
Nuclear extracts and electrophoretic mobility shift assays (EMSA)
EMSA was performed with 8–10 μg of nuclear extracts, as described34. Cells were plated and incubated with or without hOSM (10 ng/ml) for 30 min. Nuclear extracts were incubated with either the ADM sequence including putative STAT-3 Response Element (hSTRE1, forward 5′- TCACGAGCTTTTGTAAAGGGCAGCG-3′; hSTRE2, forward 5′-TCTGAAATTTCAGATAATTCCCCCC-3′) or consensus STAT-3 binding sequence (SC-2573, Santa Cruz Biotechnology) which was end-labeled with [32P]ATP for 30 min. For competition experiments, a 100-molar excess of unlabeled oligonucleotide was added to the nuclear extracts for 30 min before addition of the labeled probe. Bound and free DNA were then resolved by electrophoresis through a 5% polyacrylamide gel and exposed for autoradiography.
siRNA transfection
Cells (4.5 × 105) were transiently transfected using Lipofectamine™ RNAiMAX (Invitrogen, Carlsbad, CA, USA) with either 100 nmol of SMART pools of siRNA against STAT-3 (Lafayette, CO, USA) or negative control siRNA against GFP, according to the manufacturer's instructions. Cells were allowed to recover for 48 h before treatment with or without 10 ng/ml of hOSM for 24 h, and then analyzed by RT-PCR.
ADM mRNA expression profiles in public database
Two microarray data-sets of brain tumor were downloaded from Gene Expression Omnibus (GEO). In GSE429035 samples annotated as astrocytoma or glioblastoma patients (N = 103; astrocytoma grade II: 7, astrocytoma grade III: 19, and astrocytoma grade IV: 77) were used as brain tumor samples. Samples of epilepsy patients (N = 23) were used as non-tumor samples. In GSE1601136 samples annotated as glioma (N = 188; astrocytoma grade II: 13, astrocytoma grade III: 16, and astrocytoma grade IV: 159) were used as brain tumor samples and samples of normal adult brain (N = 8) were used as non-tumor samples. For GSE4290, probes with ABS_CALL value P were used. All probes in GSE16011 were used because there is no detection p-value in this data-set. For each data-set, probe intensities were quantile-quantile normalized within data-set. Probes were converted to gene ID in HPRD37 by averaging probe intensities for the same gene ID.
Wound-healing assay
Adherent cells were scraped off the bottom of a culture plate using a pipette tip to create a cell-free area. The cell culture was washed with PBS to remove cell debris and then incubated with OSM, full length of ADM peptide (ADM1-52), or truncated ADM peptide (ADM22-52) for 48 h in 1% FBS DMEM. The wound area was photographed after scratching for control. The number of cells migrating into the initial wound area was counted at 48 h after scratching.
Statistical analyses
Statistical analyses were performed using the Student's t-test to compare between sample groups, and one-way ANOVA with Tukey's post hoc test was used to determine differences among multiple groups (SPSS 12.0 K for Windows, SPSS Inc., Chicago, IL). Statistical significance of the data was set at P < 0.05.
Author Contributions
S.Y.L. and Y.H.C. designed research; S.Y.L., S.H.A., H.P., J.L., K.C. and Y.H.C. conducted. Research; S.Y.L., C.C., J.H.C., E.M.P. and Y.H.C. analysed data; S.Y.L. and Y.H.C. wrote the paper; S.Y.L. and Y.H.C. had primary responsibility for final content. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) Grant 2012R1A5A2A32671866 and by NRF-2013R1A1A3009978.
References
- Brandes A. A., Franceschi E., Tosoni A., Hegi M. E. & Stupp R. Epidermal growth factor receptor inhibitors in neuro-oncology: hopes and disappointments. Clin cancer Res 14, 957–960 (2008). [DOI] [PubMed] [Google Scholar]
- Maher E. A. et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev 15, 1311–1333 (2001). [DOI] [PubMed] [Google Scholar]
- Merzak A. & Pilkington G. J. Molecular and cellular pathology of intrinsic brain tumours. Cancer Metastasis Rev 16, 155–177 (1997). [DOI] [PubMed] [Google Scholar]
- Li W. & Graeber M. B. The molecular profile of microglia under the influence of glioma. Neuro Oncol 14, 958–978 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y. et al. Essential function of oncostatin m in nociceptive neurons of dorsal root ganglia. J Neurosci 24, 1941–1947 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repovic P. & Benveniste E. N. Prostaglandin E2 is a novel inducer of oncostatin-M expression in macrophages and microglia. J Neurosci 22, 5334–5343 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repovic P., Fears C. Y., Gladson C. L. & Benveniste E. N. Oncostatin-M induction of vascular endothelial growth factor expression in astroglioma cells. Oncogene 22, 8117–8124 (2003). [DOI] [PubMed] [Google Scholar]
- Mizoguchi M. et al. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol 65, 1181–1188 (2006). [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang W., Huang H. J., Liao W. S. & Fuller G. N. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab Invest 84, 941–951 (2004). [DOI] [PubMed] [Google Scholar]
- Brantley E. C. & Benveniste E. N. Signal transducer and activator of transcription-3: a molecular hub for signaling pathways in gliomas. Mol Cancer Res 6, 675–684 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich P. C. et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 374, 1–20 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J. F. et al. Stat3 as an oncogene. Cell 98, 295–303 (1999). [DOI] [PubMed] [Google Scholar]
- Buettner R., Mora L. B. & Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res 8, 945–954 (2002). [PubMed] [Google Scholar]
- Miller M. J. et al. Adrenomedullin expression in human tumor cell lines. Its potential role as an autocrine growth factor. J Biol Chem 271, 23345–23351 (1996). [DOI] [PubMed] [Google Scholar]
- Martinez A. et al. The effects of adrenomedullin overexpression in breast tumor cells. J Natl Cancer Inst 94, 1226–1237 (2002). [DOI] [PubMed] [Google Scholar]
- Ouafik L. et al. Neutralization of adrenomedullin inhibits the growth of human glioblastoma cell lines in vitro and suppresses tumor xenograft growth in vivo. Am J Pathol 160, 1279–1292 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey G. et al. Signaling network of Oncostatin M pathway. J Cell Commun Signal 7, 103–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossey S. L., Bear M. D., Kisseberth W. C., Pennell M. & London C. A. Oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines. BMC cancer 11, 125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarel M. M. & Coleman N. Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J Pathol 232, 386–390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolin C. et al. Oncostatin M promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes Cancer 3, 117–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Pardoll D. & Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 9, 798–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V. et al. Adrenomedullin is expressed in pancreatic cancer and stimulates cell proliferation and invasion in an autocrine manner via the adrenomedullin receptor, ADMR. Cancer Res 67, 2666–2675 (2007). [DOI] [PubMed] [Google Scholar]
- Hsieh F. C., Cheng G. & Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun 335, 292–299 (2005). [DOI] [PubMed] [Google Scholar]
- Shuai K. & Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 3, 900–911 (2003). [DOI] [PubMed] [Google Scholar]
- Fernandez-Sauze S. et al. Effects of adrenomedullin on endothelial cells in the multistep process of angiogenesis: involvement of CRLR/RAMP2 and CRLR/RAMP3 receptors. Int J Cancer 108, 797–804 (2004). [DOI] [PubMed] [Google Scholar]
- Zudaire E. et al. Adrenomedullin is a cross-talk molecule that regulates tumor and mast cell function during human carcinogenesis. Am J Pathol 168, 280–291 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis G. S., Pavlou M. P. & Diamandis E. P. Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol Oncol 4, 496–510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo S. et al. Interleukin-1, tumor necrosis factor and lipopolysaccharide additively stimulate production of adrenomedullin in vascular smooth muscle cells. Biochem Biophys Res Commun 207, 25–32 (1995). [DOI] [PubMed] [Google Scholar]
- Ando K., Ito Y., Kumada M. & Fujita T. Oxidative stress increases adrenomedullin mRNA levels in cultured rat vascular smooth muscle cells. Hypertens Res 21, 187–191 (1998). [DOI] [PubMed] [Google Scholar]
- Kapas S., Tenchini M. L. & Farthing P. M. Regulation of adrenomedullin secretion in cultured human skin and oral keratinocytes. J invest dermatol 117, 353–359 (2001). [DOI] [PubMed] [Google Scholar]
- Fujita Y. et al. Involvement of adrenomedullin induced by hypoxia in angiogenesis in human renal cell carcinoma. Int J Urol 9, 285–295 (2002). [DOI] [PubMed] [Google Scholar]
- Drimal J., Drimal J. Jr & Drimal D. Hypoxic stress-enhanced expression and release of adrenomedullin (AM) and up-regulated AM receptors, while glucose starvation reduced AM expression and release and down-regulated AM receptors in monkey renal cells. Physiol Res 55, 535–542 (2006). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Choi Y. H., Bernardi R., Pandolfi P. P. & Benveniste E. N. The promyelocytic leukemia protein functions as a negative regulator of IFN-gamma signaling. Proc Natl Acad Sci U S A 103, 18715–18720 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9, 287–300 (2006). [DOI] [PubMed] [Google Scholar]
- Gravendeel L. A. et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res 69, 9065–9072 (2009). [DOI] [PubMed] [Google Scholar]
- Keshava Prasad T. S. et al. Human Protein Reference Database--2009 update. Nucleic Acids Res 37, D767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information