Abstract
Background
Recently, the use of T7 RNA polymerase instead of other viral and cellular promoters is increasing due to high efficacy of transcription in the cell cytoplasm by this polymerase. In order to translate the transcripts produced by T7 RNA polymerase in mammalian cell lines, it is necessary to include Internal Ribosome Entry Site (IRES) sequences. In addition, if sequence of poly A signal would be included after interested gene, the rate of expression could be increased in the cells.
Methods
For expression of eGFP in HEK-293 and T7-BHK cells by T7 RNA polymerase, the sequence of eGFP as well as IRES sequences upstream of eGFP gene and poly A signal were inserted into a pUC57 plasmid. On the other hand, gene of T7 RNA polymerase was cloned into modified pIRES2-EGFP plasmid. Then, the constructed plasmids were transfected into HEK-293 cells. T7-BHK cell was used for control of T7 RNA polymerase activity.
Results
Our results showed that using T7 RNA polymerase for expression of foreign genes in mammalian cell lines is highly efficient.
Conclusion
Highly efficient eGFP expression in HEK-293 cells showed that T7 RNA polymerase could be used for cytoplasmic RNA transcription such as production of anti-cancer proteins and oncolytic viral genomic RNA by reverse genetics.
Keywords: T7 RNA polymerase, Cancer gene therapy, Oncolytic RNA viruses
Introduction
Stable and high level synthesis of external proteins could be achieved by usingT7 RNA polymerase activity and T7 promoter vectors in mammalian cells [1-7]. Many studies have shown that using CMV promoter as a viral promoter in plasmids is not suitable for stably expression of genes in mammalian cells. This may be due to promoter silencing in eukaryotic cells by epigenetic conditions [8, 9]. Therefore, the use of T7 promoter that is not affected by cellular silencing mechanism may be advantageous for stable expression of foreign genes in eukaryotic cells.
The enzymatic activity of T7 RNA polymerase is needed to transcribe mRNA from T7 promoter. To express foreign genes under control of T7 promoter, the sequence of IRES (Internal Ribosome Entry Site) is necessary for translation of mRNA, since cap structure is not added at 5’-end of T7 RNA polymerase transcripts. The IRES sequences could be obtained from viral genomes such as Encephalomyocarditis (EMC) virus untranslated regions. These sequences could be detected by cellular Ribosomes for translation of mRNA [10]. To increase the high transcription power for T7 RNA polymerase, three G nucleotides have been just added after T7 promoter [11]. On the other hand, if sequence of poly A signal had included after the interested gene, the rate of expression could be increased in cells. Using T7 promoter in vectors and T7 RNA polymerase activity has different application for research. For example; T7 promoter and T7 RNA polymerase have been used in reverse genetics for rescue of the negative-stranded RNA viral genome from cDNA in cytoplasm. This technique has been applied to rescue the recombinant oncolytic RNA viruses such as measles virus and Newcastle disease virus and vaccine development [11-15]. Using T7 RNA polymerase in reverse genetics for negative-stranded RNA viruses is important, since transcription of viral RNA without cap structure is essential. Thus, using T7 promoter just before first nucleotide of virus could lead to transcription of the viral RNA with correct end of genome by T7 RNA polymerase. Also, recombinant vectors consisting of T7 RNA polymerase/T7 promoter expression system could be effective in cancer gene therapies [16, 17].
Here, we describe a construction of an expression plasmid in which an eGFP was included as a reporter gene under the control of T7 Promoter and IRES sequences for assessment of T7 RNA polymerase activity in HEK-293 and T7-BHK cells (Figure 1).
Figure 1.

Schematic representation of eGFP expression plasmid under control of T7 promoter and IRES sequences.
Materials and Methods
Cell lines
HEK-293 cells and T7-BHK cells were grown in high glucose Dulbecco’s Modified Eagle’s Minimal medium (DMEM) (Gibco) containing 10% fetal bovine serum (FBS) (Gibco), the antibiotics penicillin (100 U/ml) and streptomycin (0.1 mg/ml).
Construction of plasmids
The bacteriophage T7 RNA polymerase gene was amplified by using BL-21 genome lysate as a template. The PCR amplification of T7 RNA polymerase gene was carried out by using specific primers with Platinum-pfx kit (Invitrogen). All primers for PCR amplification are listed in table 1.
Table 1.
Primers for amplification of T7 RNA polymerase gene and FT7A, green colors represent the digestion sites of primers, red colors represent T7 promoter.
| Gene | Primer sequence | |||||
|---|---|---|---|---|---|---|
| T7,Forward primer | CTA | GCTAGC | CCACCATGAACACGATTAACATCGCTAAGAACGAC | |||
| T7,Reverse primer | CCGCTCGAGTTACGCGAACGCGAAGTCCGACTC | |||||
| FT7AF,Forward primer | TAATACGACTCACTATA | GGGACTCAGATCTCGAGC | ||||
| FT7AR,Reverse primer | AAA | GTCGAC | GGACAAACCACAACTAGAATG | |||
The PCR products for T7 RNA polymerase gene was purified using Qiagen purification kit. The T7 RNA polymerase gene was digested with NheI and XhoI. Then, after purification with a gel extraction kit (Qiagen), the DNA fragment of T7 RNA polymerase (in length of 2600Kb) was cloned into modified plasmid of pIRES2-EGFP (Clontech) and recombinant vector was called pCMV-T7.
The PCR amplification of sequences of T7 promoter, IRES, eGFP gene and poly A signal (named FT7A) was carried out by using specific primers with Platinum-pfx kit (Invitrogen). Forward and reverse primers were called FT7A-F and FT7A-R, respectively. There was not any enzyme site at 5’-end of FTA-F, but the FT7A-R contained Sal I. The sequence of T7 promoter included at 5’-end of FT7A-F. pIRES2-EGFP plasmid was used as a template for amplification of FT7A sequences.
The cloning process for FT7A was done in pUC57 plasmid in form of blunt-sticky. The PCR products for FT7A sequences were purified and digested with Sal I, but pUC57 plasmid was digested with two enzymes including SmaI and SalI. Then, the DNA fragments of FT7A sequences were cloned into pUC57 plasmid and recombinant vector was called pFT7A.To confirm FT7A cloning into pUC57, digestion with EcoRV and SalI was carried out.
Transfection
To analyze the functionality of T7 RNA polymerase, two recombinant plasmids pCMV-T7 and pFT7A were co-transfected into HEK-293 cells. On the other hand, pFT7A plasmid was transfected into T7-BHK (as a positive control), which stably expressed T7 RNA polymerase. Totally, for each cells including HEK-293 and T7-BHK, cells were grown into 6 well tissue culture plates to 80% confluency and transfected with 4µg plasmids via Lipofectamine 2000 (Invitrogen).
Final concentration of each plasmid for transfection into HEK-293 cells was 2µg for pCMV-T7 and 2µg for pFT7A. Cells were incubated for 16h at 37 ˚C, and then washed once with phosphate buffer saline (PBS) and maintained in DMEM containing 10% FBS. After 2 days, fluorescence of eGFP into transfected cells was assessed.
Results
PCR amplifications and construction of plasmids
The bacteria genome containing of bacteriophage T7 RNA polymerase was isolated from BL-21, and sequences of T7 RNA polymerase gene was amplified by PCR (Figure 2). In order to express T7 RNA polymerase in HEK-293 cells, T7 RNA polymerase gene was cloned into modified pIRES2-EGFP plasmid (pCMV-T7) (Figure 3). Then, sequence of T7 RNA polymerase was confirmed by sequence analysis.
Figure 2.
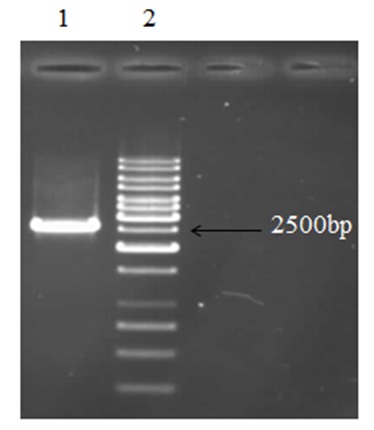
Line 2 is related to 1Kb ladder (Fermentas, cat. No.SM0311) and 2600bp band in line 1 shows amplification of T7 RNA polymerase gene.
Figure 3.
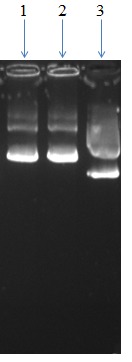
Line 3 is related with modified pIRES2-EGFP plasmid and lines 1 and 2 show recombinant pCMV-T7.
To obtain PCR product for T7 promoter, IRES, eGFP gene and poly A signal (FT7A), PCR amplification was done onto pIRES2-EGFP plasmid with specific primers (Figure 4). Then, PCR product FT7A was cloned into pUC57 plasmid. Cloning of FT7A into pUC57 was positive by double digestion with EcoRV and SalI (Figure5).
Figure 4.
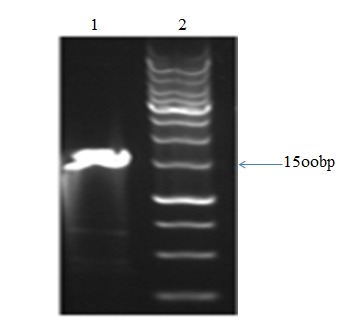
1600bp band in line 1 shows amplification of sequences including T7 promoter, IRES, eGFP gene and poly A signal (FT7A) and line 2 is related to 1Kb ladder.
Figure 5.
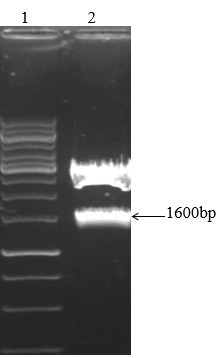
1600bp band in line 2 is related toFT7A after digestion with EcoRV and SalI.
Transfection and eGFP analysis
For analysis of T7 RNA polymerase activity in HEK-293 cells, the pCMV-T7 and pFT7A plasmids were co-transfected into HEK-293 cells. Results for T7 RNA polymerase activity were positive (Figure 6a). The eGFP fluorescence after transfection with pFT7A in T7-BHK cells which stably express T7 RNA polymerase was visible in these cells as a positive control (Figure 6b).
Figure 6.
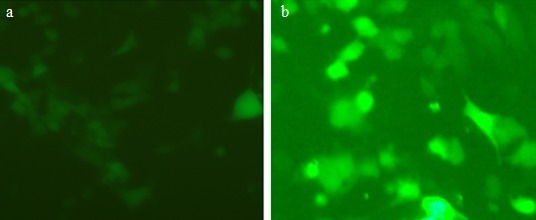
T7 RNA polymerase assay in HEK-293 cells (a) and T7-BHK cells (b) as a control was positive by using pFT7A vector.
Discussion
Transcription of mRNA in mammalian cells via CMV promoter as a viral promoter or eukaryotic promoters is dependent on cellular RNA polymerase II. Thus, expression of foreign genes by such vectors requires the entry of vectors into nucleus [1]. On the other hand, when it is necessary to constitutively express foreign genes in mammalian cells, using these promoters are problematic because they have been silent after few passages [8]. However, using T7 promoter instead of CMV promoter could be bypass gene expression silencing. Finally, gene expression by using T7 promoter and IRES sequence could be done in cytoplasm which is suitable for long transcripts.
In this study, eGFP expression from pFT7A by T7 RNA polymerase indicated that co-transfection of pFT7A and pCMV-T7 instead of stably T7 RNA polymerase expression such as T7-BHK cell line could be also efficient in eukaryotic cell lines. Therefore, this system could be used for cytoplasm RNA transcription such as production of viral RNA by reverse genetics for oncolytic RNA viruses. On the other hand, this system could be useful for cancer gene therapies. Totally, using T7 RNA polymerase and T7 promoter for expression of siRNA in cancer cells by safe bacteria delivery system has been shown that could be effective in cancer cells death [18, 19]. Our constructs including T7 RNA polymerase gene expression plasmid and foreign gene expression plasmid under control of T7 promoter (pFT7A) have the ability to express siRNA in cancer cells to target kinds of mRNA which are associated with the disease. Therefore, co-delivery of these vectors by which express T7 RNA polymerase and foreign gene for cancer cell targeting is hopeful by using viral vectors which specifically enter into cancer target cells.
Results from eGFP expression after transfection of pFT7A plasmid into T7-BHK cells indicated that using T7 promoter and IRES sequence for interested gene expression in stably cells which express T7 RNA polymerase is also efficient for increasing the rescue of oncolytic viruses by using T7 promoter before first nucleotide of virus genome.
Acknowledgments
This work was performed as part of the requirement for the fulfillment of the degree of PhD in Medical Virology of Mostafa Ghaderi at Tarbiat Modares University. We would like to thank the office of applied research of Tarbiat Modares University for its support of the project.
Footnotes
Conflicts of Interest
The authors have no conflict of interest in this study.
Authors' Contribution
Mostafa Ghaderi: PhD student who has performed as his PhD research project, Farzaneh Sabahi: Design and Management of project, and all the other authors have been involved in experiments, technical support and writing the manuscript.
REFERENCES
- 1.Brisson M, He Y, Li S, Yang J, Huang L. A novel T7 RNA polymerase autogene for efficient cytoplasmic expression of target genes. Gene therapy. 1999;6(2) doi: 10.1038/sj.gt.3300827. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Li Y, Xiong K, Wagner TE. A self-initiating eukaryotic transient gene expression system based on cotransfection of bacteriophage T7 RNA polymerase and DNA vectors containing a T7 autogene. Nucleic acids research. 1994;22(11):2114–20. doi: 10.1093/nar/22.11.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proceedings of the National Academy of Sciences. 1984;81(7):2035–9. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H, Wang C, Acsadi G, Wolff JA. High-efficiency protein synthesis from T7 RNA polymerase transcripts in 3T3 fibroblasts. Gene. 1991;109(2):193–201. doi: 10.1016/0378-1119(91)90609-f. [DOI] [PubMed] [Google Scholar]
- 5.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proceedings of the National Academy of Sciences. 1990;87(17):6743–7. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X, Huang L. Cytoplasmic expression of a reporter gene by co-delivery of T7 RNA polymerase and T7 promoter sequence with cationic liposomes. Nucleic acids research. 1993;21(12):2867–72. doi: 10.1093/nar/21.12.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieber A, Kiessling U, Strauss M. High level gene expression in mammalian cells by a nuclear T7-phage RNA polymerase. Nucleic acids research. 1989;17(21):8485–93. doi: 10.1093/nar/17.21.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teschendorf C, Warrington K, Siemann DW, Muzyczka N. Comparison of the EF-1 alpha and the CMV promoter for engineering stable tumor cell lines using recombinant adeno-associated virus. Anticancer research. 2002;22(6A):3325. [PubMed] [Google Scholar]
- 9.Choi KH, Basma H, Singh J, Cheng P-W. Activation of CMV promoter-controlled glycosyltransferase and β-galactosidase glycogenes by butyrate, tricostatin A, and 5-Aza-2′-deoxycytidine. Glycoconjugate journal. 2005;22(1-2):63–9. doi: 10.1007/s10719-005-0326-1. [DOI] [PubMed] [Google Scholar]
- 10.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends in biochemical sciences. 2008;33(6):274–83. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, et al. Rescue of measles viruses from cloned DNA. The EMBO journal. 1995;14(23):5773. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wit E, Spronken MI, Vervaet G, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. A reverse-genetics system for Influenza A virus using T7 RNA polymerase. Journal of General Virology. 2007;88(4):1281–7. doi: 10.1099/vir.0.82452-0. [DOI] [PubMed] [Google Scholar]
- 13.Yap C-C, Ishii K, Aoki Y, Aizaki H, Tani H, Shimizu H, et al. A hybrid baculovirus-T7 RNA polymerase system for recovery of an infectious virus from cDNA. Virology. 1997;231(2):192–200. doi: 10.1006/viro.1997.8537. [DOI] [PubMed] [Google Scholar]
- 14.Herfst S, de Graaf M, Schickli JH, Tang RS, Kaur J, Yang C-F, et al. Recovery of human metapneumovirus genetic lineages A and B from cloned cDNA. Journal of virology. 2004;78(15):8264–70. doi: 10.1128/JVI.78.15.8264-8270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Liu H, Liu P, Kong X. Plasmids driven minigenome rescue system for Newcastle disease virus V4 strain. Molecular biology reports. 2009;36(7):1909–14. doi: 10.1007/s11033-008-9398-x. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Li Y, Xiong K, Aizicovici S, Xie Y, Zhu Q, et al. Cancer gene therapy by direct tumor injections of a nonviral T7 vector encoding a thymidine kinase gene. Human gene therapy. 1998;9(5):729–36. doi: 10.1089/hum.1998.9.5-729. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Li Y, Xiong K, Xie Y, Aizicovici S, Snodgrass R, et al. A novel nonviral cytoplasmic gene expression system and its implications in cancer gene therapy. Cancer gene therapy. 1995;2(4):281–9. [PubMed] [Google Scholar]
- 18.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends in biotechnology. 2010;28(11):570–9. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology. 2006;24(6):697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]


