SUMMARY
The ability of p53 to regulate transcription is crucial for tumor suppression and implies that inherited polymorphisms in functional p53-binding sites could influence cancer. Here, we identify a polymorphic p53 responsive element and demonstrate its influence on cancer risk using genome-wide data sets of cancer susceptibility loci, genetic variation, p53 occupancy, and p53-binding sites. We uncover a single-nucleotide polymorphism (SNP) in a functional p53-binding site and establish its influence on the ability of p53 to bind to and regulate transcription of the KITLG gene. The SNP resides in KITLG and associates with one of the largest risks identified among cancer genome-wide association studies. We establish that the SNP has undergone positive selection throughout evolution, signifying a selective benefit, but go on to show that similar SNPs are rare in the genome due to negative selection, indicating that polymorphisms in p53-binding sites are primarily detrimental to humans.
INTRODUCTION
Common inherited genetic factors have great potential to help us better understand the origins, progression, and treatment of human cancer and to serve as important biomarkers in the clinic to identify those at increased risk for developing cancer, progressing more rapidly, and not responding to therapies. Genome-wide association studies (GWASs) have identified almost 900 single-nucleotide polymorphisms (SNPs) significantly associated with cancer susceptibility traits. However, discerning the causal SNPs responsible for the associations from the nonfunctional associated SNPs has proven challenging. Interestingly, many cancer-associated SNPs identified in GWASs are significantly enriched in noncoding functional DNA elements as defined by the ENCODE project (ENCODE Project Consortium et al., 2012). Indeed, single locus and gene-specific studies have presented strong data to support the role of polymorphic transcriptional regulatory elements in influencing the risk of cancers of the breast, kidney, colon, and connective tissues (Bond et al., 2004; Post et al., 2010; Schödel et al., 2012; Sur et al., 2012).
One of the most important and well-studied transcription factors in cancer is the p53 tumor suppressor. Three decades of intense study have clearly demonstrated that p53 is a central node of a cellular stress response pathway that is crucial in suppressing cancer formation in many tissue and cell types (Lane and Levine, 2010) and in regulating other processes such as pigmentation, fecundity, cellular metabolism, mitochondrial respiration, stem cell maintenance, and early embryonic development (Belyi et al., 2010; Junttila and Evan, 2009; Lu et al., 2009). Upon cellular stresses such as DNA damage, replicative stress, oncogene activation, hypoxia, and translational stress, p53 is activated and initiates cellular responses such as DNA repair, cell-cycle arrest, apoptosis, and senescence. p53 determines these cellular fates primarily through its ability to regulate the transcription of numerous target genes through direct, sequence-specific, DNA binding (Bieging and Attardi, 2012; Nikulenkov et al., 2012; Sperka et al., 2012). Indeed, with the advent of technologies that can screen for genome-wide p53 occupancy, coupled with the ability to measure the relative levels of almost all known transcripts, many more important p53 target genes are currently being defined (Bandele et al., 2011; Botcheva et al., 2011; Nikulenkov et al., 2012; Smeenk et al., 2011; Wei et al., 2006).
In order to regulate the vast majority of p53-target genes, p53 directly binds a DNA consensus site via its centrally located sequence-specific DNA-binding domain (DBD). Under most conditions, it binds the consensus site as a homotetramer and, once bound, recruits transcriptional coactivators to regulate transcription via an N-terminal transactivation domain (Beckerman and Prives, 2010). Its DNA consensus motif, the p53 response element (p53-RE) is composed of two decameric half-sites, RRRCWWGYYY (where W = A or T, R = purine and Y = pyrimidine), separated by a spacer of 0–13 nucleotides, and indeed a recent study suggests that p53 prefers p53-REs with half-sites separated by 0–2 nucleotides (Jolma et al., 2013). p53’s ability to bind the p53-RE and subsequently regulate transcription is crucial for its tumor suppressor function (Chao et al., 2000; Crook et al., 1994; Pietenpol et al., 1994). A reflection of this lies in the fact that approximately 50% of human cancers carry somatic mutations of the p53 gene over 80% of which are missense mutations spanning the highly conserved DBD (Freed-Pastor and Prives, 2012). Moreover, many of the same somatic DBD mutations can be found as inherited, cancer-causing mutations in extremely cancer-prone families belonging to the Li-Fraumeni syndrome (Malkin et al., 1990).
Together, these observations suggest the possibility that SNPs in key bases of functional p53-REs (p53-RE SNPs) could influence the ability of p53 to regulate transcription and result in differences in cancer susceptibility (Bandele et al., 2011; Noureddine et al., 2009). In this report, we identify and describe a SNP in a functional p53-RE that affects the ability of p53 to regulate transcription and influence cancer susceptibility and has undergone positive natural selection throughout human evolution. However, we go on to determine that SNPs in similar functional p53-REs genome-wide have been subjected to negative selection. Our data indicate that polymorphisms in functional p53 response elements are primarily detrimental but in rare instances can impart selective benefits, rising to substantial frequencies in populations and resulting in differences in cancer susceptibility among individuals.
RESULTS
One of the Strongest Cancer GWAS SNPs Resides in a Key Position of a Functional p53 Response Element
We reasoned that if SNPs in key nucleotides of functional p53-REs can lead to differential cancer risk, then at least one cancer GWAS SNP or proxy (a SNP in linkage disequilibrium [LD]) should reside in a genomic region occupied by p53, containing a strong p53-RE and in a key nucleotide of the element. To explore this possibility, we determined that the 892 SNPs known to be associated with cancer in GWAS (p values below 1 × 10−5;www.genome.gov/gwastudies, www.ncbi.nlm.nih.gov/sites/entrez?db=gap, www.hugenavigator.net/CancerGEMKB/caIntegratorStartPage.do) were in strong LD with 61,675 other SNPs in the genome (r2 ≥ 0.8). To assess which of these 62,567 cancer GWAS SNPs reside in p53-occupied genomic regions, we integrated p53 chromatin immunoprecipitation sequencing (ChIP-seq) data derived from four different laboratories (Bandele et al., 2011; Nikulenkov et al., 2012; Botcheva et al., 2011; Smeenk et al., 2011) utilizing six different cell lines derived from four different tissues: MCF7 (breast cancer), IMR90 (fibroblasts), U2OS (osteosarcoma), and lymphoblastoid cell lines (LCL; GM06993, GM11992, and GM12878). Cells were treated with seven different p53-activating agents (5-fluorouracil, Nutlin-3, RITA, actinomycin-D, etoposide, doxorubicin, and ionizing radiation), resulting in p53 ChIP-seq peaks from 11 different combinations of cell lines and treatments (Table S1). Utilizing the same peak calling algorithm (QuEST; Valouev et al., 2008) for all four independent studies, we noted a total of 20,838 significant peaks (stringent peak calling parameters, FDR q = 0.05), of which 17,118 are unique peaks (Figure 1A). With this data, we were able to determine that 86 of the 62,567 cancer GWAS SNPs reside in genomic regions occupied by p53 in at least one experiment (Figure 1B and Table S2).
Figure 1. A Cancer GWAS SNP Resides in a Key Position of a Functional p53 Response Element.
(A) A Venn diagram of all p53-occupied genomic regions from the four different studies incorporated in the genomic scan.
(B) A Circos plot displaying the cancer GWAS SNPs and their proxies screened for the presence of p53 occupancy, an above average p53-RE and location in a core nucleotide of the RE. Chromosomes are arranged in a half-circle end-to-end with chromosome’s cytobands in the outer ring. The location of the 892 cancer GWAS SNPs are displayed in the green inner ring and their 61,675 proxies are displayed in the blue ring. Grey lines identify those SNPs that are occupied by p53 in at least in one condition studied. Orange lines identify the two occupied polymorphic regions with above average p53-REs. The red line identifies the SNP that also resides in a core nucleotide of the RE.
(C) A histogram of the position weight matrix (PWM) scores of the 228 known p53-binding sites (p53-REs) that were utilized to calculate the matrix, which was incorporated in the genomic scan. Also shown are examples of below average, average, and above average p53-REs, as well as the well-described consensus p53-RE. See also Tables S1, S2, and S3.
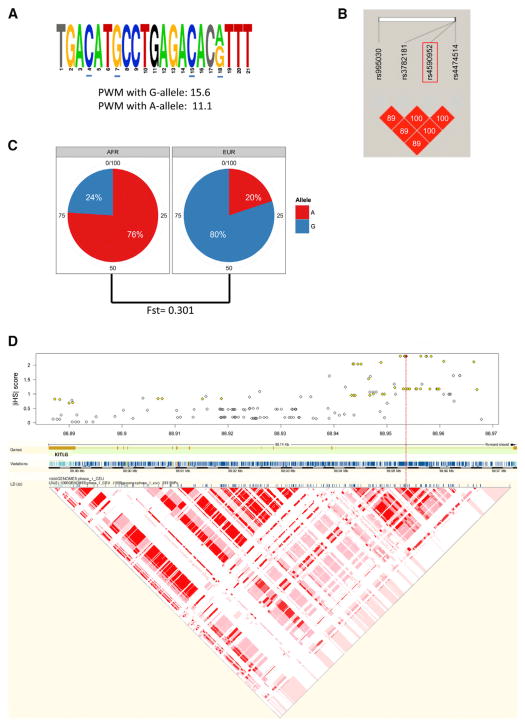
To evaluate which of the 86 cancer GWAS SNPs also reside in a strong p53-RE, we used position weight matrix (PWM) calculations computed from 228 published p53-REs to analyze the sequences surrounding these SNPs (Table S3). As seen in Figure 1C, the 228 published p53-REs have a mean PWM score of 13.8 (median of 14) ranging from 5.1 to 21.7, with sequences scored greater than the mean having strong similarity with the canonical p53-RE. Interestingly, only one of the 86 SNPs resides in a key nucleotide of a p53-RE with an above average PWM (rs4590952, G/A, PMW = 15.6/11.1). As predicted by the high PMW score, an almost canonical p53-RE is found in this region (Figure 2A). However, the PWM score decreases dramatically with the A allele because it disrupts the G nucleotide in the core CWWG motif of the second decamer (PWM 11.1, difference of 4.5). The SNP, KITLG p53-RE SNP, rs4590952 (G/A), is in LD with rs995030, rs3782181, and rs4474514 (Figure 2B), which have been shown in three GWASs (Kanetsky et al., 2009; Rapley et al., 2009; Turnbull et al., 2010) to associate with differential risk for developing testicular cancer in Caucasians with a per allele odds ratio (OR) of up to 3.07 (p = 1.0 × 10−31, Table S4). Interestingly, this is one of the highest and most statistically significant ORs noted to date in GWASs (Chanock, 2009).
Figure 2. The KITLG p53-RE SNP, rs4590952 (G/A) Is Closely Linked to One of the Strongest Cancer GWAS SNPs and Harbors Two Signatures of Recent Natural Selection.
(A) Sequence of the p53-RE in which rs4590952 resides. Nonconsensus bases are shown in gray and the spacer nucleotide is in black. Core bases are underlined in blue.
(B) A linkage disequilibrium (LD) analysis (r2 values) of the KITLG p53-RE SNP, rs4590952 (boxed in red), with the three SNPs that have been identified in three independent GWASs to associate with differential risk for developing testicular cancer in Caucasians.
(C) Pie charts of the allele frequencies of the KITLG p53-RE SNP in the African (AFR) and European (EUR) individuals from the 1000 Genomes Project. Also noted is the fixation index (F-st).
(D) A plot of the absolute values of the standardized integrated haplotype scores (iHS) for SNPs spanning the KITLG gene indicating significant allelic differences in haplotype structure indicative of positive selection. The red line denotes the location of rs4590952 (marked in red) and the yellow coloring denotes the SNPs known to be in strong LD with rs4590952. Below is a schematic depiction of the KITLG gene with exons and introns noted, as well as the genetic variations in the gene, together with an LD analysis (r2). The red color denotes variations with high r2 values. See also Table S4.
The KITLG p53-RE SNP Displays Signatures of Recent Positive Selection
A number of previously defined, functional p53 pathway SNPs display characteristic global allele frequencies and haplotype structures that suggest certain alleles have undergone selective sweeps throughout evolution (Atwal et al., 2007; Atwal et al., 2009; Beckman et al., 1994; Shi et al., 2009). These findings are consistent with the roles of this pathway in many processes subject to selective pressures, such as pigmentation, fecundity, and early embryonic development. Therefore, we next explored the possibility that the KITLG p53-RE SNP has also been subjected to natural selection pressures. Interestingly, the KITLG p53-RE SNP harbors two different signatures of recent natural selection in humans. First, in Caucasian Europeans, the frequency of the G allele with the stronger predicted p53-RE is 80%, in contrast to only 24% in Africans (Figure 2C, 1000 Genomes Project). This dramatic allele-frequency difference is significantly greater than would be expected by genetic drift alone, as indicated by a fixation index (F-st) of 0.301 (Amigo et al., 2008). Similarly, in Europeans, the haplotype(s) associated with the G allele are significantly longer and present with a higher degree of homozygosity, as indicated by an integrated Haplotype Score (iHS) of 2.3, which is among the strongest 5% of all signals of positive selection in the genome (Figure 2D) (Gautier and Vitalis, 2012) and is similar to previously reported observations (Lao et al., 2007). Together, these two signatures of natural selection provide evidence that the G allele with the stronger predicted p53-RE has undergone a selective sweep in Caucasians in a similar manner to previously defined, functional p53 pathway SNPs.
KITLG Is a Common p53 Target Gene that Can Increase Cellular Proliferation
The transcriptional activation of KITLG upon p53 activation has been clearly demonstrated in multiple studies utilizing different cell types, different p53-activating stimuli and different measurement techniques (McGowan et al., 2008; Murase et al., 2009; Smeenk et al., 2011; Terzian et al., 2010; Wei et al., 2006). Consistent with this, we observed significant p53 binding to KITLG in the region that harbors the KITLG p53-RE SNP, in 10 out of 11 different cell line and treatment combinations (Figure 3A). This observation not only provides overwhelming evidence for an in vivo association of p53 with the KITLG p53-RE, but also places it in a class of p53-REs that are bound by p53 when activated by many different mechanisms; the so-called “default” p53-REs (Table S5) (Bandele et al., 2011; Nikulenkov et al., 2012; Wei et al., 2006). Indeed, in Figure S1 (available online), we present the significant fold activation of KITLG in six diverse cell types compared to the induction of one of the most well-studied default p53 target genes CDKN1A (p21). The cells we studied were derived from germ cell, testicular, leukemia, and renal cancers, as well as primary human keratinocytes and Sertoli cells. Specifically, we measured KITLG and p21 levels using qRT-PCR before and after p53 activation by Nutlin-3, one of the most specific p53 activators. Importantly, we found a significant fold-induction of KITLG in all cells tested. Specifically, after p53 activation, we noted an average 6.8-fold induction of KITLG, ranging from 1.7-fold to 16-fold. These values were strikingly similar to the induction of p21 with an average fold induction of 5.17, ranging from 2.6-fold to 8.8-fold. Together with the results from our genomic analysis and the previously published observations, these data suggest that KITLG expression can be regulated utilizing the p53-RE in a p53-dependent manner in many cell types and under many activating stimuli.
Figure 3. The KITLG p53-RE SNP Resides in a p53 Target Gene that Can Increase Cellular Proliferation and Is Frequently Occupied by p53 in an Allele-Specific Manner.
(A) Genome browser tracks depicting the read density at rs4590952 for 10 of the 11 p53 ChIP-seqs included in this study as visualized using Quest. GWAS SNPs and rs4590952 are displayed.
(B) TRP1 and Ki67 double staining to identify proliferating melanocytes (MCs) in the epidermis. WT and p53−/− mice were irradiated at P2 with 5.6 kJ/m2 UV and the percentage of epidermal MCs determined in a minimum of three mice of each genotype at 3–5 days after treatment. (*Mann-Whitney U-test p < 0.05).
(C) Kitlg gene expression differences (± SEM) in WT and p53−/− mice 24 hr after UV exposure (P3) expressed as fold change and referenced to the nontreated P3 expression level for the respective genotype. A minimum of five pups were used for expression estimates at each time point. (#Mann-Whitney U-test p = 0.002).
(D) Visualization of the 57 unique, individual reads containing the polymorphic A/G nucleotide in the KITLG p53-RE. (E) A bar graph depicting the allelic differences in the relative amounts (± SEM) of p53-immunoprecipitated DNA compared to the input DNA for three different LCL cell lines heterozygous for the KITLG p53-RE SNP. See also Figures S1 and S2, as well as Table S5.
Because the KITLG p53-RE SNP is associated with differential cancer risk, and the action of p53 on the KITLG locus is so strikingly preserved in different cell types and upon different activating stimuli, we reasoned that the p53-dependent regulation of KITLG signaling could have a profound effect on an important hallmark of cancer such as cellular proliferation. To test this, we chose to examine the impact of p53-dependent KITLG activation on the developing pigmentary system in mice, where many of the initial roles of KIT pathway signaling were first described. Specifically, KITLG and its receptor C-KIT are a part of a pathway that is known to be central in regulating proliferation, survival and migration of primordial germ cells, melanoblasts and hematopoietic stem cells (Lennartsson and Rönnstrand, 2012). In the pigmentary system, ultraviolet radiation induces p53-Kitlg signaling in keratinocytes, which in turn affects melanocytes that express the c-Kit receptor in order to mount a cutaneous UV response such as tanning (Murase et al., 2009). Interestingly, when we compared this response in mice either wild-type (WT) or null for p53 (p53 null), we noted dramatic differences. First, we irradiated WT pups at postnatal day 2 (P2) with a melanoma-inducing 5.6 kJ/m2 of ultraviolet (UV) light (Walker et al., 2009) and observed an up to 16-fold increase in epidermal melanocyte numbers after UVR compared to nontreated mice (Figure S2A). In parallel, we examined Kitlg expression in nonirradiated WT and p53 null mice and observed that Kitlg was induced 2.15-fold between birth (postnatal day 0 [P0]) and P3 in WT mice; however, in p53 null mice it was not induced (Figure S2B). When we irradiated WT and p53 null P2 pups, we noted that melanocyte numbers were increased up to 4.6-fold that observed in p53 null mice (Figure S2C), and melanocyte proliferation was increased almost 3-fold in WT versus p53 null mice (Figure 3B). Concurrently, Kitlg expression was induced 2.5-fold in WT skin but was not induced in p53 null mice (Figure 3C). These data provide strong evidence that in its natural setting, the action of p53-Kitlg signaling results in a measurable effect on a cancer-related process such as cellular proliferation.
The KITLG p53-RE SNP Is Frequently Occupied by p53 in an Allele-Specific Manner
To explore the prediction that p53 could occupy the KITLG p53-RE in an allele-specific manner, we examined the sequence reads generated from the p53 ChIP-seqs in greater detail. From the reads, we determined that three of the four cell types were homozygous for the G allele. One of the studies incorporated heterozygous cell lines, two HapMap CEU LCL (GM06993 and GM11992), which were treated with doxorubicin (Bandele et al., 2011). In support of the hypothesis that the G allele creates a stronger p53-RE, of the 57 unique sequencing reads precipitated with p53 antibodies, 65% contained the G allele (37) and 35% the A allele (Figure 3D, p = 0.033, binomial test). In order to further investigate allelic differences in p53 binding, we used a RT-PCR allele discrimination assay for the KITLG SNP to quantify alleles in p53-immunoprecipitated DNA compared to input DNA in three heterozygous LCL cell lines: GM11992, GM11831, and GM12815. Notably, for each LCL, the G allele with the stronger p53-RE was detected at a higher level in the p53-immunoprecipitated DNA compared to A allele sequences (Figure 3E) (GM11992 = 2.8-fold G:A; GM11831 = 1.8-fold G:A, and GM12815 = 1.5-fold G:A), thereby lending further support to the model that p53 can occupy the KITLG p53-RE in an allele-specific manner.
The KITLG p53-RE SNP Results in Allele-Specific Differences in p53-Dependent Transactivation of KITLG
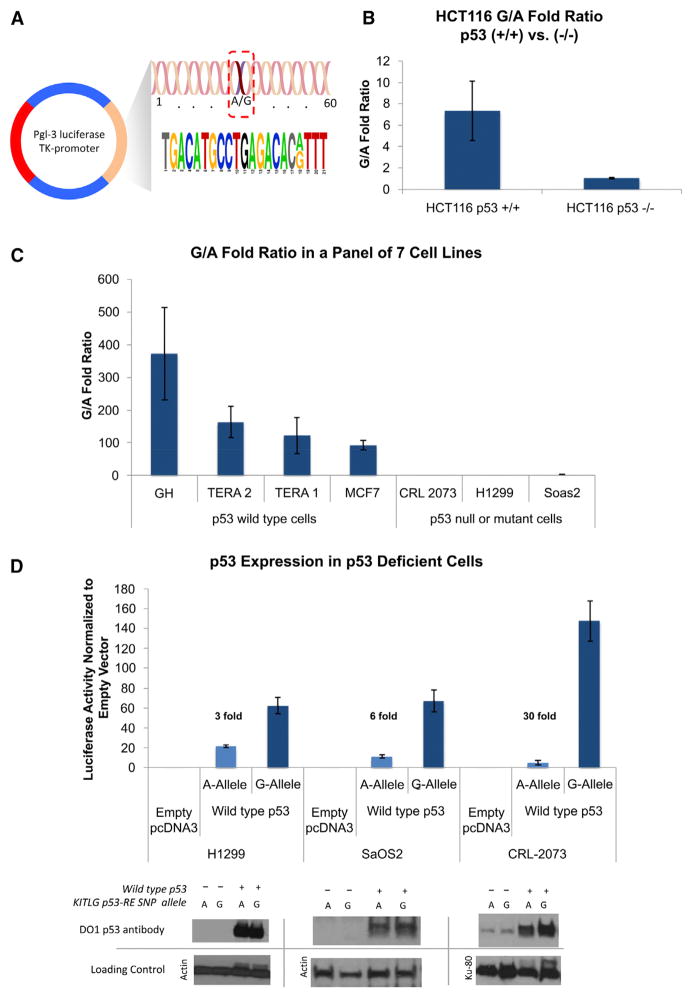
To assess whether the KITLG p53-RE SNP affects p53-dependent transcriptional activation, we cloned a 60 bp region surrounding the SNP for each allele into a pGL3 luciferase reporter vector (Figure 4A). To test p53 dependence, we transfected each of these constructs into the colorectal cancer wild-type p53 cell line (HCT116 p53+/+) and its isogenic p53 null form (HCT116 p53−/−). Interestingly, 7-fold more luciferase activity was measured from the reporter containing the G allele compared to the A allele in HCT116 p53+/+ cells (p = 0.003, Mann-Whitney Test, Figure 4B). In contrast, no significant differences in luciferase activity levels between the two variant alleles could be measured in the HCT116 p53−/− cells (Figure 4B). To extend this study to cells derived from other cancers and, importantly, to cells derived from testicular cancers, we transfected the reporter plasmids into four p53 wild-type cell lines (GH, testicular; Tera1, testicular; Tera2, testicular; and MCF7, breast) and three p53 mutant or null cell lines (CRL-2073, germ cell tumor; H1299, lung; and Soas2, osteosarcoma). In the p53 wild-type cells we measured, on average, 188-fold more luciferase activity from the G-allele-containing reporter than the A allele reporter, ranging from 93- to 373-fold (Figure 4C). Consistent with the results obtained in the HCT116 p53−/− cells, we measured, on average, a nonsignificant 1.5-fold increase in luciferase activity from the G allele reporter relative to the A allele when tested in the three p53 mutant or null cell lines, ranging from 1.1 to 2.1. In order to further confirm the p53-dependence of these effects, we attempted to rescue the lack of significant fold difference in reporter expression in these three p53 mutant/null cell lines. To do this, we cotransfected an expression vector (pcDNA3) containing wild-type p53 cDNA with both reporters. Importantly, we were able to restore wild-type p53 expression in all three cell lines and to note an average 13-fold increase in luciferase activity from the G allele reporter over the A allele reporter, ranging from 3-fold to 30-fold (Figure 4D), thus rescuing the G-allele-dependent increase in transcription.
Figure 4. The KITLG p53-RE SNP Results in Allele-Specific Differences in p53-Dependent Transactivation.
(A) The 60 base pair region surrounding the SNP, which was cloned into a pGL3 luciferase reporter vector.
(B) A bar graph depicting the fold ratio of luciferase activity from transiently transfected reporter plasmids containing either the G or A alleles into a pair of HCT116 cell lines containing either the wild-type p53 gene (+/+) or without (−/−).
(C) A bar graph depicting the fold ratio of luciferase activity from the reporter plasmids into seven diverse cancer cell lines with wild-type p53, mutant, or no p53.
(D) A bar graph depicting the fold ratio of luciferase activity in the cell lines with mutant or no p53 upon cotransfection with an expression vector containing wild-type p53 cDNA, as well as a western blot analysis of p53 levels in the transfected cell lysates. Top: western blots using a monoclonal p53 antibody (DO1). Bottom: western blots of the same filter using antibodies to either actin or KU-80 as loading controls. Error bars represent SEM of three independent experiments each performed multiple times.
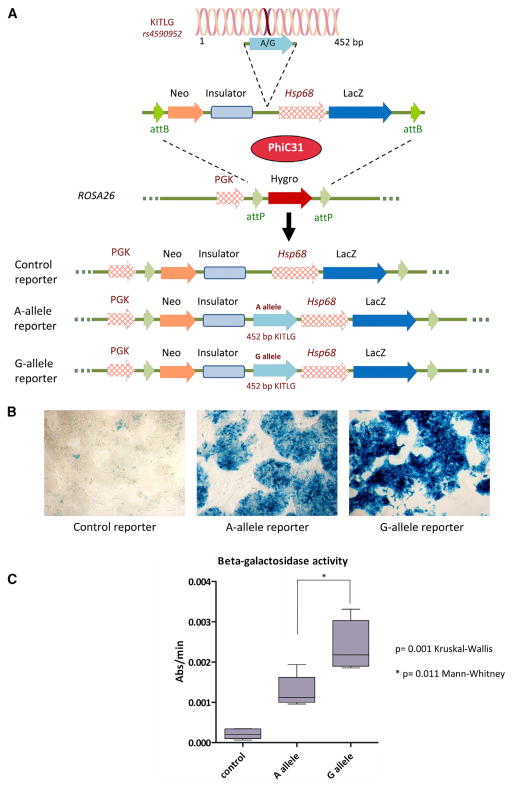
To formally assign enhancer activity to the polymorphic KITLG p53-RE region when present within the genome, an enhancer trap assay was performed using the Hsp68-LacZ reporter system in murine embryonic stem (ES) cells (Kothary et al., 1988). To facilitate the comparison between the empty control and the putative enhancer containing vectors, the test vectors were targeted to the ROSA26 locus via PhiC31 integrase-mediated cassette exchange (Chen et al., 2011) (Figure 5A). ES cell clones harboring the basal Hsp68 promoter alone resulted in only minimal beta-galactosidase expression, whereas ES cells harboring the identically positioned construct together with a 452 bp region surrounding the KITLG p53-RE region yielded robust beta-galactosidase expression, thus proving the region to have significant enhancer activity (p = 0.001 Kruskal-Wallis test) (Figures 5B and 5C). Furthermore, ES cells harboring the G allele enhancer yielded a significant 1.9-fold increase in beta-galactosidase activity over ES cells harboring the A allele (p = 0.011 Mann-Whitney test). These results demonstrate that even at a single genomic copy, the p53-RE SNP region is able to confer enhancer activity, with the G allele conferring significantly higher expression than the A allele.
Figure 5. The KITLG p53-RE Demonstrates Allele Specific Enhancer Activity when Present at Single Copy in Murine ES Cells in an Hsp68-LacZ Enhancer Trap Assay.
(A) Top: the 452 bp region surrounding the SNP, cloned from either the A or the G allele, inserted into an Hsp68-LacZ enhancer trap vector contained within a PhiC31 integrase attB flanked cassette exchange vector. Center: the PhiC31 integrase attP flanked docking site within the ROSA26 locus. PhiC31 integrase catalyzes the recombination between the attB sites on the exchange vector and the attP sites positioned within the ROSA26 locus, leading to the targeted integration of the enhancer trap construct. Bottom: the conformation of the three alleles that were used in the study.
(B) X-gal staining of ES cells harboring either the empty Hsp68-LacZ cassette (control reporter) or the KITLG p53-RE enhancer-Hsp68-LacZ cassettes (A allele reporter and G allele reporter).
(C) A box plot depicting the difference in the beta-galactosidase activity of the targeted ES cells harboring the three different constructs. Whiskers in the box plot graph represent minimum and maximum values.
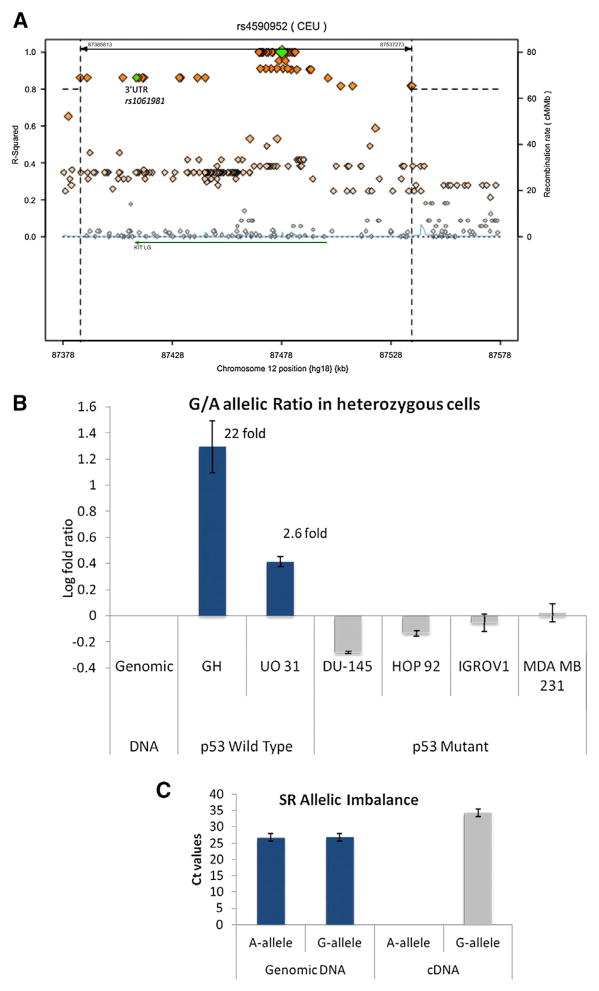
To test this further in the context of the endogenous gene, we measured the allele-specific levels of KITLG mRNA in heterozygous cells with wild-type and mutant p53. First, we identified only one heterozygous, p53 wild-type cell line from the above-mentioned four testicular cancer cells, namely GH. Therefore, in order to obtain additional heterozygous cell lines, we genotyped the NCI60 cell panel (Table S6). Fourteen of the 59 cell lines contain wild-type p53 (http://p53.free.fr/index.html), and genotyping for the KITLG-SNP identified two cell lines to be heterozygous: SR (acute lymphoblastic leukemia) and UO-31 (renal cancer). Forty-five of the cell lines are deficient for p53 activity, eight of which were determined to be heterozygous. We purified mRNA from all three logarithmically growing heterozygous cell lines with wild-type p53 and four heterozygous cell lines deficient for p53 that were chosen at random (DU145, HOP92, IGROV1, and MDAMB231). We synthesized cDNA and utilized a closely linked exonic SNP (rs1061981C/T, Figure 6A) to measure the allele-specific transcript levels, whereby the C allele associates with the G allele. Consistent with the previous outcomes, in all three cell lines with wild-type p53, a significant increase in the G allele to A allele ratio was noted in the cDNAs compared to the genomic DNAs. Specifically, 21.7-fold more G-allele-containing transcripts were measured in the testicular cancer cells GH, and 2.6-fold more in the renal cancer cells UO-31 (Figure 6B). In the SR cell line, both alleles were successfully identified in the genomic DNAs but interestingly only the G allele in the cDNA (Figure 6C). Consistent with the p53-dependence of these observations, no significant increase in the G allele to A allele ratio was noted in the cDNAs derived from the four p53 mutant cells (Figure 6B).
Figure 6. KITLG Demonstrates Allelic Imbalance in Transcript Levels in a p53-Dependent Manner.
(A) A LD regional plot depicting the 200 kb surrounding the KITLG p53-RE SNP. The diamonds represent the SNPs in the region, with the orange diamonds highlighting the SNPs in high LD with the KITLG p53-RE SNP (r2 > 0.8). The green diamonds denote the KITLG p53-RE SNP (rs4590952) and the 3′UTR SNP rs1061981. The KITLG gene is denoted by the green arrow. The plot was generated with 1000 Genomes Project data using the online tool SNAP from the Broad Institute at MIT available at: http://www.broadinstitute.org/mpg/snap/ldsearch.php. Bar graphs depict the differences in the relative transcription of KITLG from each allele of a strongly linked exonic SNP (rs1061981 C/T), whereby the C allele associates with the G allele.
(B) The log fold ratios found in cDNAs compared to genomic DNAs from the same heterozygous cell line are plotted.
(C) The same measurements were performed for the acute lymphoblastic leukemia cell line SR, and both alleles were successfully identified in the genomic DNAs but only the G allele in the cDNA. These data are presented in a bar graph plotting the cycle threshold values measured for each allele in both the genomic DNA and the cDNA. For all experiments, error bars represent SEM of three independent experiments each done in triplicate. See also Table S6.
The KITLG p53-RE SNP Is Unique in the Genome and Influences Cancer Susceptibility
Thus far, our genomic analyses and subsequent experimental validation have supported the assertion that a functional SNP in a functional p53-RE in the KITLG gene resides among the 62,567 cancer GWAS SNPs. To assess the likelihood that the KITLG p53-RE SNP’s association with the pool of cancer GWAS SNPs was by chance, we determined the frequency of similar SNPs in the genome (Figure 7A). To date, there are known to be 10,815,471 SNPs in the genome, with a minor allele frequency > 0.05% in the 1000 Genomes Project (Abecasis et al., 2012). Among these, we determined 3,123 SNPs to reside in p53-REs with PWM scores greater than or equal to the KITLG p53-RE (15.5). Of these, only six SNPs are in p53-REs that, like the KITLG p53-RE, are occupied by p53 after all seven different p53-activating treatments. Among the six frequently occupied p53-REs with SNPs, only the KITLG p53-RE SNP resides in a core nucleotide of the consensus sequence and is predicted to strongly affect p53 binding (allelic difference of PWM scores > 4.4). Therefore, among the 10,815,471 SNPs throughout the genome, only one SNP resides in a core nucleotide of a frequently occupied, strong p53-RE. The likelihood of this p53-RE SNP being found among the 62,567 cancer GWASs SNPs by chance is extremely low (p = 0.0058, Fisher’s Exact test, Figure 7B), lending strong support for a direct influence of the KITLG p53-RE SNP on cancer susceptibility.
Figure 7. The KITLG p53-RE SNP Is Unique in the Genome and Influences Cancer Susceptibility.
(A) A Circos plot displaying the known 10,815,471 SNPs with minor allele frequencies > 0.05%, which were screened to determine the number of SNPs that, like the KITLG p53-RE SNP, reside in a strong p53-RE, occupied by p53 after all seven different p53-activating treatments, and in a core nucleotide of the p53-RE. Chromosomes are arranged in a half-circle end-to-end with chromosome’s cytobands in the outer ring. The locations of the 10,815,471 SNPs are displayed in the blue inner ring. The locations of the 3,123 SNPs residing in strong p53-REs are displayed in the green inner ring. Orange lines identify those 6 SNPs in p53-REs that are occupied by p53 in after all at least in seven different p53-activating treatments. The red line identifies the only SNP that also resides in a core nucleotide of the RE, which is the KITLG p53-RE SNP.
(B) A graph depicting the genome-wide distribution of the known 10,815,471 SNPs with minor allele frequencies > 0.05%, the cancer GWAS SNPs and their proxies at a 2Mb scale. The number of SNPs is depicted in a logarithmic scale and the dotted line denotes the location of the KITLG p53-RE SNP, rs4590952. The likelihood of this p53-RE SNP being found among the cancer GWASs SNPs and proxies by chance is low (p = 0.0058, Fisher’s exact test).
(C) A genome-wide analysis provides evidence of negative selection of polymorphic p53-REs. The flowchart depicts the number of 21,904 p53-REs that are present in the genome in either p53 unoccupied regions or occupied regions after one to four or five to seven different p53-activating treatments. Also displayed is a bar graph representing the percent values of the p53-REs in each category that are polymorphic.
SNPs in Functional p53-REs Are Infrequent in the Genome Due to Negative Selection
As mentioned above and similar to the KITLG p53-RE SNP, a number of previously defined, functional p53 pathway SNPs harbor alleles that have undergone selective sweeps throughout evolution. Given this, and the fact that we are only able to identify a single strongly functional p53-RE SNP among over 10 million SNPs in the genome, we examined the possibility that functional p53-RE SNPs are infrequent in the genome, possibly due to purifying (negative) selection. To do this, we compared the frequency of p53-occupied polymorphic p53-REs, to the frequency of polymorphic p53-REs never observed to be p53 occupied in the 11 p53 ChIP-seq data sets included in our analysis. The former are most certainly enriched for functional p53 enhancers, whereas the latter are enriched for nonfunctional p53-REs. Therefore, if functional p53-REs are under negative selection pressure to exclude mutations in the consensus site, we would expect the frequencies of polymorphic p53-REs to be significantly lower in the p53-occupied REs. To test this, we identified 21,904 p53-REs genome-wide with a PWM score of 15.5 or greater. Of these, 88% (19,282) were never occupied by p53. Ten percent (2,091) were occupied by p53 in one to four different p53-activating treatments, and 2% (531) in five to seven treatments (Figure 7C). Interestingly, 15% (2,892/19,282) of the non-p53-occupied REs contain SNPs. In contrast, only 9% (194/2,091 in one to four treatments) and 7% (37/531 in five to seven treatments) of p53-occupied REs contain SNPs (Figure 7D). This dramatic decrease in the frequency of polymorphic p53-REs in the p53-occupied REs, compared to the nonoccupied REs (up to 2-fold), is highly significant (p < 1 × 10−15, Cochran-Armitage Trend test), and lends support to the hypothesis that polymorphisms in functional p53-REs are suppressed in human populations through negative selection.
DISCUSSION
In this report, we present strong evidence supporting the hypothesis that well-placed polymorphisms in functional p53-binding sites can result in differential p53-dependent transcriptional regulation and cancer risk, through the identification and characterization of the KITLG p53-RE SNP. The KITLG p53-RE SNP is strongly linked to three SNPs shown, in three independent GWASs, to associate with differential risk for developing seminomatous and nonseminomatous testicular cancer, with a per allele odds ratio of up to 3.07 (p value = 1.0 × 10−31), one of the highest and most significant findings among all cancer GWASs (Kanetsky et al., 2009; Rapley et al., 2009; Turnbull et al., 2010). Based on linkage data, individuals with the G allele, and therefore the stronger KITLG p53-RE, harbor the greater testicular cancer risk. This finding supports the intriguing hypothesis that p53-regulated paracrine growth factor signaling may be associated with promoting tumor development. Indeed, activating mutations of the KIT receptor have been shown to promote tumor formation and the receptor is currently targeted by many therapeutic agents in cancer treatments. In testicular cancer, the KIT pathway is central to its molecular pathology (Gilbert et al., 2011), and many components of this pathway are somatically mutated to activate KIT signaling. Moreover, the six SNPs identified in testicular cancer GWASs to associate with differential cancer risk reside in three genes in this pathway: KITLG, SPRY4, and BAK1. Although p53 is the most commonly mutated gene in human cancer, whereby 50% of all cancers have mutant p53, less than 3% of testicular cancers do (Lutzker, 1998; Peng et al., 1993). In many cancers that retain wild-type p53 genes, the cells have frequently attenuated p53 through other mechanisms such as the overexpression of direct inhibitors, like the amplification of the MDM2 oncogene. In dramatic contrast to these observations, p53 signaling is often found to be robust in human testicular cancers (Chresta et al., 1996; Gutekunst et al., 2011; Lutzker and Levine, 1996). Intriguingly, 80% of testicular germ cell tumors can be cured with DNA-damaging therapies, and the retention of p53 activity has been implicated in this astounding cure rate through chemotherapeutics (Bosl and Motzer, 1997; Gutekunst et al., 2011; Houldsworth et al., 1998; Masters and Köberle, 2003). Together with the data presented in this report, these observations suggest that the G allele of KITLG p53-RE allows increased p53-dependent upregulation of KITLG expression, which drives male germ cell proliferation rather than arrest in the presence of DNA damage, thus promoting tumorigenesis and thereby offering an explanation for the abnormal retention of p53 activity in testicular tumors.
Testicular cancer has been demonstrated to harbor a strong genetic component to its development (Czene et al., 2002). A very interesting observation in support of this is the fact that testicular cancer is 4- to 5-fold more prevalent in Caucasians compared to African populations (Garner et al., 2005; Holmes et al., 2008; Moul et al., 1994; Ross et al., 1979). Additional studies have gone on to provide further evidence, demonstrating that African American males have maintained the lower risk for testicular cancer over several generations, while acquiring higher risk of other cancers, such as colorectal (Chia et al., 2010; Ross et al., 1979). Here, we have shown that the G allele, which increases the affinity for p53 and associates with greater testicular cancer risk, is significantly more frequent in Caucasians than in Africans. Furthermore, among Caucasians, the G allele is harbored in a haplotype structure that suggests that it has undergone a selective sweep (Lao et al., 2007). It is fascinating that the G allele has arisen to such prevalence in Caucasians, where pigmentary system function is reduced compared to Africans. In the skin, KIT signaling serves to increase melanin production and melanocyte migration in its key role in mediating the UV protective tanning response. Interestingly, in Caucasians, variation in all other pigmentation genes that we know about results in impaired melanogenic function and concomitant reduced pigmentation. Thus, it is intriguing to hypothesize that the G allele (and its linked alleles) may have risen to such a high frequency in Caucasians to benefit UV protection in lighter-skin individuals.
Although we have provided evidence that the KITLG p53-SNP and its haplotype show signs of positive selection, when we looked at polymorphic p53 enhancers genome-wide, we observed significant signs of negative selection. Specifically, we demonstrated that the frequency of polymorphic p53-REs was significantly lower in the genomic regions occupied by p53, compared to p53-REs, for which p53 occupancy was not observable (Figure 7C). This suggests that SNPs in active p53-binding sites could have been under negative or purifying selection throughout evolution. During the past three decades of intense research, the regulation of the p53 pathway has been clearly shown to be under tight control, which is appropriate for a signaling pathway making cellular life or death decisions (Lu, 2010). Indeed, small changes in levels of target genes have been well documented to measurably change the p53 stress response, resulting in aberrant cellular fate decisions. As mentioned above, the p53 stress response has been shown not only to be important in tumor suppression, but also in many key physiological processes that could have been subjected to selection pressure throughout human evolution (Belyi et al., 2010; Junttila and Evan, 2009; Lu et al., 2009). Together, these observations suggest the possibility that mutations in p53-REs are primarily detrimental but that in rare instances, they impart selective benefits.
EXPERIMENTAL PROCEDURES
Searching for Putative p53-REs Using the PWM
We constructed a position weight matrix (PWM) for p53-REs based on 228 functional p53-RE sequences from the literature (NCBI PUBMED database) that were discovered by experimental methods investigating DNA-protein interaction, including electrophoretic mobility shift assay, DNase I footprinting, and promoter reporter gene constructs. Once a PWM model has been compiled, the PWM score for any putative p53-RE is calculated by summing the individual matrix values that correspond to the observed nucleotide at each position in that site. The potential p53-REs in a genomic sequence were detected by sliding a window along the input sequence, similar to the method we used to detect the antioxidant response elements but with an added feature that considers the spacer in p53-REs (Wang et al., 2007).
ChIP-Seq Data Analysis
The p53 ChIP-seq data are combined from three published studies and our unpublished data (M.R.C., D.S., X.W., D.A.B.). For the three published studies, the raw reads were downloaded from NCBI SRA database. All sequence reads generated from Illumina Genome Analyzer II were aligned against human reference sequence (GRCh37p5, or hg19, June 2011) using Burrows-Wheeler Alignment (BWA) Tool (Li et al., 2008).
Allele Specific Expression and Allele Specific Binding Using Real-Time PCR
The mean Ct values for each dye were calculated from the three technical replicates. Standard curves for both dyes (FAM and VIC) were constructed using serial dilutions of genomic DNA, or input DNA in the case of ChIP material. The mean Ct values were plotted against the preamplification DNA concentrations and a logarithmic regression was performed. Calculating an independent logarithmic regression for each of the dyes allowed us to quantify the fold difference in fluorescence between the alleles.
Transfection and Luciferase Assays
All transfections were done in triplicate. Forty hours posttransfection the cells were assayed. Measurements of firefly luciferase activity were normalized by Renilla luciferase activity for transfection efficiency. Mean values and standard deviations of the triplicates were calculated. When the Luciferase reporter vector was cotransfected with an expression vector (pcDNA3) containing wild-type p53 cDNA Luciferace/Renilla ratios of the cells cotransfected with the wild-type p53 were normalized to that of the Luciferace/Renilla ratios of the cells cotransfected with the empty pcDNA3 vector.
Embryonic Stem Cell Enhancer Assays
A 452 bp region of genomic KITLG, containing either the A or G alleles, was amplified by PCR and cloned upstream of an expression construct consisting of the minimal Hsp68 promoter (Hspa1b) driving the expression of a beta-galactosidase gene (LacZ). These two experimental constructs, together with the empty Hsp68-LacZ control construct, were cloned into PhiC31 integrase-mediated cassette exchange shuttle vector, pCB92 (Chen et al., 2011) downstream of a murine H19 insulator. ES cell clones haboring the enhancer-promoter-LacZ constructs at the ROSA26 locus were generated by PhiC31 integrase-mediated cassette exchange into IDG26.10-3 ES cell line as described (Hitz et al., 2007). Experiments were repeated at least three times on separate days using 5–7 technical replicates on each occasion.
Natural Selection Analysis
The integrated haplotype scores were estimated using the package “rehh”(Gautier and Vitalis, 2012), following the method described by Voight et al. (2006). Haplotype data for the CEU population were downloaded from the 1000 Genomes (phase 1 data) (Abecasis et al., 2012). To calculate the scores, only SNPs with a MAF larger than 0.05 were used. The normalization of the scores was performed with a 0.05 allele frequency window.
Supplementary Material
Acknowledgments
This work was funded in part by the Ludwig Institute for Cancer Research, the Development Fund-Oxford Cancer Research Centre-University of Oxford, the Nuffield Department of Medicine, the Clarendon Fund, the Wellcome Trust (090532/Z/09/Z), the Oxford NIHR Comprehensive Biomedical Research Centre, the Intramural Research Program of the National Institute of Environmental Health Sciences-National Institutes of Health (projects: Z01ES100475 and Z01ES046008), the Australian NHMRC, Cancer Council of Queensland and the NIAMS (projects: 1P30AR057212 and 5K01AR063203). The authors would like to acknowledge Daniel Biggs and Nicole Hortin for assistance with ES cell culture; Shuangshuang Dai for Linux computing support; Colin Goding and Will Fairbrother for helpful discussions; and Suzanne Christen, Claire Beveridge, Elisabeth Bond, and Mark Shipman for help with the preparation of this manuscript.
Footnotes
ACCESSION NUMBERS
The GEO accession number for the p53 ChIP-seq data from human lymphoblastoid cell lines reported in this paper: without treatment and treated with doxorubicin is GSE46991; treated with nutlin-3 is GSE46992; treated with ionizing radiation GSE46993.
Supplemental Information includes Extended Experimental Procedures, two figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.09.017.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amigo J, Salas A, Phillips C, Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinformatics. 2008;9:428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal GS, Bond GL, Metsuyanim S, Papa M, Friedman E, Distelman-Menachem T, Ben Asher E, Lancet D, Ross DA, Sninsky J, et al. Haplotype structure and selection of the MDM2 oncogene in humans. Proc Natl Acad Sci USA. 2007;104:4524–4529. doi: 10.1073/pnas.0610998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Böhnke A, Pempe C, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci USA. 2009;106:10236–10241. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandele OJ, Wang X, Campbell MR, Pittman GS, Bell DA. Human single-nucleotide polymorphisms alter p53 sequence-specific binding at gene regulatory elements. Nucleic Acids Res. 2011;39:178–189. doi: 10.1093/nar/gkq764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman G, Birgander R, Själander A, Saha N, Holmberg PA, Kivelä A, Beckman L. Is p53 polymorphism maintained by natural selection? Hum Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2:a001198. doi: 10.1101/cshperspect.a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging KT, Attardi LD. Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 2012;22:97–106. doi: 10.1016/j.tcb.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Bosl GJ, Motzer RJ. Testicular germ-cell cancer. N Engl J Med. 1997;337:242–253. doi: 10.1056/NEJM199707243370406. [DOI] [PubMed] [Google Scholar]
- Botcheva K, McCorkle SR, McCombie WR, Dunn JJ, Anderson CW. Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle. 2011;10:4237–4249. doi: 10.4161/cc.10.24.18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock S. High marks for GWAS. Nat Genet. 2009;41:765–766. doi: 10.1038/ng0709-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Krohn J, Bhattacharya S, Davies B. A comparison of exogenous promoter activity at the ROSA26 locus using a ΦiC31 integrase mediated cassette exchange approach in mouse ES cells. PLoS ONE. 2011;6:e23376. doi: 10.1371/journal.pone.0023376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia VM, Quraishi SM, Devesa SS, Purdue MP, Cook MB, McGlynn KA. International trends in the incidence of testicular cancer, 1973–2002. Cancer Epidemiol Biomarkers Prev. 2010;19:1151–1159. doi: 10.1158/1055-9965.EPI-10-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chresta CM, Masters JR, Hickman JA. Hypersensitivity of human testicular tumors to etoposide-induced apoptosis is associated with functional p53 and a high Bax:Bcl-2 ratio. Cancer Res. 1996;56:1834–1841. [PubMed] [Google Scholar]
- Crook T, Marston NJ, Sara EA, Vousden KH. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium Bernstein BE. Birney E, Dunham I, Green ED, Gunter C, Snyder M, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner MJ, Turner MC, Ghadirian P, Krewski D. Epidemiology of testicular cancer: an overview. Int J Cancer. 2005;116:331–339. doi: 10.1002/ijc.21032. [DOI] [PubMed] [Google Scholar]
- Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- Gilbert D, Rapley E, Shipley J. Testicular germ cell tumours: predisposition genes and the male germ cell niche. Nat Rev Cancer. 2011;11:278–288. doi: 10.1038/nrc3021. [DOI] [PubMed] [Google Scholar]
- Gutekunst M, Oren M, Weilbacher A, Dengler MA, Markwardt C, Thomale J, Aulitzky WE, van der Kuip H. p53 hypersensitivity is the predominant mechanism of the unique responsiveness of testicular germ cell tumor (TGCT) cells to cisplatin. PLoS ONE. 2011;6:e19198. doi: 10.1371/journal.pone.0019198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz C, Wurst W, Kühn R. Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference. Nucleic Acids Res. 2007;35:e90. doi: 10.1093/nar/gkm475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes L, Jr, Escalante C, Garrison O, Foldi BX, Ogungbade GO, Essien EJ, Ward D. Testicular cancer incidence trends in the USA (1975–2004): plateau or shifting racial paradigm? Public Health. 2008;122:862–872. doi: 10.1016/j.puhe.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldsworth J, Xiao H, Murty VV, Chen W, Ray B, Reuter VE, Bosl GJ, Chaganti RS. Human male germ cell tumor resistance to cisplatin is linked to TP53 gene mutation. Oncogene. 1998;16:2345–2349. doi: 10.1038/sj.onc.1201770. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Evan GI. p53—a Jack of all trades but master of none. Nat Rev Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, et al. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Brown A, Campbell R, Peterson A, Rossant J. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature. 1988;335:435–437. doi: 10.1038/335435a0. [DOI] [PubMed] [Google Scholar]
- Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- Lennartsson J, Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Tied up in loops: positive and negative autoregulation of p53. Cold Spring Harb Perspect Biol. 2010;2:a000984. doi: 10.1101/cshperspect.a000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- Lutzker SG. P53 tumour suppressor gene and germ cell neoplasia. APMIS. 1998;106:85–89. doi: 10.1111/j.1699-0463.1998.tb01323.x. [DOI] [PubMed] [Google Scholar]
- Lutzker SG, Levine AJ. A functionally inactive p53 protein in teratocarcinoma cells is activated by either DNA damage or cellular differentiation. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Masters JR, Köberle B. Curing metastatic cancer: lessons from testicular germ-cell tumours. Nat Rev Cancer. 2003;3:517–525. doi: 10.1038/nrc1120. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moul JW, Schanne FJ, Thompson IM, Frazier HA, Peretsman SA, Wettlaufer JN, Rozanski TA, Stack RS, Kreder KJ, Hoffman KJ. Testicular cancer in blacks. A multicenter experience. Cancer. 1994;73:388–393. doi: 10.1002/1097-0142(19940115)73:2<388::aid-cncr2820730225>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Murase D, Hachiya A, Amano Y, Ohuchi A, Kitahara T, Takema Y. The essential role of p53 in hyperpigmentation of the skin via regulation of paracrine melanogenic cytokine receptor signaling. J Biol Chem. 2009;284:4343–4353. doi: 10.1074/jbc.M805570200. [DOI] [PubMed] [Google Scholar]
- Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M, Kivioja T, Ignatiev I, Kel A, Taipale J, Selivanova G. Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ. 2012;19:1992–2002. doi: 10.1038/cdd.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noureddine MA, Menendez D, Campbell MR, Bandele OJ, Horvath MM, Wang X, Pittman GS, Chorley BN, Resnick MA, Bell DA. Probing the functional impact of sequence variation on p53-DNA interactions using a novel microsphere assay for protein-DNA binding with human cell extracts. PLoS Genet. 2009;5:e1000462. doi: 10.1371/journal.pgen.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HQ, Hogg D, Malkin D, Bailey D, Gallie BL, Bulbul M, Jewett M, Buchanan J, Goss PE. Mutations of the p53 gene do not occur in testis cancer. Cancer Res. 1993;53:3574–3578. [PubMed] [Google Scholar]
- Pietenpol JA, Tokino T, Thiagalingam S, el-Deiry WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintás-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, Lozano G. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, et al. UK Testicular Cancer Collaboration. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RK, McCurtis JW, Henderson BE, Menck HR, Mack TM, Martin SP. Descriptive epidemiology of testicular and prostatic cancer in Los Angeles. Br J Cancer. 1979;39:284–292. doi: 10.1038/bjc.1979.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schödel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, Tomlinson IP, Ratcliffe PJ, Mole DR. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–425. S1–S2. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tan SJ, Zhong H, Hu W, Levine A, Xiao CJ, Peng Y, Qi XB, Shou WH, Ma RL, et al. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet. 2009;84:534–541. doi: 10.1016/j.ajhg.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, Lohrum M. Role of p53 serine 46 in p53 target gene regulation. PLoS ONE. 2011;6:e17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Sur IK, Hallikas O, Vähärautio A, Yan J, Turunen M, Enge M, Taipale M, Karhu A, Aaltonen LA, Taipale J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science. 2012;338:1360–1363. doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- Terzian T, Torchia EC, Dai D, Robinson SE, Murao K, Stiegmann RA, Gonzalez V, Boyle GM, Powell MB, Pollock PM, et al. p53 prevents progression of nevi to melanoma predominantly through cell cycle regulation. Pigment Cell Melanoma Res. 2010;23:781–794. doi: 10.1111/j.1755-148X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, et al. UK Testicular Cancer Collaboration. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GJ, Kimlin MG, Hacker E, Ravishankar S, Muller HK, Beermann F, Hayward NK. Murine neonatal melanocytes exhibit a heightened proliferative response to ultraviolet radiation and migrate to the epidermal basal layer. J Invest Dermatol. 2009;129:184–193. doi: 10.1038/jid.2008.210. [DOI] [PubMed] [Google Scholar]
- Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.