Summary
RNA and DNA are simple linear polymers consisting of only four major types of subunits, and yet these molecules carry out a remarkable diversity of functions in cells and in the laboratory. Each newly-discovered function of natural or engineered nucleic acids enforces the view that prior assessments of nucleic acid function were far too narrow and that many more exciting findings are yet to come. This Perspective highlights just a few of the numerous discoveries over the past 20 years pertaining to nucleic acid function, focusing on those that have been of particular interest to chemical biologists. History suggests that there will continue to be many opportunities to engage chemical biologists in the discovery, creation, and manipulation of nucleic acid function in the years to come.
DNA makes RNA makes protein. This is the shorthand version of Francis Crick's “central dogma” of biology, which more specifically states: “the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible” (Crick 1958). Crick was referring to information that defines the precise sequence of residues within a nucleic acid or protein. He confessed at the outset of that 1958 paper: “(James) Watson said to me, a few years ago, ‘The most significant thing about nucleic acids is that we don't know what they do’”. Yet in that same paper Crick proposed that RNA does much more than serve as a passive carrier of information. He hypothesized that it functions as an “adaptor” molecule, carrying amino acids to the RNA template that directs the sequential assembly of amino acids to form proteins. He suggested that there would be (at least) one adaptor for each of the 20 amino acids, although he felt the task of joining the adaptor to its amino acid would be too challenging for RNA.
Crick's adaptor is of course tRNA, which he later said “looks like Nature's attempt to make RNA do the job of protein” (Crick 1966). Crick went further to say: “It is tempting to wonder if the primitive ribosome could have been made entirely of RNA”, and he suggested: “Possibly the first ‘enzyme’ was an RNA molecule with replicase properties” (Crick 1968). Similar comments regarding the catalytic potential of RNA also were made at that time by Woese (1967) and Orgel (1968). When Chemistry & Biology published its introductory issue in 1994, the modern ribosome was looking very much like an RNA enzyme (Noller et al., 1992), although that was still to be proven definitively based on examination of its X-ray crystal structure (Nissen et al., 2000). RNA enzymes had been discovered in nature (Kruger et al., 1982; Guerrier-Takada et al., 1983) and invented in the laboratory through test-tube evolution (Bartel and Szostak, 1993), but even a rudimentary form of a replicase was many years away (Lincoln and Joyce, 2009).
Certainly one of the most dramatic developments in chemical biology over the past 20 years has been the growing appreciation of the many complex functional roles that RNA plays in biology and can be made to play in chemical systems. Even DNA can get into the act of ligand binding and enzymatic function. The central dogma still holds, but nucleic acids are much more than carriers of information. They are both egg and chicken, and we still don't know all that they can do.
Revealing the “Dark Matter” of Biological RNAs
For decades, biologists seemed content to know that there are messenger, transfer, ribosomal, and a limited number of other RNAs in biology. Then reports of the first ribozymes (Kruger et al., 1982; Guerrier-Takada et al., 1983) hinted that the community was aiming far too low when estimating the range of functional RNAs in extant organisms. Today, more than 30 years after the discovery of catalytic RNA, it is difficult to overestimate the role of RNA in biology. Complete genome sequencing has provided a more comprehensive view of the portion of genomes that gives rise to mRNAs, even if some confusion remains about just what constitutes a translation-worthy segment of RNA. With many annotated genomes in hand, one can look to the nucleotide sequences outside the protein-coding regions for possible transcribed RNAs and puzzle over what these noncoding RNAs (ncRNAs) actually do. Similarly, RNA transcriptomics studies have yielded large collections of transcribed RNAs that apparently do not code for proteins. Is this mountain of RNAs merely junk, or are there some valuable molecules in the heap?
The strong evolutionary pressure to minimize waste in bacterial and archaeal genomes can be exploited by researchers to discover biologically relevant ncRNAs. There is little wasted space, so a gap in protein coding strongly suggests the existence of an important ncRNA. The functions of these RNAs are diverse, going far beyond tRNAs, rRNAs, and the known ribozymes. Among the most common of the ncRNAs are short RNA transcripts (sRNAs) that form complementary pairs with the untranslated regions of certain mRNAs and affect gene expression (Waters and Storz, 2009). CRISPR RNAs are a fascinating example of sRNAs that are enzymatically processed and function as viral- or plasmid-targeting systems to direct protein nucleases to cleave foreign DNA (Marraffini and Sontheimer, 2010).
Some bacterial ncRNAs function not by forming Watson-Crick base pairs to their targets, but by forming complex three-dimensional shapes that are responsible for their behavior. For example, RNA structure can be dramatically affected by temperature, and there now are numerous examples of untranslated regions of mRNAs that function as RNA thermometers to control gene expression in response to temperature change (Kortmann and Narberhaus, 2012). RNA also can form shapes that recognize various proteins, either to modulate protein function or to serve as landing sites for gene control factors. An example of the former is 6S RNA, which is a mimic of an open DNA promoter complex and inhibits RNA transcription by decoying RNA polymerase to bind the structured RNA rather than engage the promoter (Cavanagh and Wassarman, 2014). Examples of the latter occur in the 5′-untranslated region of mRNAs for ribosomal proteins, binding the protein product of the adjacent coding region as a form of feedback control (Deiorio-Haggar et al., 2013). Other complex folded RNAs from bacteria, termed “riboswitches”, will be discussed in the next section.
In eukaryotes an even greater fraction of the genome appears to give rise to ncRNAs. Some of these, such as miRNAs (Ameres and Zamore, 2013) and piRNAs (Luteijn and Ketting, 2013), are very small and function as targeting systems in a manner analogous to CRISPR. Long intergenic noncoding RNAs or lincRNAs (Ulitsky and Bartel, 2013) are a large and more mysterious collection of transcripts. Currently very little is known about these RNAs, causing some researchers to believe that many are simply the result of transcriptional noise, although others clearly have important biological functions in chromatin remodeling and DNA modification. The most extensively studied RNA of this type is Xist (Chalingné and Heard, 2014), which is essential for silencing most genes on one of the two X chromosomes of human females. The opportunities for exploration of eukaryotic lincRNA structure and function are substantial and will require considerable ingenuity in devising new approaches in RNA chemical biology.
From Aptamer to Aptazyme to Riboswitch
Someone new to the field of RNA research might question why speculation in the 1960s regarding RNA function was considered so bold at the time. Now with many recognized examples of complex RNA structures and functions in biology, the field readily embraces each newly discovered ncRNA and ponders its potential relevance to ancient RNAs. However, even in the early 1990s when much evidence was in hand that RNA can adopt complex structures and carry out sophisticated catalytic functions, considerable doubt remained whether RNA could accomplish much on its own, without the assistance of proteins.
These doubts about the functional capacity of RNA were greatly reduced when chemical biologists began to apply in vitro evolution methods to expand the boundaries of known nucleic-acid-based function (Wilson and Szostak, 1999; Joyce, 2004). Chemistry & Biology was launched in the midst of this technological revolution and became a major publisher of the discoveries that emerged. In vitro evolution employs populations of trillions of different RNA or DNA molecules that are challenged to perform a chemical task, for example, cleave DNA (Robertson and Joyce, 1990) or selectively bind a small molecule or protein target (Ellington and Szostak, 1990; Tuerk and Gold, 1990).
In vitro evolution strategies also were used to develop numerous RNAs and DNAs that form selective, high-affinity binding pockets (aptamers) for compounds ranging from drugs to fundamental metabolites (Osborne and Ellington, 1997). Many influential advances in aptamer science have been described in Chemistry & Biology, helping to reveal more about the scope of RNA function. The isolation of RNAs that bind aminoglycoside antibiotics demonstrated that RNA can adopt diverse architectures to recognize members of this important drug class (Lato et al., 1995). The first detailed analyses of aptamer structure at atomic resolution began to reveal how nucleic acids can form binding pockets for highly diverse small molecule ligands (Feigon et al., 1996; Jiang et al., 1997; Lin and Patel, 1997). Through this work, chemical biologists spearheaded the study of RNA-ligand interactions, well in advance of the discoveries of most natural aptamers. Even mirror-image aptamers (termed “Spiegelmers”) were created using in vitro evolution, first by conducting a conventional selection against the enantiomer of the target, then preparing the resulting aptamers as L-RNA or L-DNA molecules that bind the desired target (Leva et al., 2002).
An interesting technological advance, both for nature and for chemical biologists, comes from the fusion of aptamer and ribozyme functionality. Although creatures of the RNA World may have invented RNA switches billions of years ago (Breaker 2011), chemical biologists were the first scientists to demonstrate the potential for ligand-triggered switches constructed of either RNA or DNA. For example, by judiciously fusing an ATP aptamer to a structurally sensitive portion of the hammerhead ribozyme, RNA self-cleavage was made to be strongly dependent on the presence of ATP (Tang and Breaker, 1997). A similar ATP-responsive system was constructed using DNA (Levy and Ellington, 2002).
These aptamer-ribozyme constructs were investigated mostly for their intrinsic interest and for potential application as either chemical sensors (Srinivasan et al., 2004) or engineered gene control elements (Soukup and Breaker, 1999). Therefore it came as somewhat of a surprise when, only five years after the first engineered allosteric ribozyme was described, the first validated examples of riboswitches in modern cells were published (Nahvi et al., 2002). To date, more than 30 different classes of riboswitches are known to exist in nature (Breaker 2011), and certainly many more are waiting to be discovered.
Because the aptamer domain of riboswitches can be occupied by ligand analogs, these regulatory elements are intriguing targets for potential therapeutic agents (Sudarsan et al., 2005; Blount and Breaker, 2006; Deigan and Ferré-D'Amaré, 2011). Researchers are taking various approaches to develop compounds that affect riboswitch function, including designing analogs of existing ligands (Mulhbacher et al., 2010) and using high-throughput or fragment-based screening approaches (Mayer and Famulok, 2006; Warner et al., 2014).
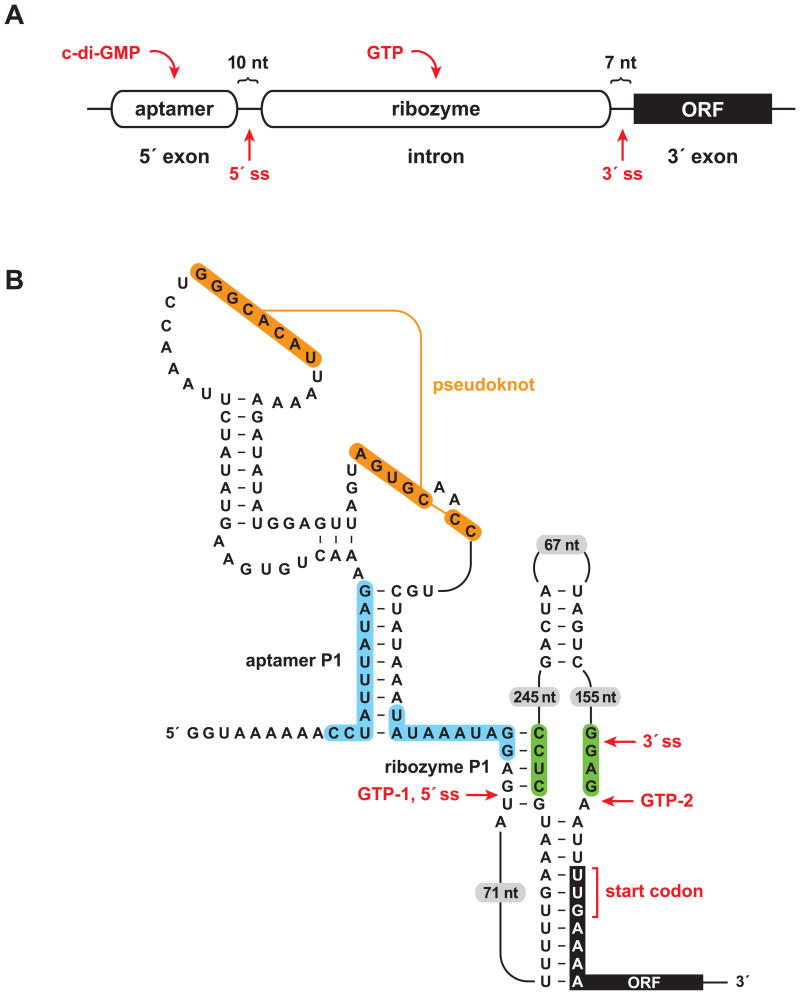
Most natural riboswitches do not control ribozyme activity, instead undergoing structural rearrangement upon ligand binding that affects various components of gene expression systems, including RNA polymerases, ribozymes, and transcription factors. However, a few cases of natural allosteric ribozymes have been discovered. One well validated example is found in Clostridium difficile, where a riboswitch contains an aptamer domain for the signaling compound c-di-GMP, which in turn regulates self-splicing by a group I ribozyme (Lee et al., 2010; Chen et al., 2011; Figure 1). This represents one of the most complex all-RNA devices yet discovered, and hints at the possibility that other complex RNA systems might exist among the vast number of RNAs of unknown function that are present in modern cells.
Figure 1.
A sophisticated RNA device. (A) Arrangement of aptamer and ribozyme domains within a naturally-occurring allosteric self-splicing group I ribozyme. The aptamer senses the bacterial second messenger c-di-GMP and the ribozyme requires guanosine or one of its phosphorylated derivatives (e.g., GTP) as a substrate to initiate the first step of splicing. ORF, open reading frame; ss, splice site. (B) Key sequence and secondary structural elements within the allosteric switch. Binding of c-di-GMP to the aptamer domain stabilizes the aptamer P1 stem, which permits formation of the ribozyme P1stem, thereby enabling splicing initiated by GTP attack at the 5′ splice site (GTP-1). This configuration allows translation of the downstream ORF. In the absence of c-di-GMP, alternative pairing (blue shading) precludes formation of the ribozyme P1 stem and allows formation of an alternative ribozyme stem (green shading). This promotes GTP to attack at a position far downstream from the normal 5′ splice site (GTP-2), thus preventing translation of the downstream ORF. Figure adapted from Lee et al., 2010.
DNA Can Be an Enzyme Too
In the fall of 1994, we (R.R.B. and G.F.J.) were attempting to develop an improved version of the self-cleaving hammered ribozyme, using in vitro selection to obtain variants with enhanced catalytic rate. The selection scheme involved tethering randomized forms of the hammerhead to a short substrate domain, which was immobilized on a solid support. The expectation was that those variants best able to bring about cleavage would preferentially detach themselves from the support and be selectively amplified. It wasn't going well because of the substantial background level of non-specific cleavage throughout the molecule. If only the rest of the molecule wasn't so susceptible to cleavage. If only it were constructed of… DNA.
The same oligonucleotides that were being used to construct the population of variant RNAs were repurposed to construct random-sequence DNAs that were linked via a single susceptible ribonucleotide to a solid support. We feared that if we provided more than one ribonucleotide, the catalytic motif would arise from that segment of RNA rather than lowly DNA. Within a period of five days the first DNA enzyme was born. The simple motif consists of two substrate-binding arms that flank a catalytic center of 15 residues, catalyzing the Pb2+-dependent cleavage of an RNA phosphodiester with a rate enhancement of 105-fold compared to the uncatalyzed reaction. At that time, and still today, there are no known evolved DNA enzymes in biology. This was a chemist's creation based on the principles of evolutionary biology and biochemistry. Naturally the paper was published in Chemistry & Biology (Breaker and Joyce, 1994).
Other DNA enzymes soon followed, including a DNA enzyme that catalyzes the joining of imidazole-activated oligodeoxynucleotides (Cuenod and Szostak, 1995), a DNA enzyme that catalyzes the Mg2+-dependent cleavage of an RNA phosphodiester (Breaker and Joyce, 1995), and a general-purpose RNA-cleaving DNA enzyme that can be directed to cleave a wide variety of target RNAs under physiological conditions (Santoro and Joyce, 1997). The latter of these, termed the “10-23” DNA enzyme, has been made to cleave c-jun mRNA in cells (Cai et al., 2012) and recently completed a successful phase I/IIa human clinical trial for the treatment of basal cell carcinoma (Cho et al., 2013).
In the Pantheon of macromolecular catalysis, protein enzymes certainly occupy the highest place. RNA enzymes come next because of their role in biology, most notably the ribosome, but also the many remarkable RNA enzymes that have been obtained by in vitro evolution (see below). DNA has its place as well, now with more than 20 examples of DNA enzymes that catalyze diverse chemical transformations. These include the phosphorylation (Li and Breaker, 1999), ligation (Sreedhara et al., 2004), deglycosylation (Sheppard et al., 2000), and hydrolytic cleavage (Chandra et al., 2009) of DNA substrates, as well as reactions involving non-nucleic-acid substrates, such as porphyrin metallation (Li and Sen, 1996), Diels-Alder cycloaddition (Chandra and Silverman, 2008), and tyrosine phosphorylation (Walsh et al., 2013).
In retrospect, it does not seem surprising that DNA can be an enzyme, given the immense combinatorics of possible DNA sequences and the power of Darwinian evolution to discover and refine those sequences that give rise to structure and function. There continues to be a sense that RNA is a more versatile catalyst than DNA, but RNA and DNA are fraternal twins, with different personalities yet highly similar composition. More surprising is that, 20 years after the discovery of DNA enzymes, no new class of evolved macromolecular catalyst has been reported. One cannot count chemically modified RNA and DNA, although there are several examples of nucleic acid enzymes that contain modified bases (Wiegand et al., 1997; Tarasow et al., 1997; Santoro et al., 2000; Lermer et al., 2002) or carry a substitution at the C2′-position (Beaudry et al., 2000). Recent advances with “xeno nucleic acids” (XNAs), which contain a backbone other than (deoxy)ribose-phosphate, appear promising and have already led to the development of XNA aptamers (Yu et al., 2012; Pinheiro et al., 2012). One can confidently predict that the first XNAzyme soon will be reported.
Pushing the Frontiers of RNA Catalysis
Many seminal discoveries pertaining to RNA catalysis have been described in Chemistry & Biology. It turns out that Crick underestimated the ability of RNA to catalyze the aminoacylation of tRNA. Suga and colleagues used in vitro evolution to obtain a 45-nucleotide RNA that charges the 3′-hydroxyl of tRNA with various activated amino acids (Saito et al., 2001; Murakami et al., 2003). An analog of the peptidyltransferase reaction also has been carried out using an in vitro evolved RNA enzyme (Zhang and Cech, 1997; Zhang and Cech, 1998). Several classic reactions of organic synthesis have been catalyzed by in vitro evolved RNAs, including Diels-Alder cycloaddition (Seelig and Jäschke, 1999), Michael addition (Sengle et al., 2001), and aldol condensation (Fusz et al., 2005), all of which were reported in Chemistry & Biology.
Chemical biologists have various motivations for developing novel RNA enzymes. One goal is to explore the catalytic potential of RNA and to understand how RNA structure gives rise to function. There are only a few known examples of RNA enzymes in biology and, other than the ribosome, all of these catalyze phosphodiester cleavage or ligation reactions. Although more naturally-occurring ribozymes are likely to be discovered, biological catalysis is overwhelmingly dominated by protein enzymes. Protein enzymologists can feast upon these diverse examples to study the relationships among sequence, structure, and function. RNA enzymologists are forced to take matters into their own hands and construct their own examples, drawing on the methods of in vitro evolution. There is added value in exploring the evolutionary process itself. One can literally track the evolutionary maturation of an enzyme, observing its phylogeny at the molecular level, including the range of permissible sequence variation.
Other motivations for developing novel RNA enzymes are more practical, for example, to provide tools for chemical synthesis, modification of biological molecules, construction of biomaterials, detection of target ligands, and therapeutic applications. These goals place additional demands on the performance of the RNA enzymes: that they be small, stable, fast, specific, and amenable to a desired set of reaction conditions; preferably all of the above. Seeking to meet those demands further drives understanding of the limits of RNA catalysis. There are some tasks that seem out of reach for RNA, such as controlling free radical chemistry, derivatizing linear alkanes, and operating in non-aqueous solvents. Yet one should not be surprised if some of these “insurmountable” challenges are achieved by the 40th anniversary of Chemistry & Biology. Greater use of catalytic cofactors likely will be beneficial in this regard. RNA has a remarkable ability to bind and position other compounds, which in turn could do the heavy lifting of catalysis. After all, this is what proteins do to augment the functionality of their RNA creators.
Finally, there is the invention of life itself, which is widely thought to have been brought about by RNA. RNA is adept at catalyzing the RNA-dependent polymerization of RNA, what Crick regarded as the first enzymatic function of life. There are many examples of in vitro evolved RNA enzymes that catalyze the RNA-templated ligation of RNA. One of these, the class I RNA ligase (Bartel and Szostak, 1993), has been through a remarkable journey of evolutionary development over the past two decades. It has been evolved to polymerize NTPs, at first just a few (Ekland and Bartel, 1996), then a full turn of the helix (Johnston et al., 2001), then several turns (Wochner et al., 2011), and now more than the length of the RNA enzyme itself (Attwater et al., 2013). Another RNA ligase has been configured so that it can produce additional copies of itself (Paul and Joyce, 2002) and undergo self-sustained Darwinian evolution (Lincoln and Joyce, 2009). Earlier this year a turbocharged version of this self-replicating enzyme was reported (Robertson and Joyce, 2014), with a doubling time of only five minutes and able to achieve 10100-fold amplification in 37.5 hours.
Can life itself be constructed in the laboratory based on RNA enzymes? If one were able to combine the best properties of the existing RNA polymerase and RNA replicase enzymes, then perhaps so. Then the self-evolving system would have the opportunity to explore the frontiers of RNA catalysis for its own selective advantage. Chemists who set those wheels in motion would become neo-biologists, spectators and interpreters of a new biology. Other chemists would pounce on that new biology as a source of natural products, targets for discovery and manipulation, and new opportunities to probe RNA structure and mechanism. It sounds like it would be a great time to be a chemical biologist, as it has been for the past 20 years.
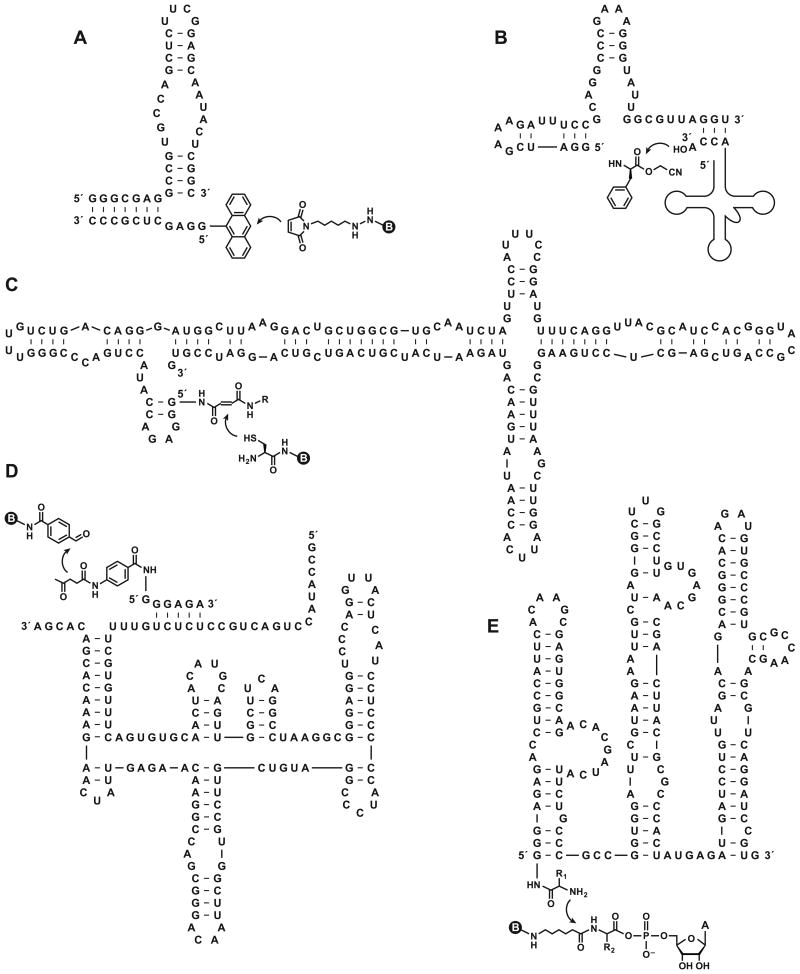
Figure 2.
What do these RNAs have in common? All are in vitro evolved ribozymes that catalyze biologically relevant chemical transformations and all were published in Chemistry & Biology. (A) The “39M38tr” ribozyme, which catalyzes Diels-Alder cycloaddition between biotin maleamide and anthracene that is tethered to the 5′ end of the ribozyme via an alkyl linker (Seelig and Jäschke, 1999). (B) The “Fx3 (Flexizyme)” ribozyme, which catalyzes 3′-aminoacylation of tRNA using the cyanomethyl ester of phenylalanine or other amino acids (Murakami et al., 2003). (C) The “UV5” ribozyme, which catalyzes Michael addition between biotin cysteine and fumaramide that is tethered to the 5′ end of the ribozyme (Sengle et al., 2001). (D) The “11D2” ribozyme, which catalyzes aldol condensation between biotin-linked benzaldehyde-4-carboxamide and levulinic amide that is tethered to the 5′ end of a separate oligonucleotide (Fusz et al., 2005). (E) The “R180” ribozyme, which catalyzes peptide bond formation between an aminoacyl 5′-adenylate and an amino acid that is tethered to the 5′ end of the ribozyme via a disulfide linkage (Zhang and Cech, 1998; Sun et al., 2002). Curved arrow indicates the site of reaction. Circled B indicates a biotin moiety.
Acknowledgments
RNA science in the Breaker laboratory is supported by NIH and by the Howard Hughes Medical Institute. Research in the Joyce laboratory is supported by grants from NASA, NIH, and the Simons Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ronald R. Breaker, Email: ronald.breaker@yale.edu.
Gerald F. Joyce, Email: gjoyce@scripps.edu.
References
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Attwater J, Wochner A, Holliger P. In-ice evolution of RNA polymerase ribozyme activity. Nat Chem. 2013;5:1011–1018. doi: 10.1038/nchem.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Beaudry A, DeFoe J, Zinnen S, Burgin A, Beigelman L. In vitro selection of a novel nuclease-resistant RNA phosphodiesterase. Chem Biol. 2000;7:323–334. doi: 10.1016/s1074-5521(00)00110-1. [DOI] [PubMed] [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Breaker RR, Joyce GF. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem Biol. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- Cai H, Santiago FS, Prado-Lourenco L, Wang B, Patrikakis M, Davenport MP, Maghzal GJ, Stocker R, Parish CR, Chong BH, et al. DNAzyme targeting c-jun suppresses skin cancer growth. Sci Transl Med. 2012;4:139ra82. doi: 10.1126/scitranslmed.3003960. [DOI] [PubMed] [Google Scholar]
- Cavanagh AT, Wassarman KM. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu Rev Microbiol. 2014 Apr 10; doi: 10.1146/annurev-micro-092611-150135. e-pub ahead of print. 2014. [DOI] [PubMed] [Google Scholar]
- Chalingné R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014 Jun 14; doi: 10.1016/j.febslet.2014.06.023. e-pub ahead of print. 2014. [DOI] [PubMed] [Google Scholar]
- Chandra M, Silverman SK. DNA and RNA can be equally efficient catalysts for carbon-carbon bond formation. J Am Chem Soc. 2008;130:2936–2937. doi: 10.1021/ja7111965. [DOI] [PubMed] [Google Scholar]
- Chandra M, Sachdeva A, Silverman SK. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat Chem Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AGY, Sudarsan N, Breaker RR. Mechanism for gene control by a natural allosteric group I ribozyme. RNA. 2011;17:1967–1972. doi: 10.1261/rna.2757311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EA, Moloney FJ, Cai H, Au-Yeung A, China C, Scolyer RA, Yosufi B, Raftery MJ, Deng JZ, Morton SW, et al. Safety and tolerability of an intratumorally injected DNAzyme, Dz13, in patients with nodular basal-cell carcinoma: a phase 1 first-in-human trial (DISCOVER) Lancet. 2013;381:1835–1843. doi: 10.1016/S0140-6736(12)62166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- Crick FHC. The genetic code — yesterday, today, and tomorrow. Cold Spring Harb Symp Quant Biol. 1966;31:3–9. [PubMed] [Google Scholar]
- Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Cuenoud B, Szostak JW. A DNA metalloenzyme with DNA ligase activity. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D'Amaré AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiorio-Haggar K, Anthony J, Meyer MM. RNA structures regulating ribosomal protein biosynthesis in bacilli. RNA Biol. 2013;10:1180–1184. doi: 10.4161/rna.24151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Bartel DP. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Feigon J, Diekmann T, Smith FW. Aptamer structures from A to ξ. Chem Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- Fusz S, Eisenführ A, Srivatsan SG, Heckel A, Famulok M. A ribozyme for the aldol reaction. Chem Biol. 2005;12:941–950. doi: 10.1016/j.chembiol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Jiang L, Suri AK, Fiala R, Patel DJ. Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex. Chem Biol. 1997;4:35–50. doi: 10.1016/s1074-5521(97)90235-0. [DOI] [PubMed] [Google Scholar]
- Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Joyce GF. Directed evolution of nucleic acid enzymes. Annu Rev Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lato SM, Boles AR, Ellington AD. In vitro selection of RNA lectins: using combinatorial chemistry to interpret ribozyme evolution. Chem Biol. 1995;2:291–303. doi: 10.1016/1074-5521(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermer L, Roupioz Y, Ting R, Perrin DM. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J Am Chem Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- Leva S, Lichte A, Burmeister J, Muhn P, Jahnke B, Fesser D, Erfurth J, Burgstaller P, Klussmann S. GnRH binding RNA and DNA Spiegelmers: a novel approach toward GnRH antagonism. Chem Biol. 2002;9:351–359. doi: 10.1016/s1074-5521(02)00111-4. [DOI] [PubMed] [Google Scholar]
- Levy M, Ellington AD. ATP-dependent allosteric DNA enzymes. Chem Biol. 2002;9:417–426. doi: 10.1016/s1074-5521(02)00123-0. [DOI] [PubMed] [Google Scholar]
- Li Y, Breaker RR. Phosphorylating DNA with DNA. Proc Natl Acad Sci USA. 1999;96:2746–2751. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sen D. A catalytic DNA for porphyrin metallation. Nat Struct Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- Lin CH, Patel DJ. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to ATP. Chem Biol. 1997;4:817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- Mayer G, Famulok M. High-throughput-compatible assay for glmS riboswitch metabolite dependence. Chembiochem. 2006;7:602–604. doi: 10.1002/cbic.200500490. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLOS Pathog. 2010;6:e10000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Saito H, Suga H. A versatile tRNA aminoacylation catalyst based on RNA. Chem Biol. 2003;10:655–662. doi: 10.1016/s1074-5521(03)00145-5. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Paul N, Joyce GF. A self-replicating ligase ribozyme. Proc Natl Acad Sci USA. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro V, Taylor A, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew S, McLaughlin S, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF. Highly efficient self-replicating RNA enzymes. Chem Biol. 2014;21:238–245. doi: 10.1016/j.chembiol.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kourouklis D, Suga H. An in vitro evolved precursor tRNA with aminoacylation activity. EMBO J. 2001;20:1797–1806. doi: 10.1093/emboj/20.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF. RNA cleavage by a DNA enzyme with extended chemical functionality. J Am Chem Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- Seelig B, Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem Biol. 1999;6:167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- Sengle G, Eisenführ A, Arora PS, Nowick JS, Famulok M. Novel RNA catalysts for the Michael reaction. Chem Biol. 2001;8:459–473. doi: 10.1016/s1074-5521(01)00026-6. [DOI] [PubMed] [Google Scholar]
- Sheppard TL, Ordoukhanian P, Joyce GF. A DNA enzyme with N-glycosylase activity. Proc Natl Acad Sci USA. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedhara A, Li Y, Breaker RR. Ligating DNA with DNA. J Am Chem Soc. 2004;126:3454–3460. doi: 10.1021/ja039713i. [DOI] [PubMed] [Google Scholar]
- Srinivasan J, Cload ST, Hamaguchi N, Kurz J, Keene S, Kurz M, Boomer RM, Blanchard J, Epstein D, Wilson C, et al. ADP-specific sensors enable universal assay of protein kinase activity. Chem Biol. 2004;11:499–508. doi: 10.1016/j.chembiol.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sun L, Cui Z, Gottlieb RL, Zhang B. A selected ribozyme catalyzing diverse dipeptide synthesis. Chem Biol. 2002;9:619–628. doi: 10.1016/s1074-5521(02)00141-2. [DOI] [PubMed] [Google Scholar]
- Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon–carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SM, Sachdeva A, Silverman SK. DNA catalysts with tyrosine kinase activity. J Am Chem Soc. 2013;135:14928–14931. doi: 10.1021/ja407586u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner KD, Homan P, Weeks KM, Smith AG, Abell C, Ferré-D'Amaré AR. Validating fragment-based drug discovery for biological RNAs: lead fragments bind and remodel the TPP riboswitch specifically. Chem Biol. 2014;21:591–595. doi: 10.1016/j.chembiol.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand TW, Janssen RC, Eaton BE. Selection of RNA amide synthases. Chem Biol. 1997;4:675–683. doi: 10.1016/s1074-5521(97)90223-4. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- Woese C. The Genetic Code: The Molecular Basis for Genetic Expression. Harper & Row; New York: 1967. pp. 179–195. [Google Scholar]
- Yu H, Zhang S, Chaput JC. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem. 2012;4:183–187. doi: 10.1038/nchem.1241. [DOI] [PubMed] [Google Scholar]
- Zhang B, Cech TR. Peptide bond formation by in vitro selected ribozymes. Nature. 1997;390:96–100. doi: 10.1038/36375. [DOI] [PubMed] [Google Scholar]
- Zhang B, Cech TR. Peptidyl-transferase ribozymes: trans reactions, structural characterization and ribosomal RNA-like features. Chem Biol. 1998;5:539–553. doi: 10.1016/s1074-5521(98)90113-2. [DOI] [PubMed] [Google Scholar]