Abstract
Planarians are flatworms capable of regenerating any missing body region. This capacity is mediated by neoblasts, a proliferative cell population that contains pluripotent stem cells. Although population-based studies have revealed many neoblast characteristics, whether functionally distinct classes exist within this population is unclear. Here, we used high-dimensional single-cell transcriptional profiling from over a thousand individual neoblasts to directly compare gene expression fingerprints during homeostasis and regeneration. We identified two prominent neoblast classes that we named ζ (zeta) and σ (sigma). Zeta-neoblasts encompass specified cells that give rise to an abundant postmitotic lineage including epidermal cells, and are not required for regeneration. By contrast, sigma-neoblasts proliferate in response to injury, possess broad lineage capacity, and can give rise to zeta-neoblasts. These findings present a new view of planarian neoblasts, in which the population is comprised of two major and functionally distinct cellular compartments.
Introduction
Adult stem cells play crucial roles in processes such as tissue turnover and regeneration, but regulatory mechanisms involved in the maintenance and lineage specification of stem cells remain poorly understood. Adult planarians maintain a population of dividing cells with broad differentiation potential, presenting the opportunity to study these processes in vivo.
The seemingly inexhaustible regenerative capacity of planarians has fascinated biologists for well over a century (Reddien and Sánchez Alvarado, 2004). Planarians can repair extensive wounds and replace entire organs such as the brain. Planarian regeneration is mediated by a population of small, proliferative cells (~8µm in diameter) termed “neoblasts” that are dispersed throughout mesenchymal body regions. The neoblast population is essential for regeneration, and contains pluripotent stem cells (“cNeoblasts”) that can give rise to all somatic cell types in the adult body (Wagner et al., 2011).
Studies using general mitotic markers (Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010), whole-body irradiation (Guedelhoefer and Sánchez Alvarado, 2012; Reddien et al., 2005b; Wolff and Dubois, 1948), and BrdU labeling (Eisenhoffer et al., 2008; Newmark and Sánchez Alvarado, 2000), have yielded population-level analyses of neoblast proliferation, dynamics, and collective cellular output. Furthermore, transcriptional analyses have identified many neoblast-enriched transcripts (Blythe et al., 2010; Eisenhoffer et al., 2008; Galloni, 2012; Labbe et al., 2012; Onal et al., 2012; Resch et al., 2012; Shibata et al., 2012; Solana et al., 2012; Wagner et al., 2012) including those encoding several Sox and Pou transcription factors, gene families with important roles in stem cell maintenance in other organisms. Neoblasts also express transcripts for the PIWI proteins Smedwi-1 and Smedwi-2 (Reddien et al., 2005b), the Bruno-like protein Bruli (Guo et al., 2006), and a Tudor protein (Solana et al., 2009). These proteins are typically found in association with nuage, an electron-dense perinuclear organelle present in germ cells, which plays a role in transposon silencing and maintenance of genome integrity (Voronina et al., 2011). Interestingly, several of these nuage-related genes are required for maintenance of the neoblast population (Guo et al., 2006; Reddien et al., 2005b; Salvetti et al., 2005; Solana et al., 2009).
Previous studies have thus provided important insights into the general features of neoblasts. However, they also reinforced the historic notion that neoblasts represent a single homogeneous stem cell population. Recently, gene expression signatures specific to differentiated tissues were shown to mark small numbers of lineage-specified neoblasts during regeneration (Cowles et al., 2013; Currie and Pearson, 2013; Lapan and Reddien, 2011; Scimone et al., 2011) and in homeostasis (Hayashi et al., 2010; Moritz et al., 2012). In addition, neoblasts can be subdivided with respect to an otherwise uncharacterized surface antigen (Moritz et al., 2012) or a trans-splicing sequence (Rossi et al., 2014). However, as a consequence of small numbers and/or lack of functional characterization, it remains unknown whether the above phenomena reflect the presence of substantial cellular classes within the neoblast population.
Multi-dimensional single-cell transcriptional profiling is a powerful approach to distinguish true cellular heterogeneity from biological noise and has facilitated the deconvolution of heterogeneous cell populations across a wide range of biological systems. Here, we for the first time used this approach on neoblasts, comparing gene expression fingerprints from over a thousand individual cells under different conditions. We show that neoblasts can be divided into multiple prominent classes, characterized by co-regulated transcript sets and displaying distinct regenerative properties. We propose that neoblasts are a heterogeneous pool of cells, containing both pluripotent and prominent lineage-committed subsets.
Results
Selection of markers for single neoblast profiling
To molecularly profile individual neoblasts of Schmidtea mediterranea, we performed parallel single-cell qPCR on the Biomark (Fluidigm) platform (Figure S1A–F), analyzing 96 genes from each cell. Genes were selected from a de novo neoblast transcriptome (accession SRP042226) and included nuage-related neoblast markers (smedwi-1, smedwi-2, smedwi-3, Smed-vasa-1, Smed-vasa-2, Smed-tdrd1L2, pumilio, and Smed-bruli (Guo et al., 2006; Palakodeti et al., 2008; Reddien et al., 2005b; Salvetti et al., 2005; Solana et al., 2009; Wagner et al., 2012)), cell cycle regulators (Smed-rb, Smed-pcna, Smed-mcm2 and Smed-cyclinB (Reddien et al., 2005a; Salvetti et al., 2000; Zhu and Pearson, 2013)), markers of post-mitotic planarian cell types (Smed-prog-1, Smed-prog-2, Smed-porcn-a and Smed-mhc1 (Eisenhoffer et al., 2008; Pearson et al., 2009; Wagner et al., 2012)), reference genes (Smed-g6pd, Smed-ubiquilin, Smed-luc7-like, and Smed-clathrin), and 16 other genes with enriched expression in neoblasts (Wagner et al., 2012) (Figure S1H, Table S1). The majority of selected genes consisted of the 55 most abundantly expressed transcription factors and transcriptional regulators in our neoblast transcriptome.
Global expression patterns of selected genes were assessed by whole-mount in situ hybridization and by RNAseq analysis of isolated cell populations (Figure S1H). These analyses showed that although the selected transcripts were all present in neoblasts, they were not necessarily enriched in these cells.
Gene expression profiling divides neoblasts into two major classes
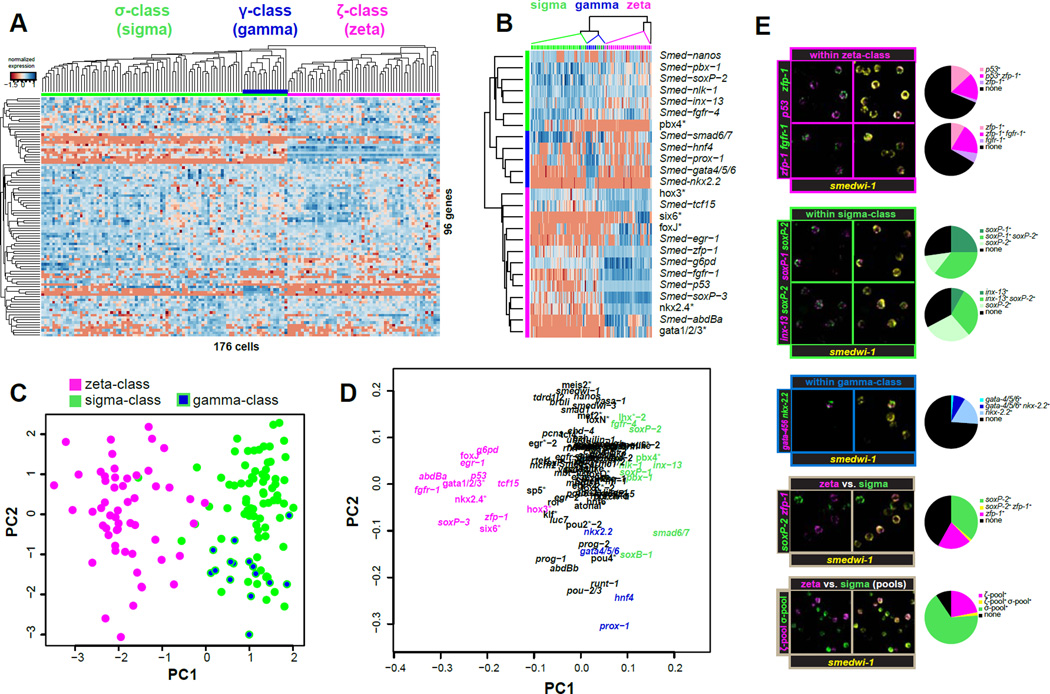
We used fluorescence activated cell sorting (FACS) (Hayashi et al., 2006) to isolate individual neoblasts with 4C DNA content (X1(4C)) from the prepharyngeal region of intact worms for single-cell transcriptional analysis (Figure S1A–D). Hierarchical clustering (HC) of the cells based on their gene expression profiles revealed that neoblasts comprise two major, roughly equally sized populations (Figure 1A, Figure S1G). One population, the zeta-class (written as “zeta-class” or “ζ-class”), marked in magenta, expressed high levels of a discrete set of genes (e.g., Smed-zfp-1, Smed-g6pd, Smed-fgfr-1, Smed-p53, Smed-soxP-3, and Smed-egr-1); the other population, the sigma-class (written as “sigma-class” or “σ-class”), marked in green, expressed low levels of those genes, but had elevated expression of Smed-soxP-1, Smed-soxP-2, Smed-soxB-1, Smed-smad-6/7, Smed-inx-13, Smed-pbx-1, Smed-fgfr-4, and Smed-nlk-1. Within these two major classes, at least one subclass was identified: the gamma-class (“γ-class”) marked in blue, was readily discerned as a subclass within the sigma class and expressed high levels of Smed-gata4/5/6, Smed-nkx2.2-like, Smed-hnf4, and Smed-prox-1 (see Figure S1G for description of further subclasses).
Figure 1. Single cell transcriptional profiling reveals neoblast classes.
A. Normalized expression and hierarchical Clustering (HC) of 176 individual X1(4C) cells isolated by FACS and profiled for 96 transcripts by qPCR. High and low expression relative to a reference sample is indicated by blue and red shades, respectively. Clustering of cells and genes was guided by Pearson correlation. Top colored bar indicates class membership: ζNeoblasts (magenta), σNeoblasts (green), and γ-subclass (blue).
B. Simplified heatmap based on the 25 most informative genes for class membership, as determined by ANOVA. Left color bar indicates transcript class enrichment. Transcripts marked with asterisks are annotated based upon top BLASTx hit.
C. Principle Component Analysis (PC A) of the full qPCR results on 176 cells from the X1(4C) gate. Each dot represents a cell, colored according to its class as determined by HC. Cells are plotted against the first two principle components (PCs). PC1 separates the two classes, indicating that this is the primary source of variation in the dataset. PC2 largely separates the γ-subclass from the remainder of σ-cells.
D. Contributions of each transcript to the first two PC vectors. Genes are colored according to class enrichment.
E. Fluorescent in situ hybridizations (FISH) on isolated X1 cells. Images show a representative confocal plane for each probe pair. Pie charts quantify the percentage of smedwi-1 positive cells labeled with single, both, or neither FISH probe. >500 cells were analyzed per probe pair. Use of pooled FISH probes (bottom panel) improves detection and reduces the proportion of unclassifiable cells (black pie wedges).
Feature reduction by ANOVA revealed a reduced set of markers (primarily transcription factors) with high differential expression between the classes (Figure 1B), and HC based on the 25 most discriminating genes correctly assigned the majority of cells to their classes. Principle Component Analysis (PCA) was used as an independent method to reduce data complexity, and identified the differences between the sigma- and zeta-neoblasts as the primary source of variance in the expression data (Figure 1C). Moreover, the subset of transcripts contributing the majority of the variance was similar to that discovered by ANOVA (Figure 1D).
Fluorescent in situ hybridization (FISH) on FACS-isolated 4C cells, using smedwi-1 as a ubiquitous neoblast marker, confirmed the largely overlapping expression of transcripts within each class, as well as the non-overlapping expression of transcripts between classes (Figure 1E). Because many transcripts are expressed at low levels and because no single transcript can reliably label all members of a class, we assembled RNA probe pools for improved class detection by FISH. Balancing signal intensity, class specificity, and neoblast specificity, we pooled zfp-1, fgfr-1, soxP-3, and egr-1 probes for zeta-neoblasts, and soxP-1 and soxP-2 probes for sigma-neoblasts. Indeed, probe mixtures displayed non-overlapping expression and improved overall class detection (Figure 1E).
Neoblast classes are not defined by cell cycle state
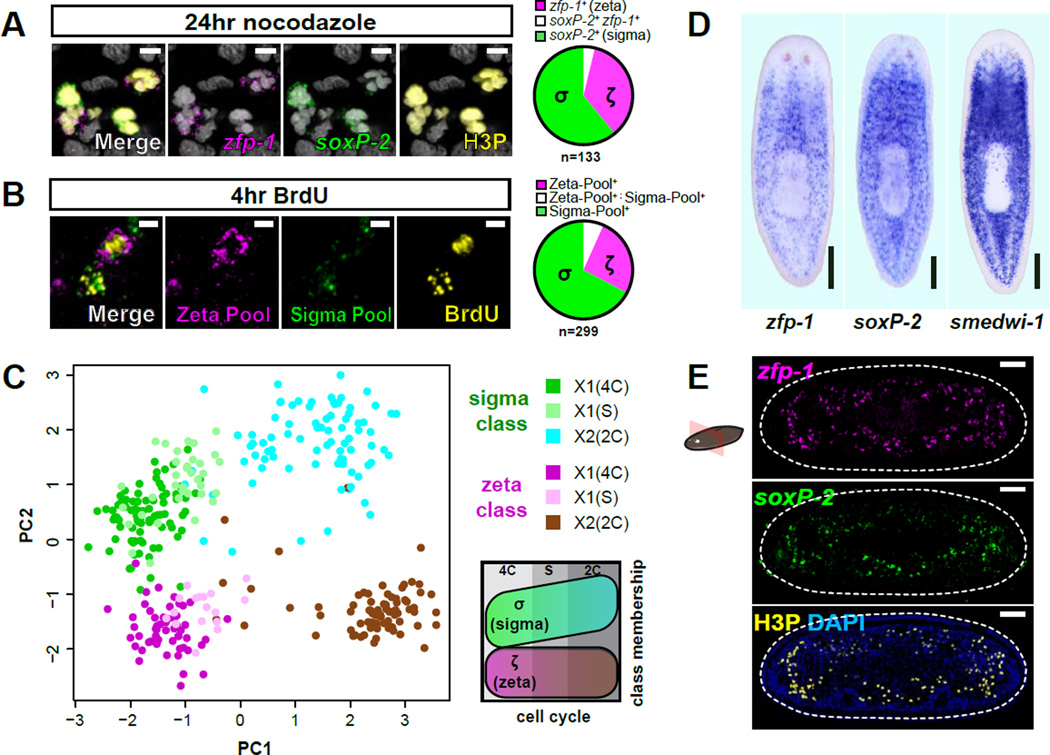
The two identified neoblast classes could reflect different cell cycle states within an otherwise homogenous 4C cell population, namely G2-phase and M-phase. FISH analysis of animals treated with the M-phase blocker Nocodazole (Figure S2D), however, showed that cells of each class were co-labeled with the mitotic marker H3P (histone H3 phosphorylated on Serine 10), indicating that both classes are present among M-phase cells (Figure 2A, Figure S2E). Similarly, each class was rapidly co-labeled with bromodeoxyuridine (BrdU), a thymidine analog that is incorporated into newly synthesized DNA (Figure 2B, Figure S2F), indicating that both classes are also present during S-phase.
Figure 2. Neoblast classes do not reflect anatomical localization or cell cycle state.
A. Representative confocal plane from prepharyngeal region after Nocodazole treatment. FISH of ζ-class (zfp-1) and σ-class (soxP-2) shows colabeling with phosphorylated histone H3 (H3P) immunofluorescence (IF). Pie chart indicates class distribution for 133 mitotic cells (from 5 individual animals) that displayed detectable levels of class marker transcripts (n=308 total H3P+ cells analyzed). Scale bars, 5µm. Nuclei are labeled by DAPI (grey).
B. Representative confocal plane from a planarian subjected to a 2hr BrdU pulse and 2hr chase and labeled by pooled class FISH probes and BrdU IF. Pie chart indicates class distribution for 299 BrdU+ cells (from 3 individual animals) with detected class marker expression (n=402 BrdU+ cells analyzed). Scale bars, 5µm.
C. PCA of qPCR data from individual neoblasts isolated from cell cycle-restricted FACS gates. Each dot represents a cell, colored based on its FACS gate and class membership (determined by HC). PC1 separates cell cycle stages whereas PC2 separates the classes. Schematic illustrates the distribution of cell cycle stages and classes over the plot area.
D. Whole-mount ISH comparing spatial distributions of ζNeoblasts (zfp-1) and σ Neoblasts (soxP-2) to the global neoblast population (smedwi-1). Ventral views, anterior up; scale bar, 200µm.
E. Transverse tissue section labeled by whole-mount double-FISH shows the spatial distribution of CNeoblasts (zfp-1) and σNeoblasts (soxP-2). Mitotic cells are labeled by IF for H3P. Dotted line indicates animal boundary. Dorsal up; scale bar, 50µm.
We next used FACS to isolate additional cell-cycle-restricted populations according to DNA content: 2C irradiation-sensitive cells (“X2 cells”, which encompass a mixture of cells in G0 or G1-phases and early post-mitotic cells) and cells with DNA content between 2C and 4C (cells in S-phase, “X1(S)”, Figure S1D). Single-cell profiling showed that although levels of several nuage-related neoblast markers (such as smedwi-1, vasa-1, vasa-2, tdrd1L2, and bruli) and all cell cycle markers (rb, pcna, mcm2, cyclinB) were correlated with cell cycle stage, the σ- and ζ-class remained prominent in all three stages analyzed (Figure S2G–I). In addition, PCA showed that the primary variance vector (PC1) of the gene expression fingerprints correlated with cell cycle stage, but the second independent vector (PC2) correlated with class membership (Figure 2C).
Together these data indicate that the σNeoblasts and the ζNeoblasts reflect two separate populations that remain present throughout the cell cycle. This represents the first identification of major cell cycle-independent classes within the neoblast population.
Neoblast classes are not spatially restricted
We tested whether the two major neoblast classes are correlated with anatomical position using FISH probes with expression restricted to the smedwi-1+ population (zfp-1 for zeta and soxP-2 for sigma) (Figure 2D Figure S2A–C). Cells of both classes displayed a similar smedwi-1-like distribution (Figure 2D), without obvious differential localization along the anterior-posterior axis, the medial-lateral axis, or the dorsal-ventral axis (Figure 2E, Figure S2A). These observations indicate that the two classes represent two intermingled, distinct cell classes of roughly equal abundance.
Both neoblast classes are present during regeneration
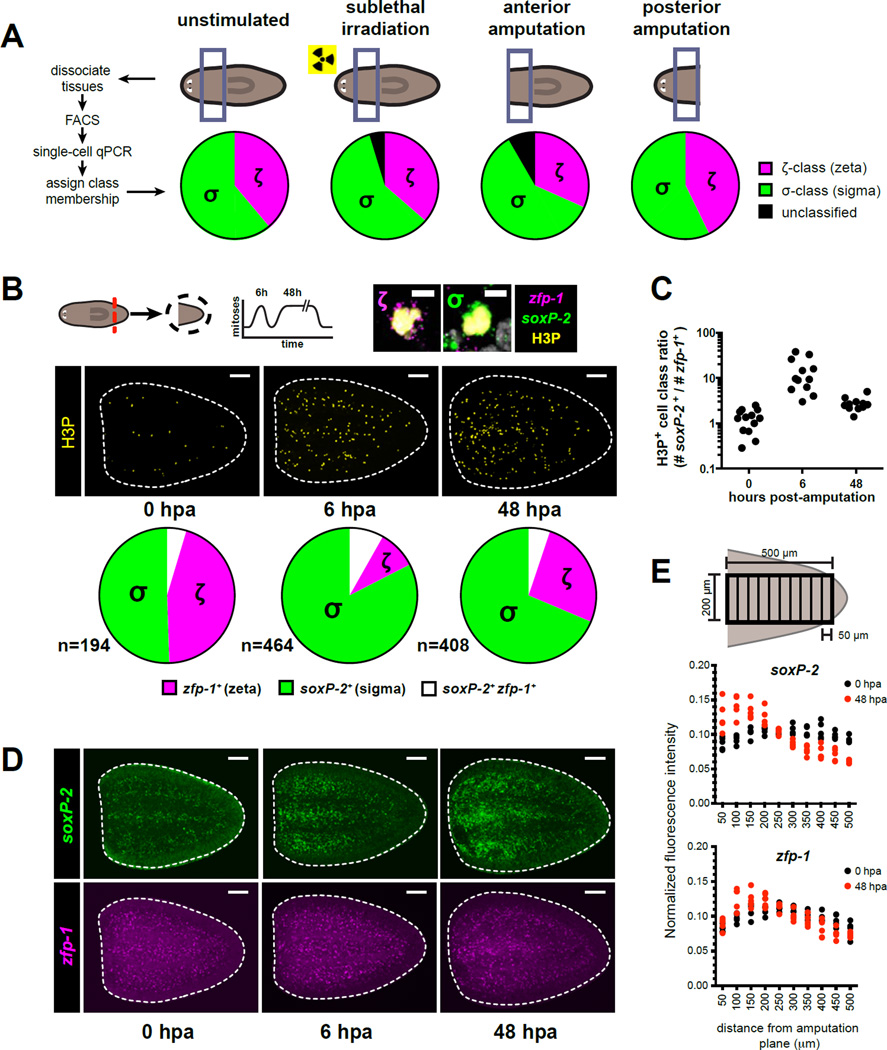
To assess the role of neoblast classes during regeneration, we first determined their presence and abundance during three regenerative challenges: regeneration of anterior or posterior body regions (assessed 48 hours after amputation), or repopulation of the neoblast compartment (assessed 10 days after sublethal irradiation). Profiling of single X1(4C) cells revealed several global changes in the levels of individual transcripts, such as upregulation of several nuage-related neoblast markers (smedwi-1, smedwi-2, smedwi-3, vasa-1, tdrd1L2), and downregulation of rtel-1 and junli-1 (Figure S3A–D). However, HC and PCA each identified the σ- and the ζ-classes in the irradiated sample and in both amputation samples, without significant changes in their relative abundances (Figure 3A).
Figure 3. Neoblasts undergoing early responses to injury are enriched for the σ-class.
A. X1 cells from the prepharyngeal region were profiled by single-cell qPCR for 96 transcripts on day 10 after sublethal irradiation (1,250 Rads), or on day 2 after anterior or posterior amputation. Pie charts illustrate distribution of neoblast class assignments (determined by HC) for >90 individual cells per condition.
B. Representative confocal projections through tail fragments fixed 0, 6, and 48hr post-amputation (hpa) labeled by H3P IF together with double FISH for zeta (zfp-1) and sigma (soxP-2) class markers (panel D). Dotted lines: tail fragment boundary. Amputation plane is left. Pie charts indicate proportions of H3P+ cells expressing detectable levels of soxP-2 (σ-class), zfp-1 (ζ-class), or both transcripts as green, magenta, and white pie wedges. Total number of cells represented in each pie chart is indicated. Zoomed examples of ζ-class and σ-class mitotic cells are displayed at the upper right. Scale bars, 100µm (tail fragments) and 5µm (zoomed panels).
C. Dots show the proportions of soxP-2+ to zfp-1+ H3P+ cells for each specimen at each post-amputation timepoint described in (B), indicating similar trends across all specimens included in the analysis.
D. Confocal projections of FISH labeling for amputated tail fragments described in (B). Scale bars, 100µm.
E. Quantification of FISH signal intensities for ζ- and σ-class markers displayed in (D) at 0hr and 48hr post-amputation. Normalized FISH signal intensities (see Supplemental Methods) are plotted for >4 individual tail fragments across 50 µm bins extending from the amputation plane towards the posterior.
The σNeoblasts dominate the response to wounding
The regenerative response in S. mediterranea involves two mitotic waves: a global increase in mitosis that peaks at 6 hours after wounding, and a more localized mitotic response at 48 hours after wounding (Wenemoser and Reddien, 2010). In addition, neoblasts localize to wounds approximately 48 hours after injury (Wenemoser and Reddien, 2010). Although both neoblast classes were stably present throughout regeneration, σNeoblasts were substantially overrepresented among mitotic cells 6hr after amputation (P<0.0001, 2-tailed Fisher's Exact Test) (Figure 3B,C). At 48 hours, the σ-class still represented the largest fraction of cells undergoing mitosis. We conclude that the increase in neoblast proliferation early after wounding can be largely attributed to mitotic activity of σNeoblasts. Furthermore, at 0 hours post-amputation, cells from both classes were approximately equally distributed across tail fragments (Figure 3D,E). At 6 to 48 hours post-amputation, cells of the ζ-class were still evenly distributed throughout the amputated fragment. By contrast, σ-class cells were enriched near the wound (Figure 3D,E), indicating that wound-site accumulation of neoblasts mainly involves σNeoblasts. Together, these experiments indicate that the early cellular responses to wounds are primarily mediated by σNeoblasts.
zfp-1 RNAi specifically eliminates the ζNeoblasts
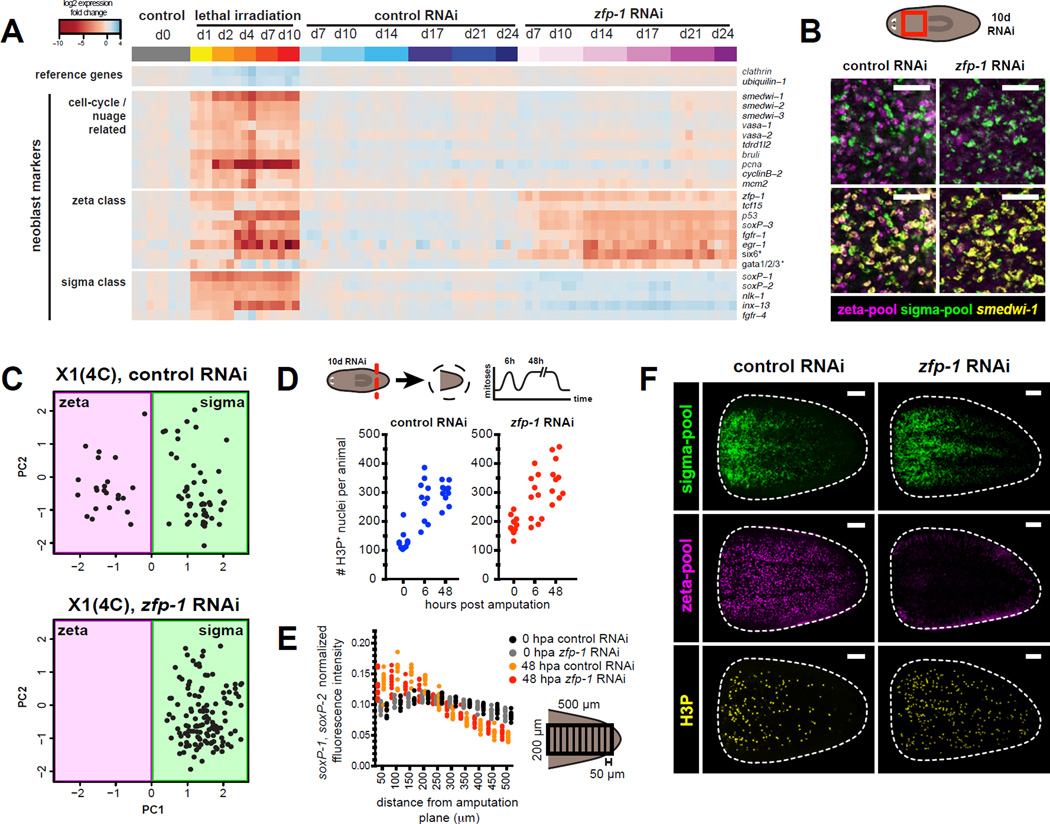
zfp-1 expression is restricted to neoblasts (Figure S2B,C) and is specific to the ζ-class (Figure 1A,B). Notably, zfp-1 RNAi causes animal lethality, with only a modest reduction in neoblast numbers (Figure S4A–D and (Wagner et al., 2012)). We characterized the effect of zfp-1 RNAi on the neoblast population for over three weeks using qPCR. Significant zfp-1 transcript reduction was evident within 10 days of RNAi (Figure 4A), followed by a reduction of other ζ-class transcripts. By contrast, general neoblast markers and σ-class transcripts remained unaffected for up to 24 days of zfp-1 RNAi. FISH (Figure 4B, Figure S4E,F) and RNAseq (Figure S4G) confirmed that zfp-1 RNAi resulted in the specific depletion of zfp-1 and other ζ-class transcripts.
Figure 4. zfp-1 RNAi eliminates the ζ-class without affecting the σ-class.
A. qPCR time course of transcriptional changes following zfp-1 RNAi. Top bar indicates treatment and timepoint: control RNAi in blue, zfp-1 RNAi in magenta, and lethal irradiation in orange. Biological replicates (10,000 live cells each) were isolated by FACS. Heatmap shows log2 fold changes in expression levels (relative to day 0) after global median normalization.
B. Whole-mount FISH for ζ- and σ-class probe pools together with smedwi-1. Shown is a confocal projection from the prepharyngeal region of the animal fixed after 10 days of RNAi. Scale bars, 50µm.
C. Analysis of single-cell expression data obtained by qPCR of 96 transcripts. Dots represent individual X1 cells from animals exposed to 10 days of zfp-1 or control RNAi, projected onto PC1 and PC2 vectors obtained by PC A of untreated X1 cells. Background shades indicate class membership.
D. Tail fragments cut from 10-day zfp-1 (RNAi) and control RNAi animals were fixed at 0hr, 6hr, and 48hr post-amputation. Numbers of mitotic events observed in confocal projections through entire tail pieces are plotted.
E–F. Effects of zfp-1 RNAi on wound-induced accumulation of neoblasts. ζ- and σ-class cells are labeled with pooled FISH probes together with IF for H3P. E. FISH signal intensities are quantified as in Figure 3E F. Representative confocal projections from tail fragments fixed 48 hours post-amputation. Amputation plane is at left. Scale bars, 100µm.
We next isolated X1(4C) cells from zfp-1 (RNAi) animals for single-cell qPCR analysis and used PCA to determine class membership. Both control and zfp-1 (RNAi) samples contained cells with a σ-class signature (Figure 4C). By contrast, ζ-class cells were absent from the zfp-1 (RNAi) sample, leaving a population consisting entirely of σNeoblasts. Based on these FISH, RNAseq, and bulk and single-cell qPCR data, we conclude that zfp-1 RNAi leads to specific depletion of ζNeoblasts, while leaving the σNeoblasts largely unaffected.
σNeoblasts function normally in the absence of ζNeoblasts
The ability to specifically deplete ζNeoblasts by zfp-1 RNAi presents an opportunity to study the properties of σNeoblasts in isolation. We thus examined the regenerative response in amputated tail fragments from zfp-1 (RNAi) animals. The peaks in mitoses at 6 and 48 hours after wounding (mediated primarily by σNeoblasts) were not significantly altered in zfp-1 (RNAi) animals (Figure 4D). Furthermore, σNeoblast localization to wounds 48 hours after amputation was similar to that of control animals (Figure 4E,F). These data demonstrate that σNeoblasts undergo mitosis in the absence of the ζNeoblasts and remain capable of mounting a normal regenerative response.
Neoblasts maintain lineage potential in the absence of ζNeoblasts
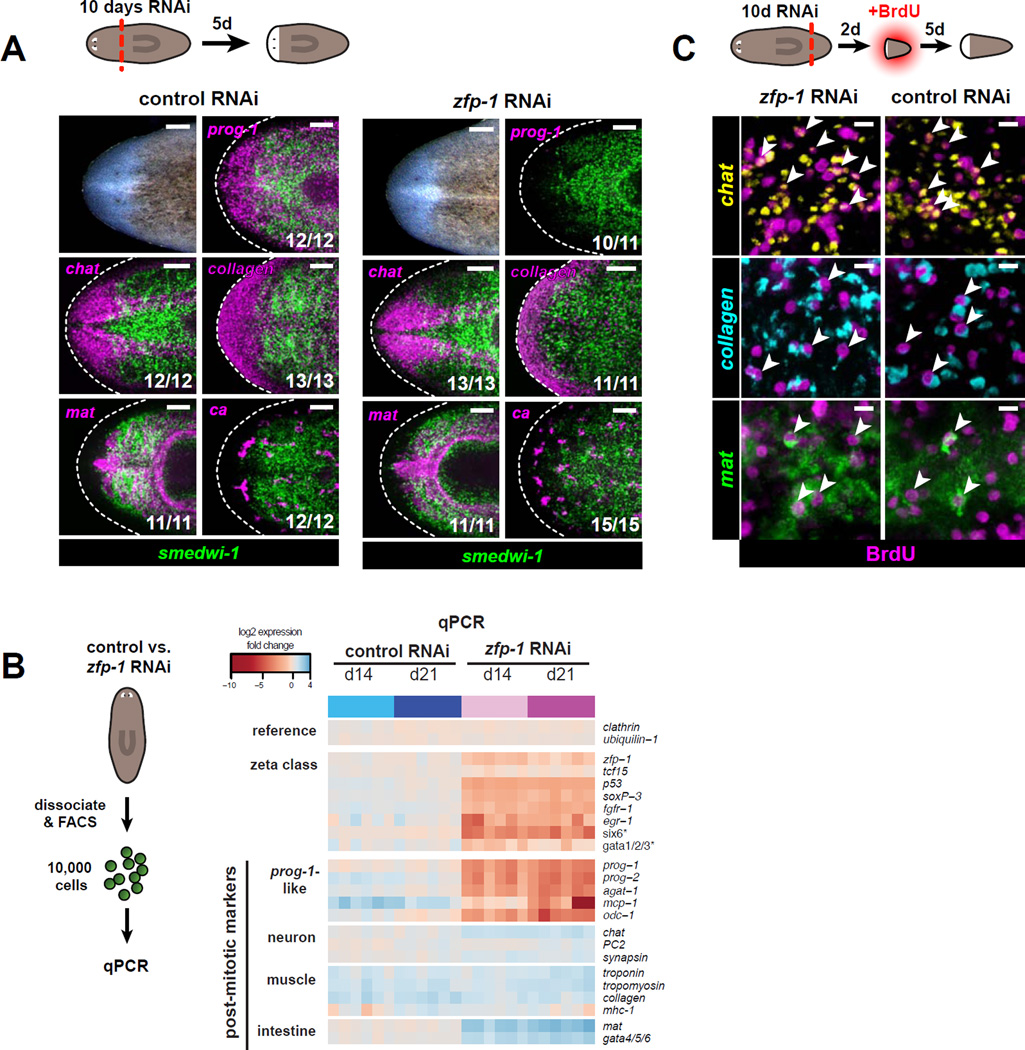
Strikingly, animals devoid of ζNeoblasts (10 days of zfp-1 RNAi) were still able to generate a blastema following amputation (Figure 5A). We used whole-mount FISH to examine the tissue composition of zfp-1(RNAi) head blastemas. Previous work (Eisenhoffer et al., 2008) described several related post-mitotic cell populations (neoblast progeny) characterized by expression of specific marker genes (e.g., prog-1 and AGAT-1). These “ prog-1-related” cell populations are located subepidermally, experience rapid turnover, and share common genetic requirements (Pearson and Sánchez Alvarado, 2010; Scimone et al., 2010). Consistent with earlier observations (Wagner et al., 2012), we found that prog-1 and all other markers of these related populations were specifically depleted upon zfp-1 RNAi during regeneration as well as during homeostasis (Figure 5A,B, Figure S5A). By contrast, markers of other tissues remained detectable in zfp-1 (RNAi) regenerating animals; brain (chat), intestine (mat), muscle (collagen-2), protonephridia (ca), eyes (ovo-1), and pharynx (mhc-1) were all present and appeared morphologically normal (Figure 5A,B, Figure S5C). These new structures could have been formed either by newly dividing and differentiating cells, or from pre-existing post-mitotic cells. To distinguish between these possibilities, we assessed de novo incorporation of BrdU into blastemas. Five days post-amputation, each of the differentiated tissues analyzed contained BrdU+ cells (Figure 5C), indicating that these tissues were newly derived from a ζ-class-depleted neoblast population. Together, these data indicate that the σNeoblasts, as a population, can form a broad range of tissues without contribution from the ζ-class.
Figure 5. Neoblasts maintain the ability to form blastemas and regenerate multiple tissue types in the absence of the ζ-class.
A. Regenerated head blastemas from 10-day RNAi animals were analyzed 5 days post-amputation. Tissues are labeled by RNA probes for ca (nephridia), chat (CNS), collagen (muscle), mat (intestine), prog-1, and smedwi-1 (neoblasts). Numbers of animals with displayed tissue patterns are indicated. Dotted lines indicate animal boundary. Anterior, left. Scale bars, 100 µm.
B. Transcriptional changes in ζ-class transcripts and post-mitotic tissue markers following zfp-1 RNAi, analyzed by qPCR. Biological replicates (10,000 live cells each) were isolated by FACS after control RNAi (blue) or zfp-1 RNAi (magenta). Heatmap shows log2 fold changes in expression levels (relative to day 0) after global median normalization.
C. Tail fragments cut from 10-day RNAi animals were soaked in BrdU starting 2 days post-amputation and fixed 5 days later. Specimens were assessed by triple FISH for chat (CNS), collagen (muscle), and mat (intestine), together with BrdU IF. Representative confocal projections through head blastemas regenerated following zfp-1 or control RNAi are shown. BrdU-positive cells expressing each tissue marker are indicated by arrowheads. Scale bars, 10 µm.
ζNeoblasts are involved in epidermal maintenance and regeneration
To identify any tissues that might derive from ζNeoblasts, we analyzed 5-day regenerating head blastemas of zfp-1(RNAi) animals by RNAseq. As expected, prog-1-related markers were severely reduced and transcripts characteristic of eye, brain, intestine, protonephridia, and muscle were unaltered (Figure S6A). Notably, several transcripts associated with epidermis, cilia, and secretory cells were reduced in the zfp-1 (RNAi) blastemas.
We selected 20 transcripts that were severely (> 16-fold) reduced in zfp-1 (RNAi) blastemas and highly expressed in the controls for whole-mount ISH. Sixteen transcripts revealed interpretable expression patterns (Figure 6A, Figure S6B). Nine of the transcripts were detected in sparse subepidermal cells identical or similar to prog-1-expressing cells. The other seven transcripts were present in the epidermis, either globally, or localized to specific epidermal domains. FISH analysis confirmed the epidermal localization of several of these transcripts (Figure 6B) and demonstrated that expression of these genes failed to be fully re-established in zfp-1 (RNAi) blastemas (Figure 6C).
Figure 6. The ζ-class is involved in epidermal regeneration.
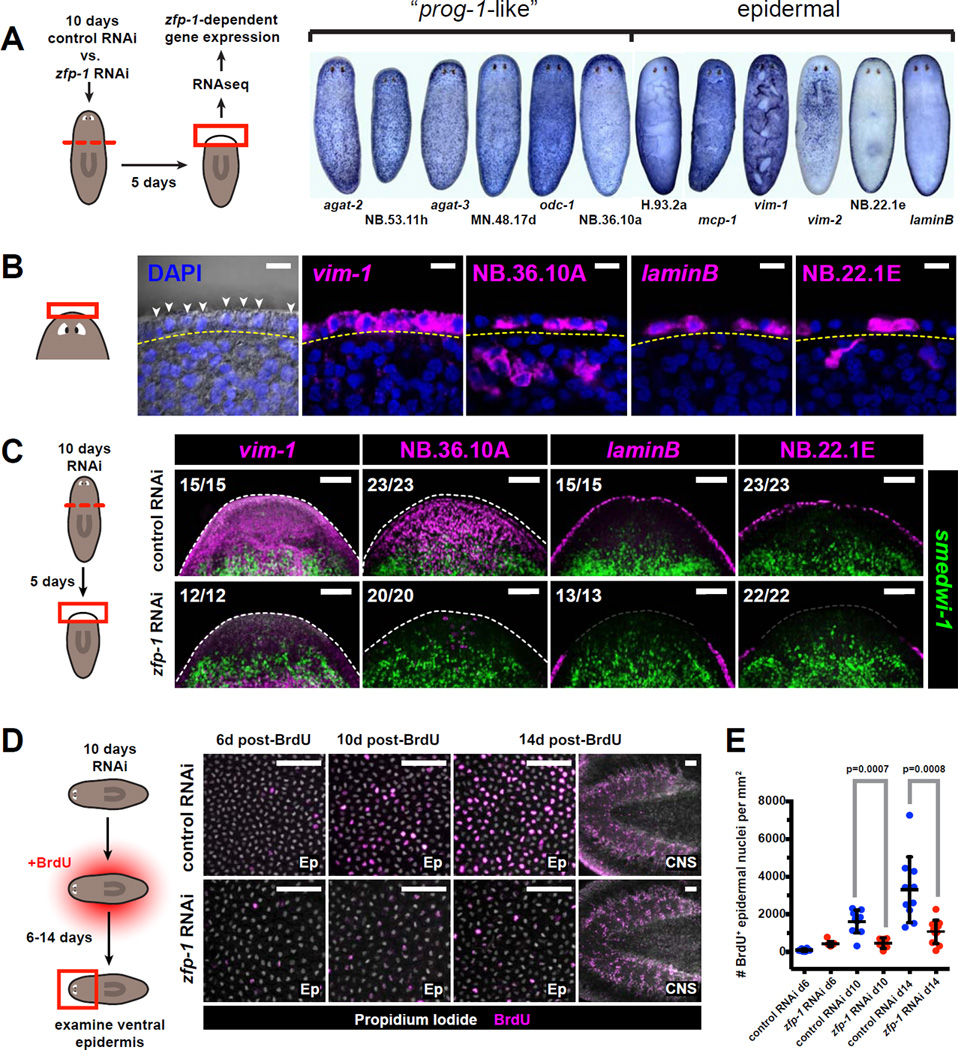
A. Schematic for RNAseq strategy used to identify transcripts differentially expressed in zfp-1 (RNAi) vs. control blastemas. Whole-mount ISH patterns for several transcripts identified by this strategy are shown at right.
B. Several genes identified in (A) are expressed in the planarian epidermis. Left panel shows epidermal nuclei (arrowheads) in the head rim, distal to the basement membrane (yellow dotted line) as visualized by differential interference contrast (DIC) microscopy. Right panels are confocal planes from animals labeled by FISH. Labeled cells are found distal to the basement membrane. Scale bars, 10µm.
C. Tissue fragments cut from 10-day RNAi animals were fixed 5 days post-amputation and assessed by double-FISH. Total numbers of animals with displayed tissue patterns are indicated. Dotted lines indicate animal boundary. Scale bars, 100µm.
D. Effects of zfp-1 RNAi on epidermal cell turnover. Images depict the ventral epidermis from anterior regions of RNAi animals 6–14 days after BrdU labeling. Rightmost images depict CNS tissues, in which cell turnover is not affected by zfp-1 RNAi. Scale bars, 50µm.
E. Numbers of BrdU+ epidermal cells per mm2 are plotted for each timepoint and RNAi condition. Dots represent individual animals. Lines and error bars indicate mean and standard deviation. P-values are based on unpaired Student’s t-test (two-tailed).
These results raise the possibility that ζNeoblasts are required for epidermal regeneration. Assessment of global epidermal morphology using the lectin ConcanavalinA (Zayas et al., 2010) showed that the epidermal layer of zfp-1 (RNAi) blastemas was disorganized and contained fewer and thinner cells compared to the control (Figure S6C,D). Prolonged zfp-1 RNAi in the absence of injury resulted in similar epidermal defects as seen upon abrogation of cell turnover by lethal irradiation (Figure S6E–G), suggesting that the epidermal phenotype of zfp-1(RNAi) could be caused by reduced epidermal cell renewal.
We used BrdU labeling to quantify new cell entry into the epidermis. Two weeks after BrdU incorporation, significantly fewer BrdU+ cells were detected in the epidermis of zfp-1 (RNAi) animals as compared to controls (Figure 6D,E). These results confirm that the ζNeoblasts give rise to a lineage that at least in part includes prog-1-related cells, and ultimately is involved in the maintenance and/or differentiation of an epidermal cell type.
The σ-class is collectively pluripotent and regenerates the ζ-class
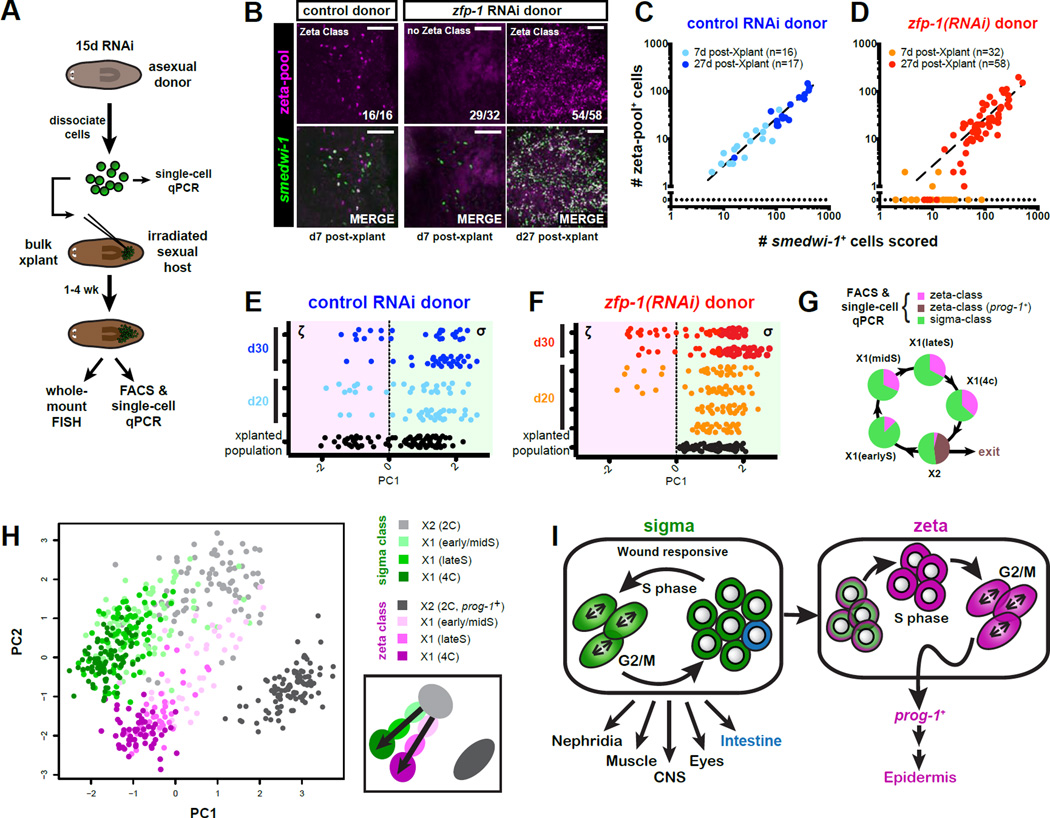
To assess the long-term renewal capabilities of the σ-class we performed bulk transplantation of RNAi-treated neoblasts into neoblast-depleted hosts (outlined in Figure 7A), and assessed outcomes of over a hundred separate transplantations by FISH. Neoblasts from both control and zfp-1 (RNAi) donors proliferated robustly (Figure 7B–D), and grafts derived from control donors maintained a consistent ratio of ζ- to σ-class cells at all timepoints (Figure 7B,C). By contrast, the vast majority of zfp-1(RNAi)-derived grafts (29/32) lacked any detectable ζ-class gene expression at 7 days post-transplantation (the 3 remaining grafts contained 1 or 2 ζ-class cells) (Figure 7B,D), whereas by 27 days post-transplantation, most of these neoblast grafts (54/58) did contain ζ-class cells. These data demonstrate that zfp-1(RNAi)-derived grafts re-established ζ-class cells post-transplantation (P<0.0001, 2-tailed Fisher’s Exact Test). Single-cell transcriptional profiling of several neoblast grafts confirmed that hosts transplanted with zfp-1(RNAi) donor cells, initially received only σNeoblasts but could re-establish a ζ-class as well as prog-1+ neoblast progeny (Figure 7E,F, Figure S7A,B). These data indicate that σ-class cells have long-term renewal capacity and can generate ζ-class cells.
Figure 7. The σ-class regenerates the ζ-class.
A. Schematic representation of transplantation procedure. Animals were exposed to control or zfp-1 RNAi for 15 days. A subset of isolated cells was analyzed by single-cell qPCR. Remaining cells were transplanted into lethally irradiated hosts and allowed to expand up to 30 days. Transplanted populations were assessed by single-cell transcriptional profiling or whole-mount FISH.
B. Representative confocal images of neoblast grafts derived from control or zfp-1 (RNAi) donor cells. Hosts were fixed 7 or 27 days post-transplantation, and labeled by FISH probes for smedwi-1 and the ζ-class. ζ-class gene expression was undetected in the majority (29/32) of day 7 grafts from zfp-1 (RNAi) donors, but was robustly detected in grafts (54/58) assessed at day 27. Scale bars, 50µm.
C–D. Log-scale plots of scored smedwi-1+ cells plotted against numbers of cells that co-expressed ζ-class markers for transplant grafts described in (B). Each dot represents an individual graft. Line represents linear regression fit of the control RNAi data.
E–F. PCA of individual X1 cells from transplants at 0, 20, and 30 days post-transplantation. Each row represents a host, each dot represents a cell. Cells from control (blue, E) and zfp-1 (RNAi) grafts (red/orange, F), are plotted against the first PC value, which reflects class membership. Positive PC1 value assigns a cell to the σ-class; a negative value to the ζ-class. zfp-1 (RNAi) grafts contain only σ-class cells at day 0, but at later time points ζ-class cells are detected.
G. Cell-cycle restricted X1 cells were isolated using different FACS gates (see Fig. S7C) and profiled by qPCR for 96 genes. Pie charts indicate class membership (determined by HC).
H. PCA of single-cell qPCR data from different cell cycle-restricted FACS gates. Each dot represents a cell. Colors indicate FACS gate and class membership. PC1 separates according to cell cycle stage; PC2 indicates class membership. Schematic depicts the distribution of cell cycle stages and proposed class transitions. (See also Figure S7C,G)
I. Model of the neoblast population. Two major classes, the σ-class and the ζ-class, represent functionally separate neoblast compartments. σNeoblasts are able to self-renew, and collectively gives rise to a wide range of tissue types. Over the course of S-phase, a subset of the σNeoblasts gains markers specific for theζ-class. These cells give rise to the prog-1-related lineages and to epidermal cells.
If σNeoblasts are indeed the source of ζNeoblasts, transition states between the two classes should be detectable in intact worms. We therefore analyzed changes of the class markers during cell cycle progression. The irradiation-sensitive 2C cells (located in the “X2” FACS gate) are a mixture of cycling cells in G1-phase and post-mitotic cells early after their last division. Single-cell transcriptional profiling indicated that the large majority of ζ-class cells in the X2 gate co-expressed high levels of postmitotic markers prog-1 and prog-2, and reduced levels of neoblast markers smedwi-1, vasa-1, and bruli (Figure S2H), suggesting that the majority of ζ-class cells in the X2 gate might in fact have exited the cell cycle and will not go through another round of division. In order to explore this possibility further we divided S-phase cells into three stages by DNA content (Figure S7C) and performed single-cell profiling on cells from each stage (Figure S7H–L). Two independent lines of evidence suggest that ζNeoblasts are indeed derived from σNeoblasts during S-phase in intact animals. First, the fraction of ζNeoblasts in the stage just entering S-phase (stage with the lowest DNA content, “X1(earlyS)”) was significantly smaller than the fraction of ζNeoblasts in later S-phase stages (“X1(midS)” and “X1(lateS)”) (P<0.05, 2-tailed Fisher’s Exact Test ) (Figure 7G). This trend was replicated by class membership analysis of a BrdU-labeled cell population as it progressed through the cell cycle (Figure S7D,E). Second, PCA analysis showed that early S-phase ζ-class cells were more similar to the X2 σ-class cells than to the X2 ζ-class cells, and that the ζ-class identity became more resolved during progression through S-phase stages (Figure 7H, Figure S7G). Together, these data suggest that the ζ-class cells represent a population of cells that continually arise from σNeoblasts, and that their shift in transcriptional profile begins directly upon entry into S-phase.
Discussion
Planarian neoblasts are an attractive system for the in vivo study of adult stem cell renewal and differentiation in the context of tissue regeneration. To date, most studies of neoblasts have been conducted on a cell population-level. Although this strategy has resulted in the identification of important genes and stimuli affecting the neoblast population as a whole, such observations reflect the average of all the cells constituting the population. Understanding the regenerative process, however, requires an appreciation of the roles and capabilities of individual neoblasts. Recent technical developments have dramatically lowered gene expression detection limits and allowed the fields of developmental biology and stem cell biology to move their focus from population-based analyses to single cells (Junker and van Oudenaarden, 2014; Livak et al., 2013). Single-cell profiling approaches that utilize highly parallel measurements allow for the rapid analysis of many transcripts in hundreds of cells (Dalerba et al., 2011; Guo et al., 2010), and are well equipped to handle the biological noise present in the levels of individual transcripts (Raj and van Oudenaarden, 2008). Here, we present the first highly parallel gene expression analysis of single cells from the planarian neoblast population, and validate this approach by describing the biological roles of the cell types identified.
Neoblast heterogeneity
Apart from noise, variance revealed by single-cell profiling can reflect a wide range of transient biological phenomena, including differences in cell cycle state, nutritional state, or stress. Several notable features, however, suggest that the cellular classes reported here represent genuine and stable subtypes of neoblasts. First, the classes are distinguished by co-regulated groups of transcripts detectable by qPCR as well as in vivo by FISH. This makes it highly unlikely that their detection would be the result of experimental or biological noise. Second, multiple classification methods (HC and PCA) independently identified the same classes, indicating that their discrimination is robust. Third, the classes are present throughout the cell cycle, and thus are not the result of cell-cycle-dependent transcriptional variation of a single cell type. Fourth, the classes are prominent, each involving a large fraction of the neoblast population. This makes it unlikely that they represent a fringe effect with minor relevance to general neoblast biology. Finally, the classes possess distinct regeneration and lineage potential features.
This study has thus identified two major classes of neoblasts. The first class, the zeta-neoblasts, expresses a large cohort of transcripts including zfp-1. The second class, the sigma-neoblasts, expresses low levels of transcripts characteristic of zeta-neoblasts, but higher levels of several other genes including soxP-1 and soxP-2. Cells of both major classes are present throughout the cell cycle, are distributed throughout the body, and remain abundant in homeostasis and during regeneration. Together these features indicate that zeta-neoblasts and sigma-neoblast classes are prominent and integral aspects of the planarian neoblast system.
Determinants of neoblast classes
Cell-cycle-related genes, and transcripts associated with nuage fluctuated throughout the cell cycle and showed some co-regulation; however variation in these genes was not informative for neoblast class membership. This conclusion is consistent with previous neoblast single-cell qPCR data (Hayashi et al., 2010), which also found that analyzing the presence of nuage-related neoblast markers did not distinguish subsets among neoblasts.
By contrast, a subset of the selected transcription factors was able to distinguish clear subpopulations among the neoblasts. Notably, expression of some of these genes (zfp-1, soxP-2) was restricted to the neoblast compartment (Figure S2B,C), whereas others were also expressed in specific differentiated tissues (Figure S1H). Although each class is defined by a unique expression fingerprint of distinct transcription factor genes, some common themes are also evident. For example, each class is characterized by expression of specific Sox transcription factor family members (e.g., soxP-1 and soxP-2 for sigma versus soxP-3 for zeta), and each class also expresses distinct FGF receptors (fgfr4 for sigma and fgfr-1 for zeta). Future studies will be required, however, to explore the roles these genes play in defining the distinct biological features of the two classes.
The zeta-neoblast class
Examination of zfp-1(RNAi) blastemas, which lacked zeta-neoblasts, revealed that most tissues were regenerated normally. Zeta-neoblasts however were required for formation of several related cell types (“prog-1-related cells”) previously identified as the major post-mitotic populations following mitotic exit (Eisenhoffer et al., 2008). These markers have since been widely used to assess the process of neoblast differentiation (Pearson and Sánchez Alvarado, 2010; Scimone et al., 2010; Wagner et al., 2012; Wagner et al., 2011), but it has remained unknown to which lineage these cell types belong.
Several lines of evidence now suggest that this lineage is related to an epidermal cell type. First, zfp-1 RNAi resulted in depletion of several epidermal transcripts, severe perturbations to epidermal cell density and morphology, and significant reductions in epidermal cell differentiation. Second, we identified several transcripts for which expression spanned both prog-1-like cells and epidermal cells, potentially marking transition states between these cells (Figure 6B). Furthermore, expression of three out of six established prog-1-related markers extended into the epidermis (Figure S5B). Third, the dynamics of a single BrdU pulse as it passed through post-mitotic cell populations are consistent with prog-1+ cells existing as cellular intermediates in an epidermal cell lineage (Figure S6H,I). These data suggest that the zeta-class and the prog-1-related population, at least in part represent different stages of epidermal cell differentiation.
The sigma-neoblast class
Our data showed that most established functional attributes of the neoblast population were in fact properties of the sigma-neoblast class. Sigma-neoblasts mediated the early regenerative responses to injury (proliferation and migration), and the fact that the early wound response remained unaltered upon elimination of the zeta-neoblasts by zfp-1 RNAi confirms that the sigma-neoblasts are the primary class involved in the early wound response. The sigma-neoblasts were also able to generate a blastema containing most major cell types. In addition, they were able to self-renew in the absence of the zeta-neoblasts, and could reconstitute the zeta-class after it had been depleted by zfp-1 RNAi. This shows that the sigma-neoblasts are collectively pluripotent.
We propose that the ability to generate zeta-class cells is a general feature of sigma-neoblasts, which extends to non-stimulated conditions. It remains possible that, when appropriately challenged, zeta-neoblasts might also self-renew and even transition into a sigma-neoblast state. However, based on the expression analysis of cells in different stages of S-phase it seems likely that under non-stimulated conditions zeta-class neoblasts are generated from the 2C sigma-class population, whereas the majority of the 2C cells derived from zeta-class mitoses exit the cell cycle permanently. We therefore propose a model in which zeta-neoblasts continuously depend on the sigma-class for their production (Figure 7I).
Previous work has established that clonogenic, pluripotent stem cells (“cNeoblasts”) exist in the adult planarian body and, at the single cell level, can give rise to diverse cell types (Wagner et al., 2011). cNeoblasts, however, can only be identified retrospectively, based on the presence and composition of resulting stem cell clones. Study of cNeoblast expression profiles and other features therefore remains a challenge. Our data suggest that the cNeoblast is likely contained within the sigma-class. Further examination and subdivision of the sigma-class, will therefore aid in the identification of markers and features of the cNeoblast.
Evidence for further neoblast heterogeneity
The major classes described in this study are an important step in evaluating neoblast complexity, but neither the sigma-class nor the zeta-class can be considered homogeneous cell populations. For example, one subset of the zeta-class cells co-expressed higher levels of both AbdBb and a six6-related transcript whereas another subset expressed high levels of AbdBa, meis-2, and a gata1/2/3-related transcript (Figure S1G), suggesting the presence of subclasses. Within the sigma-class, the gamma-neoblasts form a clear subpopulation characterized by high expression of prox-1, hnf4, gata4/5/6, and nkx2.2 (Figure 1A–E). Furthermore, additional heterogeneity might be established by genes not included in this study.
As the zeta-neoblasts likely represent progenitors to restricted lineages, we hypothesize that other subclasses will also be lineage precursors. Interestingly, three of the four transcripts marking the gamma-neoblast subclass (hnf4, gata4/5/6, and nkx2.2) have been previously linked to the planarian intestine (Forsthoefel et al., 2012; Wagner et al., 2011). In addition, RNAi of hnf4 or gata4/5/6 resulted in reduced expression of gata4/5/6 and nkx-2.2 in neoblasts without significantly affecting genes representative of the other classes (Figure S4G), suggesting that these genes indeed form a co-regulated unit. Co-regulation and co-expression of these factors in the same cells within the neoblast population strongly suggests that gamma-neoblasts represent an intestinal progenitor state. Future studies will be needed, however, to establish direct evidence for this lineage relationship.
New insights into planarian regenerative mechanisms
The identification of neoblast classes presented the opportunity to re-evaluate various neoblast properties, such as the wound response, that previously had been ascribed to the neoblast population as a whole. The finding that early regenerative responses to injury are in fact properties of sigma-neoblasts (but not zeta-neoblasts) indicates that wound detection, wound-induced proliferation, and migration to sites of injury are all properties of only a subset of the neoblast population. Therefore the proliferative and migratory capacities of responding neoblasts have probably been underestimated.
Single-cell profiling also provides useful insights into dynamic properties of the neoblast compartment during regeneration. It has been unclear, for example, whether initial neoblast responses to wounds are generic or customized to specific types of injury. Single-cell qPCR analysis allows for the assessment of the class composition of the total neoblast population, and thereby presents an opportunity to evaluate shifts in these classes in response to various regenerative stimuli. Our preliminary data (Figure S3B–D) suggest that such shifts can indeed be detected, suggesting that the neoblast response can in fact be tailored to the identity of the missing tissue. Such results illustrate the value of single-cell profiling in furthering our understanding of regenerative mechanisms.
Because the planarian neoblast system has the potential to maintain itself seemingly indefinitely and can give rise to every cell type in the body, it forms a remarkably versatile stem cell system. Neoblasts have been studied for over a century by traditional mixed-population studies, and have largely been treated as a single homogenous cell type. Examination of neoblasts at the single cell level, however, facilitates a more accurate understanding of planarian stem cell dynamics, and of the roles specific types of neoblasts can play in complex regenerative decisions. We conclude that the neoblasts can be divided into multiple prominent categories of cells that display unique properties in regeneration. These findings are significant for the interpretation of all neoblast studies and for furthering our understanding of regenerative biology.
Experimental Procedures
Planarian Culture
Schmidtea mediterranea clonal strain ClW4 was maintained as previously described (Newmark and Sánchez Alvarado, 2000). Animals were cultured in the dark at 25°C in 1X Montjuic salts, fed homogenized beef liver paste every 1–2 weeks and expanded through continuous cycles of amputation or asexual fissioning and regeneration.
Gene Cloning
All planarian gene sequences were cloned from cDNA into the pGEM vector (Promega) and verified by Sanger sequencing. For RNAi experiments, gene sequences were subcloned using a Gateway BP reaction (Invitrogen) into pPR244, a vector containing two inducible T7 promoters and two T7 terminators flanking the Gateway cassette (Reddien et al., 2005a). Gene-specific dsRNA was expressed from these plasmids in E.coli strain HT115.
RNAi
Schmidtea mediterranea strain ClW4 was maintained as previously described (Newmark and Sánchez Alvarado, 2000). HT115 E. coli with genes in the pPR244 vector (Reddien et al., 2005a) were induced with 1mM IPTG for 2 hr, centrifuged, and resuspended in 66% homogenized beef liver (in water) at 1/300th initial culture volume. Liver-bacteria suspensions were fed to planarians 3–4 times over the course of 7–10 days. Negative RNAi controls used unc-22, a C. elegans gene lacking significant similarity to the S. mediterranea genome.
Drug Treatments and Irradiation
Nocodazole (Sigma) was administered to planarians by soaking animals for ~24 hours in a 400ng/mL solution dissolved in 1X Montjuic salts supplemented with 1% DMSO. BrdU (Sigma) was administered by soaking for specified pulse lengths in 1X Montjuic salts containing 25mg/mL BrdU and 3% DMSO; animals were then incubated in either 1X Montjuic salts or Instant Ocean dissolved at 5g/L. Irradiation doses were delivered to animals using dual Gammacell-40 137Cesium sources.
Whole-Mount in situ Hybridization
Fixations and whole-mount in situ hybridizations (ISH) were performed as previously described (Pearson et al., 2009). Some hybridized RNA probes were detected using anti-DNP-HRP Conjugate (Perkin Elmer). HRP enzyme was inactivated overnight between labelings by 154mM sodium azide, in PBSTx(0.1%). Most specimens were counterstained with DAPI (Sigma, 1µg/mL in PBSTx).
Single-Cell Quantitative RT-PCR
Amplified cDNA libraries were generated from lysates of individual FACS-isolated cells using SuperScriptIII/ PlatinumTaq mix (Invitrogen) and a mixture of outer primers specific to 96 planarian genes (See Figure S1). Single-cell qPCR experiments were performed with inner (nested) primers on the BioMark platform (Fluidigm) using 96.96 DynamicArray IFC chips (Fluidigm) according to the manufacturer’s instructions. For additional details, see Supplemental Experimental Procedures.
Cell Transplantations
Bulk transplantation of X1(FS) FACS-sorted cells from 15 day control RNAi or zfp-1 (RNAi) donors were performed similarly as previously described (Wagner et al., 2011). Host animals to receive transplants were starved in the presence of Gentamycin for at least seven days prior to onset of experiments, and were exposed to 6,000 rads of irradiation two days prior to transplantation. X1(FS) cells collected by flow cytometry as in (Wagner et al., 2011) were spotted onto glass cover slips (coverslips pretreated with 2% dimethyldichlorosilane (Sigma) in chloroform). Under 10X magnification, cells were loaded by mouth pipetting into the tip of pulled borosilicate glass microcapillaries (Sutter) treated with 0.1% polyvinylpyrrolidone (Sigma). Loaded cells were injected into the post-pharyngeal or post-gonopore region of cold-immobilized host animals at 1.5–2.5 psi (Eppendorf FemtoJet). Individual animals that received a transplant were maintained in 6 cm Petri dishes (3 worms/dish) in the dark in the presence of Gentamycin; water and dishes were changed every 3–4 days.
Supplementary Material
Highlights.
Neoblasts, planarian regenerative cells, comprise large distinct cellular classes
ζ-class neoblasts are lineage-specified precursors that give rise to epidermis
σ-class neoblasts collectively possess many stem cell-like features
Acknowledgements
We thank members of the Reddien lab for comments on the manuscript. J.C.vW. is supported by the Human Frontier Science Program (HFSP). P.W.R. is an Investigator of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We acknowledge support from the NIH (R01GM080639).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.C.vW, D.E.W., P.W.R.: overall design, J.C.vW, D.E.W.: performed all experiments presented in figures 1–7 and supplement. J.C.vW, D.E.W., P.W.R.: wrote manuscript.
References
- Blythe MJ, Kao D, Malla S, Rowsell J, Wilson R, Evans D, Jowett J, Hall A, Lemay V, Lam S, et al. A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS ONE. 2010;5:e15617. doi: 10.1371/journal.pone.0015617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles MW, Brown DDR, Nisperos SV, Stanley BN, Pearson BJ, Zayas RM. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development. 2013;140:4691–4702. doi: 10.1242/dev.098616. [DOI] [PubMed] [Google Scholar]
- Currie KW, Pearson BJ. Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development. 2013;140:3577–3588. doi: 10.1242/dev.098590. [DOI] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea . Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Developmental Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M. Global irradiation effects, stem cell genes and rare transcripts in the planarian transcriptome. Int J Dev Biol. 2012;56:103–116. doi: 10.1387/ijdb.113455mg. [DOI] [PubMed] [Google Scholar]
- Guedelhoefer OC, Sánchez Alvarado A. Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development. 2012;139:3510–3520. doi: 10.1242/dev.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. A bruno-like Gene Is Required for Stem Cell Maintenance in Planarians. Developmental Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Shibata N, Okumura R, Kudome T, Nishimura O, Tarui H, Agata K. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ. 2010;52:131–144. doi: 10.1111/j.1440-169X.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- Labbe RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DDR, Ross EJ, Voisin V, Bader GD, Blencowe BJ, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. dlx and sp6-9 Control Optic Cup Regeneration in a Prototypic Eye. PLoS genetics. 2011;7:e1002226. doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz S, Stockle F, Ortmeier C, Schmitz H, Rodriguez-Esteban G, Key G, Gentile L. Heterogeneity of planarian stem cells in the S/G2/M phase. Int J Dev Biol. 2012;56:117–125. doi: 10.1387/ijdb.113440sm. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Developmental Biology. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Onal P, Grün D, Adamidi C, Rybak A, Solana J, Mastrobuoni G, Wang Y, Rahn H-P, Chen W, Kempa S, et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO. 2012;31:2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu Y-C, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Developmental Dynamics. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Developmental Cell. 2005a;8:635–649. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005b;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annual Review of Cell and Developmental Biology. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Resch AM, Palakodeti D, Lu Y-C, Horowitz M, Graveley BR. Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PLoS ONE. 2012;7:e34447. doi: 10.1371/journal.pone.0034447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Ross EJ, Jack A, Sanchez Alvarado A. Molecular cloning and characterization of SL3: a stem cell-specific SL RNA from the planarian Schmidtea mediterranea. Gene. 2014;533:156–167. doi: 10.1016/j.gene.2013.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Deri P, Batistoni R. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev Dyn. 2000;218:603–614. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1016>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Meisel J, Reddien PW. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea . Development. 2010;137:1231–1241. doi: 10.1242/dev.042051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Hayashi T, Fukumura R, Fujii J, Kudome-Takamatsu T, Nishimura O, Sano S, Son F, Suzuki N, Araki R, et al. Comprehensive gene expression analyses in pluripotent stem cells of a planarian, Dugesia japonica . Int J Dev Biol. 2012;56:93–102. doi: 10.1387/ijdb.113434ns. [DOI] [PubMed] [Google Scholar]
- Solana J, Kao D, Mihaylova Y, Jaber-Hijazi F, Malla S, Wilson R, Aboobaker A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome biology. 2012;13:R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Lasko P, Romero R. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Developmental Biology. 2009;328:410–421. doi: 10.1016/j.ydbio.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harbor Perspectives in Biology. 2011:3. doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic Neoblasts Are Pluripotent Adult Stem Cells That Underlie Planarian Regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien P. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Developmental Biology. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E, Dubois F. Sur la migration des cellules de régénération chez les planaires. Revue Swisse Zool. 1948;55:218–227. [Google Scholar]
- Zayas RM, Cebrià F, Guo T, Feng J, Newmark PA. The use of lectins as markers for differentiated secretory cells in planarians. Developmental Dynamics. 2010;239:2888–2897. doi: 10.1002/dvdy.22427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Pearson BJ. The Retinoblastoma pathway regulates stem cell proliferation in freshwater planarians. Dev Biol. 2013;373:442–452. doi: 10.1016/j.ydbio.2012.10.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.