Abstract
Fusion of inducible degradation signals, so-called degrons, to cellular proteins is an elegant method of controlling protein levels in vivo. Recently, a degron system relying on the plant hormone auxin has been described for use in yeast and vertebrate cells. We now report the construction of a series of vectors that significantly enhance the versatility of this auxin-inducible degron (AID) system in Saccharomyces cerevisiae. We have minimized the size of the degron and appended a series of additional epitope tags, allowing detection by commercial antibodies or fluorescence microscopy. The vectors are compatible with PCR-based genomic tagging strategies, allow for C- or N-terminal fusion of the degron, and provide a range of selection markers. Application to a series of yeast proteins, including essential replication factors, provides evidence for a general usefulness of the system. © 2013 The Authors. Yeast published by John Wiley & Sons, Ltd.
Keywords: degron, auxin, budding yeast, protein degradation, protein stability
Introduction
Molecular biology offers a variety of tools to manipulate proteins for in vivo functional analysis: deletion of the relevant gene is the most efficient way of completely removing a protein of interest, but the procedure is generally irreversible and not applicable to essential genes. In higher eukaryotes, downregulation of gene expression by RNAi-mediated depletion of the mRNA is most commonly used today, but this technology exhibits a rather slow response, is still prone to off-target effects and is unavailable in simple model organisms such as budding yeast (Carthew, 2001). Furthermore, its efficiency is strongly dependent on protein stability. Hence, interfering directly at the protein level may sometimes be more useful because it might allow for quicker and reversible responses. Several systems have been described that manipulate protein localization to achieve a conditional regulation, such as the use of steroid hormone-binding or rapamycin-dependent dimerization domains to control nuclear localization (Haruki et al., 2008; Picard et al., 1988). However, they all rely on the protein's activity being restricted to a specific cellular compartment.
The most generally applicable method of controlling the activity of a protein is therefore its targeted depletion by means of an inducible degradation signal, or degron. Two degradation systems have been characterized in S. cerevisiae, based on either the N-end rule or an auxin-inducible degron (Dohmen et al., 1994; Nishimura et al., 2009). The original N-end rule degron system relies on the exposure of a destabilizing amino (N)-terminal residue upon a temperature shift, which induces ubiquitylation and subsequent proteasomal degradation. This method is highly efficient and reversible, but limited by its requirement for N-terminal tagging. Moreover, the procedure induces a heat shock response and in most cases requires a change of the carbon source in the growth medium, both of which might cause unwanted physiological changes in the cells.
The recently described auxin-inducible degron (AID) system uses a plant hormone-induced degradation signal to control protein levels (Nishimura et al., 2009). In its natural context, auxin (indole-3-acetic acid; IAA) induces degradation of a family of short-lived transcriptional repressors, the IAA proteins, by mediating the interaction of a degron domain in the target protein with the substrate recognition domain of an F-box protein, TIR1 (Hayashi, 2012). Productive interaction in the presence of auxin leads to ubiquitylation of the target by recruitment of an SCF-type ubiquitin ligase (E3) and finally proteasomal degradation. The SCF complex, consisting of a cullin subunit, a catalytic RING finger protein (RBX1), the adaptor SKP1 and an F-box protein as a substrate recognition subunit, is highly conserved among eukaryotes (Zimmerman et al., 2010), such that the plant F-box protein TIR1 is able to form an active E3 with the remaining SCF components from other organisms. Hence, constitutive expression of TIR1 allows a reconstitution of the AID system in yeast or vertebrate cells. Proteins of interest are fused to an auxin-dependent degron sequence derived from IAA17. This AID tag can in principle be placed at the N- or carboxy (C)-terminus of the target, thus making the system more flexible than the N-end rule degron. Degradation is reversible and quick, measured in minutes rather than hours. Furthermore, due to the lack of an auxin-responsive system in animals or yeast, the hormone as well as the F-box protein are otherwise biologically silent and cause no measurable physiological changes in the absence of a target, thus minimizing possible side-effects of the treatment (Nishimura et al., 2009).
In budding yeast, the AID system has been successfully applied to several proteins resident in different cellular compartments (Nishimura et al., 2009; Watase et al., 2012). In these studies, the AID-tag comprised full-length IAA17, a 229 amino acid protein. While AID-tagged GFP constructs were detected either by means of fluorescence or by an anti-GFP antibody, an antibody raised against IAA17 itself served for detection of the AID-tagged yeast proteins. However, a direct comparison between basal protein levels of tagged and untagged targets, which would reveal any auxin-independent effects of the AID-tag, has not been reported.
In an effort to enhance the utility of the AID system for functional studies in S. cerevisiae, we have now explored variations of the tag by reducing its size in exchange for a series of short epitopes for which commercial antibodies are available. In combination with an expanded set of selection markers, this series of tagging cassettes provides enhanced flexibility in applying the AID system by facilitating the detection of AID-tagged proteins, comparing their stability with those of untagged or endogenous proteins, and exerting a better control over initial protein levels.
Materials and methods
Plasmids
Features of the tagging cassettes generated in this study are summarized in Table 1; all oligonucleotides are listed in Table S1 (see Supporting information). Plasmids pNHK53 (OsTIR1), pSM409 (IAA17–hphNT1), pMK43 (IAA17–KanMX4), pSM410 (IAA17–HIS3MX6) and pMK38 (PCUP1–IAA17–KanMX4) were obtained from the National BioResource Project–Yeast (Japan). pFA–Nat–NT2 and pAW8–6xFLAG were from EUROSCARF, and pYM3 from M. Knop (Knop et al., 1999).
Table 1.
Aid-tagging plasmids used in this study
| Name | IAA17 | Position | Extension | Selection | Parent vector |
|---|---|---|---|---|---|
| pHyg–AID1–114 | 1–114 | C-terminal | – | hphNT | pSM409 |
| pHyg–AID31–114 | 31–114 | C-terminal | – | hphNT | pSM409 |
| pHyg–AID* | 71–114 | C-terminal | – | hphNT | pSM409 |
| pHyg–AID1–114–8myc | 1–114 | C-terminal | 8myc | hphNT | pSM409 |
| pHyg–AID31–114–9myc | 31–114 | C-terminal | 9myc | hphNT | pSM409 |
| pHyg–AID*–9myc | 71–114 | C-terminal | 9myc | hphNT | pSM409 |
| pHyg–AID*–6FLAG | 71–114 | C-terminal | 6FLAG | hphNT | pSM409 |
| pHyg–AID*–6HA | 71–114 | C-terminal | 6HA | hphNT | pSM409 |
| pHyg–AID*–GFP | 71–114 | C-terminal | GFP | hphNT | pSM409 |
| pKan–AID*–9myc | 71–114 | C-terminal | 9myc | kanMX | pSM409 |
| pNat–AID*–9myc | 71–114 | C-terminal | 9myc | natNT | pSM409 |
| pHis–AID*–9myc | 71–114 | C-terminal | 9myc | HIS3MX | pSM409 |
| pKan–PCUP1–AID*(N) | 71–114 | N-terminal | – | kanMX | pMK38 |
| pKan–PCUP1–AID*(N)–9myc | 71–114 | N-terminal | 9myc | kanMX | pMK38 |
| pKan–PCUP1–9myc–AID*(N) | 71–114 | N-terminal | 9myc | kanMX | pMK38 |
| pKan–PRFA1–9myc–AID*(N) | 71–114 | N-terminal | 9myc | kanMX | pMK38 |
Construction of C-terminal tagging cassettes
Truncations of IAA17 were amplified by PCR and cloned as KpnI/XmaI fragments into pSM409 to replace the full-length IAA17 sequence (AID1–114, oligos 1772/1773; AID31–114, oligos 1788/1773; AID*, 1787/1773). For fusion of epitope tags to AID, oligo 1773 was replaced by 1774 in the cloning, and PCR products encoding the tags were subsequently inserted as XmaI/AscI fragments downstream of the AID sequence (AID*–9myc, oligos 1795/1790; template, pNHK53; AID*–6FLAG, oligos 2071/2072; template, pAW8–6xFLAG; AID*–6HA, oligos 2069/2070; template, pYM3; AID*–eGFP, 2073/2074; template, Clontech).
Exchange of selection markers
Different yeast selection markers were subcloned into pHyg–AID*–9myc as AscI/SacI fragments to replace HphNT1 (KanMX4 from pMK43; NatNT2 from pFA-NatNT2; HIS3MX6 from pSM410).
Construction of N-terminal tagging cassettes
AID* and AID*–9myc were amplified from pHyg–AID*–9myc (oligos 1996/1993 and 1996/1992, respectively) and inserted into pMK38 as EcoRI/XhoI fragments, replacing the IAA17 sequence, thus yielding pKan–PCUP1–AID*(N) and pKan–PCUP1–AID*(N)–9myc. For construction of pKan–PCUP1–9myc–AID*(N), the 9myc tag was amplified from pNHK53 (oligos 2064/2065) and inserted as an EcoRI fragment upstream of AID* in pKan–PCUP1–AID*(N), followed by site-directed mutagenesis to create NheI (oligos 2039/2040) and PmlI (oligos 2185/2186) sites flanking the CUP1 promoter. The CUP1 promoter was replaced by an NheI/PmlI fragment bearing the RFA1 promoter, amplified from genomic DNA (oligos 2189/2190) to yield pKan–PRFA1–9myc-AID*(N).
Yeast strains
All experiments were carried out in the DF5 strain background (Finley et al., 1987). All strains used are listed in Table S2 (see Supporting information). Standard procedures were followed for yeast cultivation and transformation (Sherman, 1991). For use of the CUP1 promoter, 100 µM CuSO4 was added to the growth medium. Geneticin was used at 200 µg/ml (for kanMX4 selection); hygromycin B at 300 µg/ml (for hphNT1); and nourseothricin at 100 µg/ml (for natNT2). Auxin was used at 1 mm unless otherwise indicated. TIR1 strains were created by integration of pNHK53 (encoding OsTIR1 under control of the ADH1 promoter) into the URA3 locus (Nishimura et al., 2009). All strains carrying a deletion or epitope-tagged allele of RAD53 were constructed in an sml1Δ background. Gene deletions and tags were introduced by means of PCR-generated cassettes (Longtine et al., 1998), using the primers listed in Table S1 (see Supporting information).
Antibodies
Antibodies are indicated in the respective figure legends. Rabbit polyclonal antibodies against Rfa1 and Rad53 were from S. Brill and J. Diffley, respectively. Monoclonal antibodies were from Molecular Probes (PGK), Roche (GFP), Sigma (FLAG) or Cancer Research UK (MYC, 9E10; HA, 12CA5).
Auxin sensitivity assays
Sensitivity to auxin was determined by spotting serial dilutions of exponentially growing yeast cultures on YPD plates containing the indicated concentrations of auxin and/or hydroxyurea and treatment by UV irradiation using a Stratalinker 2400 (Stratagene) where indicated, as described previously (Stelter and Ulrich, 2003).
Detection of proteins
Total yeast lysates were prepared by trichloroacetic acid (TCA) precipitation: aliquots of ca. 4 × 107 cells were resuspended in 500 µl water and incubated with 75 µl 1.85 m NaOH, 7.5% β-mercaptoethanol for 15 min on ice, followed by addition of 75 µl of 55% w/v TCA and incubation for 10 min on ice. After centrifugation at 16 000 × g for 10 min, the supernatant was removed and the pellet resuspended in 40 µl HU buffer (8 m urea, 5% SDS, 200 mm Tris–HCL, pH 6.8, 1 mm EDTA, 0.1% bromophenol blue) and incubated at 65 °C for 10 min. Proteins were analysed by SDS–PAGE/western blotting.
Flow cytometry
Cells were fixed in 70% ethanol overnight and washed twice in 50 mm sodium citrate, pH 7.0. After incubation with 0.1 mg/ml DNase-free RNAse A for 1 h at 50 °C, followed by addition of 100 U Proteinase K (from T. album; Sigma) and further incubation for 1 h at 50 °C, DNA was stained with 16 µg/ml propidium iodide and the cells were analysed by flow cytometry, using a FACsCalibur instrument (BD Biosciences) and the FloJo application.
Results
Effect of the AID-tag and its truncations on basal protein levels
Fusion of the full-length IAA17 protein (229 amino acids, ∼ 25 kDa) results in the addition of a relatively large tag, which might interfere with the function or expression of some, especially small target proteins, independent of their auxin-induced degradation. We therefore aimed to reduce the size of the AID tag without affecting the inherent stability of the fusion partner or its efficiency in mediating auxin-induced degradation. The IAA proteins share four highly conserved domains (I–IV, Figure 1A), of which domain II has been demonstrated to act as the TIR1 interaction site (Abel et al., 1994; Worley et al., 2000). Further analysis revealed a 13 amino acid consensus sequence among IAA proteins that was crucial for degradation, although additional regions in the N-terminal half of the proteins, spanning amino acids 68–111 in IAA17, were found to be required for efficient degradation in plant cells (Ramos et al., 2001). Similar results were obtained with truncations of three other IAA family proteins in a reconstituted yeast system (Havens et al., 2012).
Figure 1.
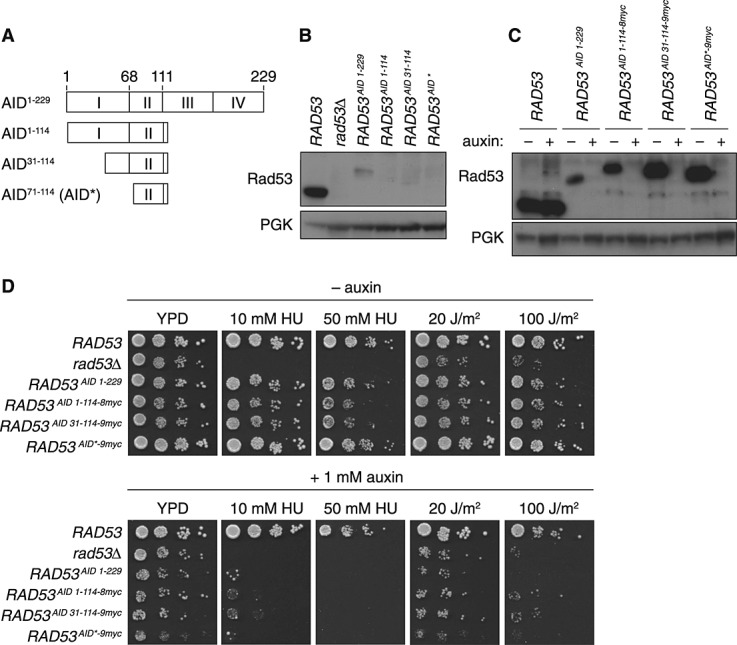
Truncation and tagging of the C-terminal AID degron. (A) Protein structure of IAA17, corresponding to the full-length AID tag, and truncations examined in this study. (B) Protein levels of Rad53 carrying full-length and truncated versions of the AID tag, examined by western blotting of total lysates; PGK served as a loading control. (C) Protein levels of Rad53 carrying the full-length AID tag in comparison with truncated versions extended by a C-terminal 9myc tag. Where indicated, cultures were treated with 1 mm auxin for 1 h before preparation of the lysates. (D) Degradation of Rad53 by means of the AID tag renders cells sensitive to hydroxyurea (HU) and ultraviolet (UV) radiation. Growth inhibition was monitored on YPD plates containing auxin and/or HU or treated with the indicated UV doses as noted
Based on these notions, we constructed three different truncations of IAA17 (Figure 1A): the N-terminal half of the protein (amino acids 1–114) and two additional N-terminal truncations (amino acids 31–114 and 71–114). The relevant sequences were used to replace the original C-terminal AID-tag in the context of a hygromycin B (HphNT1)-resistance cassette. For our initial analysis, we chose the checkpoint protein Rad53 as a target, because its activity is easy to follow by phenotypic assays and antibodies are available for monitoring protein levels. Rad53 was tagged in its genomic locus, and expression of the fusion proteins in the absence of auxin was compared to the levels of native Rad53. Surprisingly, the full-length AID-tag (1–229) considerably reduced Rad53 protein levels and the truncated tags resulted in almost undetectable levels (Figure 1B). Although not all target proteins were similarly affected by the tag, the same auxin-independent effect was observed with several other proteins that we tested (our unpublished data). Western blot analysis in yeast strains, with or without TIR1, revealed that the F-box protein was not responsible for the auxin-independent destabilization of Rad53 (see Supporting information, Figure S1).
Addition of epitopes for detection of the AID tag
In order to facilitate the analysis of AID-tagged proteins for which no antibody is available, we appended multiple myc epitopes to the AID sequences, allowing for easy detection with commercially available antibodies. Unexpectedly, their presence stabilized Rad53 in the context of all three truncated AID tags to levels much higher than those achieved by the original full-length IAA17. Importantly, the truncated AID–myc tags all conferred sensitivity to auxin treatment, indicating that they are fully functional as degrons (Figure 1C). Further truncations resulted in a loss of auxin responsiveness (data not shown), indicating that the region spanning amino acids 71–114 is close to the minimal size required for function. We therefore concentrated our further analysis on this construct, henceforth named AID*.
Deletion of RAD53 results in sensitivity to agents causing replication stress or DNA damage, such as hydroxyurea (HU) and ultraviolet (UV) radiation (Branzei and Foiani, 2006). These phenotypes were used as a measure of in vivo Rad53 activity (Figure 1D). As expected from the reduction in protein levels, the full-length AID-tag caused measurable sensitivity to high doses of HU or UV, and the same was observed with the AID1–114–8myc and AID31–114–9myc tags. However, strains bearing RAD53AID*–9myc displayed essentially wild-type sensitivity to both agents in the absence of auxin, suggesting that the tag does not interfere with Rad53 function. Addition of auxin resulted in sensitivities close to those of a rad53Δ strain for all the tags analysed. Taken together, these results suggest that AID*–9myc exhibits a robust auxin response and can serve as a useful reagent for manipulating protein stability in vivo.
Variation of epitopes for detection of the AID* tag
To further increase the versatility of the auxin system, we created AID* fusions with three additional C-terminal tags: 6HA, 6FLAG and GFP. As a target protein for analysis we chose the replication factor Rfa1, since this protein was less sensitive than Rad53 to the destabilizing effect of the full-length AID tag. All the fusion proteins exhibited similar steady-state levels and were sensitive to auxin (Figure 2A). The respective strains had similar growth rates, except for RFA1AID*–6HA, which exhibited a slight growth defect (Figure 2B). This indicates that most of the tags did not interfere with the essential function of Rfa1. It should be noted that the TIR1 protein also carries a 9myc epitope, which provides a convenient internal standard for expression levels of myc-tagged target proteins.
Figure 2.
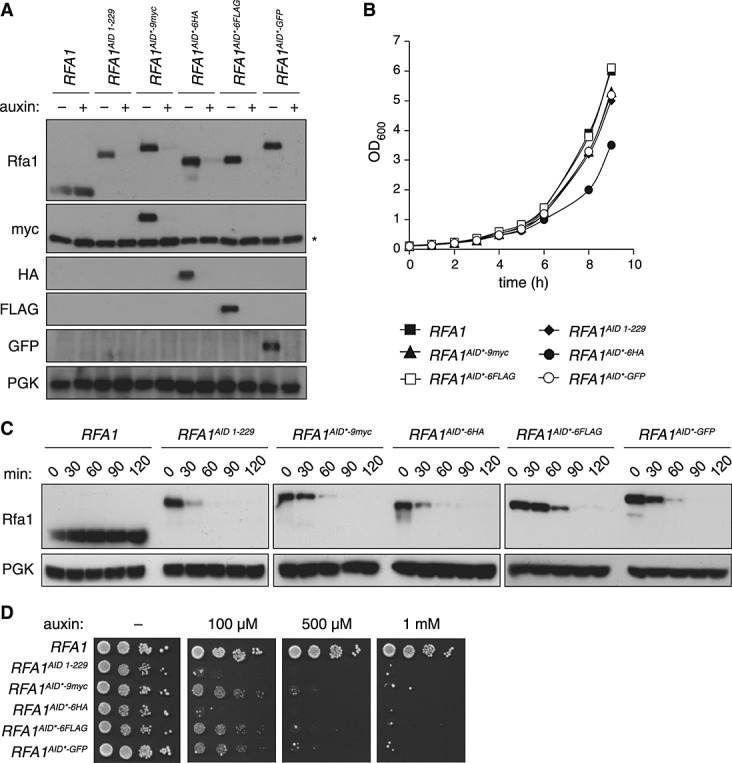
Variation of epitopes for detection of the AID* tag. (A) Protein levels of Rfa1 carrying a C-terminal AID* tag extended by myc, HA, FLAG or GFP, examined by western blotting with the respective antibodies. Where indicated, cultures were treated with 1 mm auxin for 1 h before preparation of the lysates. Note that the anti-myc blot also detects myc-tagged TIR1 (marked *). (B) Growth curves of the strains described in (A), cultured at 30 °C in YPD medium. (C) Degradation time course of Rfa1 carrying the tags described in (A), after treatment with 1 mm auxin. (D) Growth inhibition by degradation of tagged Rfa1, monitored on YPD plates containing the indicated concentrations of auxin
In order to assess potential effects of the tags on auxin-induced degradation rates, exponentially growing cells were treated with 1 mm auxin, and samples were collected every 30 min for western blot analysis. Rfa1AID*–9myc, Rfa1AID*–6FLAG and Rfa1AID*–GFP were degraded somewhat more slowly than Rfa1AID and Rfa1AID*–6HA, but after 90 min degradation appeared to be complete in all strains (Figure 2C).
As reported, the full-length AID-tag is responsive to even low auxin concentrations (Nishimura et al., 2009). In order to test the responsiveness of our new constructs, we examined the growth of the tagged RFA1 strains on plates with different auxin concentrations. As Rfa1 is an essential protein, efficient degradation will result in a loss of growth. Consistent with the differences in degradation rates, RFA1AID and RFA1AID*–6HA were the most sensitive and failed to grow even at very low auxin concentrations; however, RFA1AID*–9myc, RFA1AID*–6FLAG and RFA1AID*–GFP responded well to higher doses of auxin (Figure 2D). These results imply that all four constructs can be efficiently used as auxin-dependent degrons, although higher auxin concentrations may be needed for some of the constructs.
Variation of selection markers for the AID* tag
In order to further enhance the flexibility of the AID*–9myc tag, we combined it with additional selection markers, KanMX4, NatNT2 and HIS3MX6, conferring resistance to the antibiotics geneticin and nourseothricin or histidine prototrophy, respectively. As shown in Figure 3, Rfa1AID*–9myc was efficiently expressed and responsive to auxin in all strains.
Figure 3.
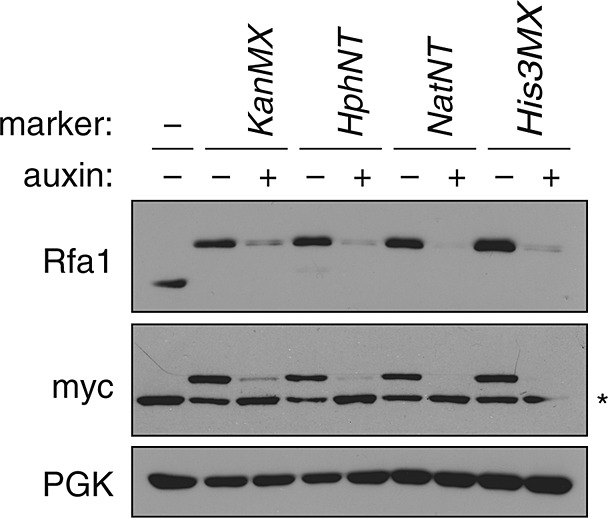
Variation of selection markers for the AID* tag. Protein levels of Rfa1AID*–9myc carrying the indicated selection markers were monitored in the presence and absence of auxin, as described in Figure 1C; *, myc-tagged TIR1
Construction of N-terminal AID* tags
C-terminal tagging is the most straightforward method for protein detection in yeast, as integration of the tagging cassette downstream of the open reading frame does not affect the upstream regulatory sequences. However, if protein function is affected by the tag, N-terminal tagging is a useful alternative, even though it requires the introduction of a promoter together with the tag. Nishimura et al. (2009) prepared N-terminal AID-tagging constructs based on the copper-inducible CUP1 promoter. In order to combine the 9myc epitope with the AID* tag in the N-terminal setting, we explored the arrangements illustrated in Figure 4A. The tagged protein AID*–9mycRfa1 (construct I) was detected at the expected size, but the appearance of additional bands with enhanced mobility that were not responsive to auxin treatment indicated that the N-terminal AID* tag tends to become cleaved or degraded from the rest of the protein (Figure 4B). This property seriously limits the usefulness of this construct. In contrast, 9myc–AID*Rfa1 (construct IIa) was stable, and any detectable bands of higher mobility were still responsive to auxin treatment (Figure 4B). Construct IIb, which lacks the endogenous start codon on the target protein, can easily be generated by a variation of the oligonucleotide used for amplification of the tagging cassette in order to prevent its use as an alternative translational start site. Auxin sensitivity assays in the presence and absence of copper indicated the efficiency of degradation, although again construct I proved less useful, as it conferred a growth defect even in the absence of auxin (Figure 4C).
Figure 4.
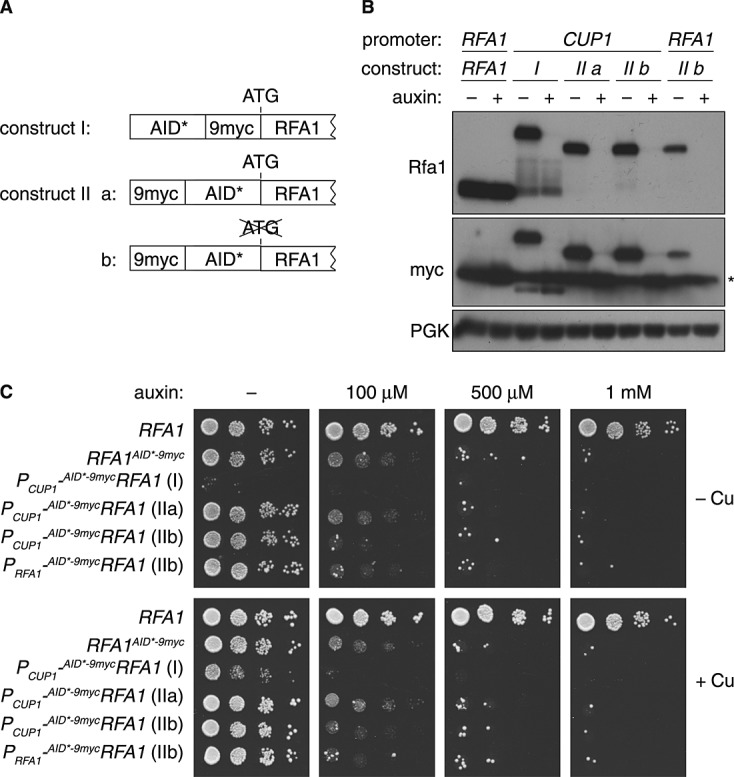
Construction of N-terminal AID* tags. (A) Schematic representation of different N-terminal AID* constructs. (B) Protein levels of Rfa1 carrying the N-terminal AID* tags under control of the RFA1 or CUP1 promoter were analysed as described in Figure 1C; *, myc-tagged TIR1. Culture medium contained 0.1 mM CuSO4. (C) Growth inhibition by degradation of N-terminally tagged Rfa1, monitored as in Figure 2D in the presence or absence of 0.1 mM CuSO4
Although the activity of the CUP1 promoter can be controlled to some degree by the addition of CuSO4 to the media, use of the endogenous promoter might be preferable for target proteins whose expression is regulated in a more complex manner. We therefore introduced restriction sites (NheI/PmlI) into the N-terminal 9myc–AID* cassette by site-directed mutagenesis, which facilitates the insertion of alternative promoters. When 9myc–AID*RFA1 was expressed from its own promoter, the protein accumulated to a lower level than the wild-type, again reflecting the slight auxin-independent destabilizing influence of the degron, but effects on viability were comparable to the constructs bearing the CUP1 promoter (Figure 4B, C).
Versatility of the AID* tag
The versatility of the C-terminal AID*–9myc tagging module was probed by applying it to a range of essential replication factors: Orc6, Pol1, Pol2, Pol31 and Rfc1 (Figure 5A). In order to explore the effects of different cellular compartments, kinetochore-associated Ask1 and cytoplasmic Sec14 were also tested, as well as Ubc13, Mms2 and Yen1, which partition between the cytoplasm and the nucleus (Figure 5B). All of the fusion proteins were degraded in response to auxin, as detected by means of anti-myc antibody staining, although degradation of the highly abundant Sec14 protein was incomplete. Again, the presence of myc-tagged TIR1 served as an internal standard for protein levels, although it interfered with the detection of the targets if they were of a similar size.
Figure 5.
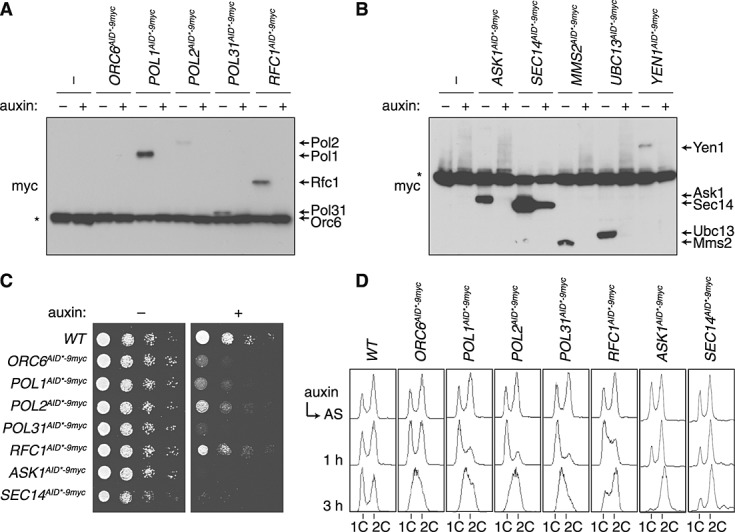
Effects of auxin on various AID*-tagged proteins. (A, B) Levels of the indicated proteins carrying the C-terminal AID*–9myc tag after incubation of the respective strains in the presence and absence of 1 mm auxin for 1 h. Myc-tagged TIR1 (marked *) was used as a loading control. (C) Growth inhibition by degradation of the tagged proteins, monitored on YPD plates containing 1 mm auxin where indicated. (D) Cell cycle profiles of the strains used in (A), collected at the indicated times after addition of 1 mm auxin; AS, asynchronous culture
Where essential genes were concerned, inspection of overall growth on auxin-containing plates revealed subtle differences in auxin sensitivity, with POL31AID*–9myc, ORC6AID*–9myc, ASK1AID*–9myc and SEC14AID*–9myc causing nearly complete growth inhibition (Figure 5C), while RFC1AID*–9myc afforded the least effective inhibition. Placement of the degron at the N-terminus of Rfc1 did not improve growth arrest, suggesting that proliferation is possible even with very low concentrations of this protein (see Supporting information, Figure S2). Surprisingly, auxin treatment of SEC14AID*–9myc resulted in complete growth inhibition despite the relatively inefficient degradation (Figure 5B, C). Flow cytometry was used to assess the efficiency of the degron in the course of a single cell cycle, as depletion of any of the replication factors should result in cell cycle arrest in G1 or early S phase. When asynchronous populations were treated with auxin, the degron-tagged alleles of replication proteins indeed caused a substantial accumulation in G1/early S phase, although again with different kinetics: POL1AID*–9myc, POL2AID*–9myc and POL32AID*–9myc all responded with a tight arrest within 1 h of auxin addition, resulting in an S phase accumulation after 3 h. ORC6AID*–9myc showed a delayed response and, as already observed in the plate assay, the arrest of RFC1AID*–9myc appears less efficient and more transient, as cells had largely progressed to G2/M after 3 h (Figure 5D). ASK1AID*–9myc cells quickly accumulated in G2/M, consistent with the function of this protein in chromosome segregation. Taken together, these results show that the AID* tag is responsive to auxin in a variety of contexts, although the strength of the resulting phenotype may depend on the identity of the target.
Discussion
The series of vectors described in this study (Table 1) represents a significant expansion of the previously described auxin-inducible degron system (Nishimura et al., 2009). We have minimized the size of the AID degron without significant loss in efficiency, and we constructed and analysed combinations of AID with a set of common epitope tags for detection by antibodies or fluorescence microscopy. This now enables improved control over basal and regulated protein levels in the cell, even when no antibody against the protein of interest is available. The importance of such control is illustrated by our finding that the AID tag by itself is prone to destabilizing the target protein in a TIR1-independent manner (Figure 1B; see also Supporting information, Figure S1). This observation is surprising, as no expression problems were reported previously for constructs spanning the N-terminal half of IAA17, fused to a luciferase gene at the C-terminus (Ramos et al., 2001). Moreover, problems with partial degradation, such as the cleavage of the AID* tag from AID*–9mycRfa1 (Figure 4B), would not have been detectable by means of antibodies generated against the AID tag itself that were used in previous studies (Nishimura et al., 2009). Our system is compatible with the PCR-mediated genomic tagging procedure commonly used in budding yeast, and restriction sites allow for a relatively straightforward exchange of the promoter if the tag is fused N-terminally. Added flexibility is provided by a number of different selection markers.
The new AID*-based vectors have not eliminated potential problems inherent in the auxin-based system. For example, variations in the efficiency of degradation as well as the basal levels of the tagged proteins of interest still need to be considered. However, there are a number of ways in which the system could potentially be improved even further. In at least one case, Watase et al. (2012) have overcome the problem of inefficient proteolysis by applying the AID tag to a target protein that was inherently destabilized by a temperature-sensitive mutation. Another, more general, strategy was explored in Schizosaccharomyces pombe, where a fusion of TIR1 to the SCF adaptor protein Skp1 overcame a lack of compatibility of the plant F-box protein with the yeast cullin-based ubiquitin ligase (Kanke et al., 2011). Other options might be the use of alternative auxin-responsive IAA proteins with shorter half-lives (Abel et al., 1994; Ouellet et al., 2001) or different auxin receptors, such as AFB2, which appear to promote enhanced degradation compared to TIR1 in a yeast-based system (Havens et al., 2012). Tandem fusion of multiple AID sequences (Kubota et al., 2013) or combination with a heat-inducible N-end rule degron might also enhance degradation efficiency, and the use of repressible promoters would overcome the inherent problem of protein resynthesis acting against degradation. Finally, it should be noted that the series of constructs described here can easily be adapted for use in other organisms.
Acknowledgments
We would like to thank Masato Kanemaki for helpful discussions, sharing unpublished results and generously providing plasmids. We thank Steve Brill and John Diffley for antibodies. This study was supported by Cancer Research UK.
Supporting information on the internet
Supporting information may be found in the online version of this article.
Figure S1. TIR1 is not responsible for the auxin-independent destabilisation of Rad53 by the AID-tags. Protein levels of Rad53 carrying variations of the AID-tag in comparison with the native protein were examined in strains either containing or lacking TIR1 as indicated. All cultures were grown in the absence of auxin before preparation of the lysates, and proteins were detected by Western blotting with the indicated antibodies.
Figure S2. The position of the AID*-tag has little influence on degradation efficiency in the case of RFC1. (A) Western blot analysis showing protein levels of C- and N-terminally AID*-tagged Rfc1 in the presence or absence of 1 mM auxin. 0.1 mM CuSO4 was added where indicated to enhance expression of PCUP1-9myc-AID*RFC1. (B) Auxin sensitivity assays of the indicated strains in the presence or absence of 1 mM auxin and 0.1 mM CuSO4 (Cu).
Table S1. Sequences of oligonucleotides used in this study
Table S2. Yeast strains used in this study
References
- Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA. 1994;91:326–330. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Carthew RW. Gene silencing by double-stranded RNA. Curr Opin Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K. The interaction and integration of auxin signaling components. Plant Cell Physiol. 2012;53:965–975. doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- Kanke M, Nishimura K, Kanemaki M, et al. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011;12:8. doi: 10.1186/1471-2121-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, et al. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, et al. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, et al. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin–RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. TIR1 is not responsible for the auxin-independent destabilisation of Rad53 by the AID-tags. Protein levels of Rad53 carrying variations of the AID-tag in comparison with the native protein were examined in strains either containing or lacking TIR1 as indicated. All cultures were grown in the absence of auxin before preparation of the lysates, and proteins were detected by Western blotting with the indicated antibodies.
Figure S2. The position of the AID*-tag has little influence on degradation efficiency in the case of RFC1. (A) Western blot analysis showing protein levels of C- and N-terminally AID*-tagged Rfc1 in the presence or absence of 1 mM auxin. 0.1 mM CuSO4 was added where indicated to enhance expression of PCUP1-9myc-AID*RFC1. (B) Auxin sensitivity assays of the indicated strains in the presence or absence of 1 mM auxin and 0.1 mM CuSO4 (Cu).
Table S1. Sequences of oligonucleotides used in this study
Table S2. Yeast strains used in this study


