Abstract
Summary
Given the biological function of SOX6 and recent genome-wide association finding, we performed a fine-mapping association analyses to investigate the relationship between SOX6 and BMD both in Caucasian and Chinese populations. We identified many single-nucleotide polymorphisms (SNPs) within or near the SOX6 gene to be significantly associated with hip bone mineral density (BMD).
Introduction
SOX6 gene is an essential transcription factor in chondrogenesis and cartilage formation. Recent genome-wide association studies (GWAS) detected a SNP (rs7117858) located at the downstream of SOX6 significantly associated with hip BMD.
Methods
Given the biological function of SOX6 and the GWAS finding, we considered SOX6 as a new candidate for BMD and osteoporosis. Therefore, in this study, we performed a fine-mapping association analyses to investigate the relationship between SNPs within and near the SOX6 gene and BMD at both hip and spine. A total of 301 SNPs were tested in two independent US Caucasian populations (2,286 and 1,000 unrelated subjects, respectively) and a Chinese population (1,627 unrelated Han subjects).
Results
We confirmed that the previously reported rs7117858-A was associated with reduced hip BMD, with combined P value of 2.45×10−4. Besides this SNP, we identified another 19 SNPs within or near the SOX6 gene to be significantly associated with hip BMD after false discovery rate adjustment. The most significant SNP was rs1347677 located at the intron 3 (P=3.15×10−7). Seven additional SNPs in high linkage disequilibrium with rs1347677 were also significantly associated with hip BMD. SNPs in SOX6 showed significant skeletal site specificity since no SNP was detected to be associated with spine BMD.
Conclusion
Our study identified many SNPs in the SOX6 gene associated with hip BMD even across different ethnicities, which further highlighted the importance of the SOX6 gene influencing BMD variation and provided more information to the understanding of the genetic architecture of osteoporosis.
Keywords: Association, BMD, Osteoporosis, SOX6
Introduction
Osteoporosis is a serious public health problem. It is a metabolic skeletal disease mainly characterized by low bone mass and microarchitectural deterioration of bone tissue, with the consequent increase in the risk of fragility fractures [1]. Clinically, bone mineral density (BMD) is the single best predictor of osteoporotic fractures, and has been widely used as a reference standard for the description of osteoporosis [2, 3]. BMD is a highly heritable quantitative trait. Previous studies have demonstrated that approximately 50% to 80% of the variation in BMD can be explained by genetic factors [4–6].
SOX6 gene (SRY-box 6) encodes a member of the D subfamily of sex determining region y-related transcription factors, which is characterized by a conserved DNA-binding domain termed the high-mobility group box. SOX6 is an essential transcription factor in chondrogenesis and cartilage formation [7–10]. This gene was found to cooperate with Col2a1 in chondrogenesis [11] and mediate BMP2 in the activation and maintenance of chondrogenesis during murine fracture healing [12]. SOX6 was also found to have significant differential expression during osteoblast development [13]. Interestingly, in human, recent genome-wide association studies (GWAS) [13, 14] and a replication study [15] have reported that an SNP (rs7117858) located at the downstream of the SOX6 gene was significantly associated with hip BMD. Given the biological function of SOX6 and this GWAS finding, we thought that the SOX6 gene could be a new candidate for elucidating the genetic impact on BMD. We further raise a question that whether other SNPs within the SOX6 gene are also associated with BMD. Investigating the SNPs within the SOX6 gene may provide further insights to understand the relationship between this gene and BMD. In addition, replication of genetic associations in independent populations is essential to evaluate a positive finding and to further explore the role of these variants in the complex traits. Therefore, the aim of this study was to conduct a fine-mapping association analyses to investigate the relationship between SNPs located within and near the SOX6 gene and BMD. Our study was performed in three sample sets from two ethnicities, including two US Midwestern Caucasian populations and a Midwestern Chinese Han population, in order to see whether the variants identified are common or ethnicity-specific.
Materials and methods
Subjects
The study was approved by the required Institutional Review Board or Research Administration of the involved institutions. Signed informed consent documents were obtained from all study participants before entering the study.
Caucasian samples 1 and 2
Caucasian samples 1 and 2 contained 2,286 and 1,000 unrelated adults, respectively. These unrelated subjects from both samples were identified from our established and expanding database containing more than 10,000 subjects. All of the chosen subjects were US Caucasians of Northern European origin living in Midwestern area. Subjects with chronic diseases and conditions that might potentially affect bone mass, structure, or metabolism were excluded. These diseases/conditions included chronic disorders involving vital organs (heart, lung, liver, kidney, brain), serious metabolic diseases (diabetes, hypo- and hyper-parathyroidism, hyperthyroidism, etc.), other skeletal diseases (Paget’s disease, osteogenesis imperfecta, rheumatoid arthritis, etc.), chronic use of drugs affecting bone metabolism (hormone replacement therapy, corticosteroid therapy, anti-convulsant drugs), and malnutrition conditions (such as chronic diarrhea, chronic ulcerative colitis, etc.), etc. In addition, subjects taking anti-bone resorptive or bone anabolic agents/drugs, such as bisphosphonates were also excluded from this study. The purpose of these exclusions was to minimize the influence of known environmental and therapeutic factors on bone variation. BMD values at hip and spine were measured using Hologic 4500W machines (Hologic Inc., Bedford, MA, USA) that were calibrated daily. The coefficient of variation (CV) values of the dual-energy X-ray absorptiometry (DXA) measurements for spine and hip BMDs were approximately 1.98% and 1.87%, respectively.
Chinese sample
The Chinese sample consisted of 1,627 unrelated subjects. The subjects were recruited from Midwestern Chinese Han adults living in Xi’an and Changsha cities. The exclusion criteria were the same as with Caucasian samples. BMD at hip and spine was measured using the same model Hologic 4500W machines (Hologic Inc., Bedford, MA, USA) under the same strict protocols used in the Caucasian sample. The CV values of the DXA measurements for spine and hip BMDs were approximately 1.01% and 1.33%, respectively.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. For Caucasian sample 1 and Chinese sample, SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), according to the Affymetrix protocol. Briefly, approximately 250 ng of genomic DNA was digested with restriction enzyme NspI or StyI. Digested DNA was adaptor-ligated and polymerase chain reaction (PCR)-amplified for each sample. Fragment PCR products were then labeled with biotin, denatured, and hybridized to the arrays. Arrays were then washed and stained using Phycoerythrin on Affymetrix Fluidics Station, and scanned using the GeneChip Scanner 3000 7G to quantitate fluorescence intensities. Data management and analyses were conducted using the Genotyping Command Console. Only samples with a minimum call rate of 95% were included. Due to efforts of repeat experiments, all samples met this criteria and the final mean call rate reached a high level of 98.93% for Caucasian sample 1 and 98.96% for Chinese sample. For the Caucasian sample 2, SNP genotyping was performed using the Affymetrix Human Mapping 500K array set, which has been finished in our previous experiments [16]. SNPs that deviated from Hardy–Weinberg equilibrium (HWE, P<0.0001) and had a minor allele frequency <0.01 were discarded in each sample set. Thus, 301 SNPs located within and near the SOX6 gene were included for subsequent association analyses. The basic characteristics of these SNPs are summarized in Supplementary Table 1.
Statistical analyses
Before association analyses, principal component analysis implemented in EIGENSTRAT [17] was used to correct for potential population stratification that may lead to spurious association results. The first ten principal components emerging from the EIGENSTRAT analyses, along with sex, height, weight and age, were used as covariates to adjust the raw BMD values in each sample. The residues were used for association analyses. For the Caucasian sample 1 and the Chinese sample, a linear regression implemented in PLINK [18] was fitted to test for association assuming an additive inheritance model. For the Caucasian sample 2, imputation was used to evaluate associations for the same SNPs across study populations using different Affymetrix arrays. Thus, IMPUTE program [19] was utilized to impute the genotypes of SNPs detected on Array 6.0 but not on 500K array set based on HapMap data (release 22). To ensure the reliability of the imputation, all of those imputed SNPs have reached a calling threshold of 0.90, i.e., a 90% probability that an imputed genotype is true. SNPTEST [19] was used to test for associations in this sample.
Meta-analysis calculations were done using the METAL software package (http://genome.sph.umich.edu/wiki/METAL_Documentation) taking into account sample size and direction of effect. SNAP was used to characterize linkage disequilibrium (LD) and depict the regional association plot [20]. A raw P value of <0.05 in our study was considered nominally significant, which were further subjected to a false discovery rate (FDR) of Benjamini and Hochberg procedure [21] to account for multiple comparisons.
Results
The basic characteristics of the study subjects are presented in Table 1. We summarized the major association results of SOX6 with hip BMD in Table 2. After association tests and multiple testing corrections, 20 SNPs showed significant results with hip BMD, with P values ranging from 9.12×10−4 to 3.15×10−7 (Table 2). The directions of effect for each significant SNP were perfectly consistent in Caucasian samples 1, Caucasian sample 2, and Chinese sample. Among these 20 SNPs, 15 SNPs located within the SOX6 gene, the other 5 SNPs located at the downstream of the SOX6 gene.
Table 1.
Basic characteristics of the study subjects
| Trait | Caucasian sample 1 | Caucasian sample 2 | Chinese sample |
|---|---|---|---|
| Number | 2,286 | 1,000 | 1,627 |
| Age (years) | 51.37 (13.76) | 50.23 (18.24) | 34.49 (13.24) |
| Weight (kg) | 75.27 (17.54) | 80.16 (17.79) | 60.12 (10.48) |
| Height (cm) | 166.35 (8.47) | 170.83 (9.74) | 164.25 (8.16) |
| Female/male | 1727/558 | 501/499 | 825/802 |
| Hip BMD (g/cm2) | 0.968 (0.175) | 0.973 (0.156) | 0.920 (0.134) |
| Spine BMD (g/cm2) | 1.025 (0.157) | 1.032 (0.164) | 0.947 (0.127) |
Data are shown as mean (standard deviation, SD).
Table 2.
Significant association results for 20 SNPs in SOX6 with hip BMD
| SNP | Position | Genic position | A1/A2 | Caucasian sample 1
|
Caucasian sample 2
|
Chinese sample
|
Pcombine | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | BETA | SE | P value | Freq | BETA | SE | P value | Freq | BETA | SE | P value | |||||
| rs1531903 | 15625402 | Downstream | C/G | 0.160 | 0.0089 | 0.0049 | 7.09×10−2 | 0.159 | 0.0132 | 0.0074 | 7.35×10−2 | 0.181 | 0.0120 | 0.0051 | 1.89×10−2 | 6.55×10−4 |
| rs7117858 | 15651038 | Downstream | G/A | 0.206 | 0.0100 | 0.0045 | 2.72×10−2 | 0.220 | 0.0063 | 0.0067 | 3.46×10−1 | 0.181 | 0.0151 | 0.0051 | 3.20×10−3 | 2.45×10−4 |
| rs7108738 | 15666660 | Downstream | C/A | 0.177 | 0.0092 | 0.0048 | 5.45×10−2 | 0.169 | 0.0105 | 0.0076 | 1.70×10−1 | 0.153 | 0.0199 | 0.0055 | 3.27×10−4 | 4.92×10−5 |
| rs16931831 | 15686130 | Downstream | T/C | 0.167 | 0.0094 | 0.0048 | 5.14×10−2 | 0.174 | 0.0105 | 0.0071 | 1.40×10−1 | 0.179 | 0.0152 | 0.0051 | 3.02×10−3 | 1.84×10−4 |
| rs11827785 | 15687087 | Downstream | C/A | 0.199 | 0.0108 | 0.0046 | 1.83×10−2 | 0.212 | 0.0072 | 0.0068 | 2.90×10−1 | 0.180 | 0.0142 | 0.0051 | 5.60×10−3 | 2.07×10−4 |
| rs7118395 | 16155641 | Intron6 | C/T | 0.499 | 0.0085 | 0.0037 | 2.15×10−2 | 0.492 | 0.0009 | 0.0053 | 8.60×10−1 | 0.294 | 0.0125 | 0.0043 | 3.82×10−3 | 7.83×10−4 |
| rs2028162 | 16204051 | Intron4 | G/A | 0.260 | 0.0136 | 0.0042 | 1.19×10−3 | 0.242 | 0.0062 | 0.0070 | 3.71×10−1 | 0.302 | 0.0090 | 0.0043 | 3.39×10−2 | 1.30×10−4 |
| rs1837096 | 16219450 | Intron3 | G/A | 0.199 | 0.0161 | 0.0046 | 4.73×10−4 | 0.205 | 0.0179 | 0.0067 | 7.60×10−3 | 0.303 | 0.0093 | 0.0043 | 2.89×10−2 | 1.49×10−6 |
| rs12798980 | 16234929 | Intron3 | C/G | 0.197 | 0.0166 | 0.0046 | 3.44×10−4 | 0.204 | 0.0173 | 0.0067 | 1.04×10−2 | 0.305 | 0.0090 | 0.0042 | 3.41×10−2 | 1.75×10−6 |
| rs10832576 | 16236880 | Intron3 | A/T | 0.199 | 0.0170 | 0.0046 | 2.33×10−4 | 0.205 | 0.0175 | 0.0067 | 9.38×10−3 | 0.305 | 0.0080 | 0.0043 | 6.14×10−2 | 2.45×10−6 |
| rs12274377 | 16247709 | Intron3 | A/T | 0.425 | −0.0079 | 0.0037 | 3.56×10−2 | 0.422 | −0.0133 | 0.0057 | 2.00×10−2 | 0.420 | −0.0065 | 0.0041 | 1.12×10−1 | 7.32×10−4 |
| rs1347677 | 16252988 | Intron3 | G/T | 0.196 | 0.0173 | 0.0047 | 2.33×10−4 | 0.200 | 0.0205 | 0.0067 | 2.37×10−3 | 0.304 | 0.0094 | 0.0043 | 2.69×10−2 | 3.15×10−7 |
| rs11023907 | 16281025 | Intron3 | T/G | 0.196 | 0.0120 | 0.0048 | 1.24×10−2 | 0.205 | 0.0161 | 0.0068 | 1.73×10−2 | 0.300 | 0.0080 | 0.0043 | 6.62×10−2 | 1.36×10−4 |
| rs10219384 | 16285545 | Intron3 | T/C | 0.424 | −0.0085 | 0.0037 | 2.28×10−2 | 0.410 | −0.0142 | 0.0055 | 1.06×10−2 | 0.412 | −0.0073 | 0.0040 | 7.01×10−2 | 1.93×10−4 |
| rs297366 | 16286125 | Intron3 | C/T | 0.425 | −0.0082 | 0.0037 | 2.70×10−2 | 0.413 | −0.0134 | 0.0055 | 1.54×10−2 | 0.412 | −0.0077 | 0.0040 | 5.42×10−2 | 2.19×10−4 |
| rs297326 | 16346308 | Intron1 | T/G | 0.289 | 0.0040 | 0.0040 | 3.21×10−1 | 0.269 | 0.0183 | 0.0060 | 2.46×10−3 | 0.487 | 0.0089 | 0.0040 | 2.42×10−2 | 7.98×10−4 |
| rs297324 | 16345860 | Intron1 | A/G | 0.291 | 0.0047 | 0.0040 | 2.38×10−1 | 0.272 | 0.0185 | 0.0059 | 1.81×10−3 | 0.505 | 0.0076 | 0.0040 | 5.46×10−2 | 9.12×10−4 |
| rs297339 | 16360859 | Intron1 | A/G | 0.289 | 0.0044 | 0.0041 | 2.89×10−1 | 0.271 | 0.0191 | 0.0059 | 1.37×10−3 | 0.494 | 0.0099 | 0.0040 | 1.45×10−2 | 3.29×10−4 |
| rs10832606 | 16416136 | Intron1 | C/T | 0.292 | 0.0041 | 0.0040 | 3.02×10−1 | 0.270 | 0.0193 | 0.0059 | 1.19×10−3 | 0.488 | 0.0081 | 0.0039 | 3.90×10−2 | 7.86×10−4 |
| rs11023944 | 16417484 | Intron1 | A/C | 0.293 | 0.0044 | 0.0040 | 2.68×10−1 | 0.271 | 0.0195 | 0.0059 | 1.02×10−3 | 0.505 | 0.0075 | 0.0039 | 5.41×10−2 | 8.25×10−4 |
Freq, frequency is shown for allele A1. For the Caucasian sample 1 and the Chinese sample, a linear regression implemented in PLINK was fitted to test for association assuming an additive inheritance model. For the Caucasian sample 2, IMPUTE program was utilized to impute the genotypes of SNPs detected on Array 6.0 but not on 500K array set based on HapMap data (release 22). SNPTEST was used to test for associations in this sample. Meta-analysis was done using the METAL software package taking into account sample size and direction of effect (Pcombine).
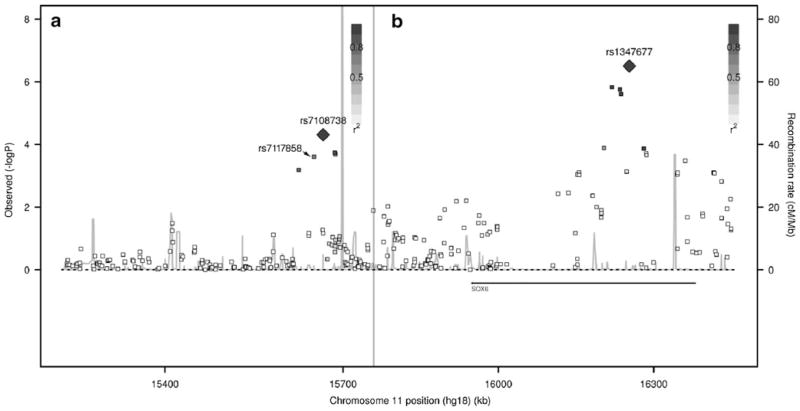
The most significant SNP was rs1347677 located at the intron 3 of SOX6, with the P values of 2.33×10−4, 2.37×10−3, and 2.69×10−2 in Caucasian sample 1, Caucasian sample 2, and Chinese sample, respectively. After meta-analysis by METAL software package, the combined P value achieved a highly significant level of 3.15×10−7. The rs1347677-T was associated with reduced hip BMD values with the effect size (beta) of −0.0173, −0.0205, and −0.0094 in Caucasian sample 1, Caucasian sample 2, and Chinese sample, respectively. According to the FASTSNP program (http://fastsnp.ibms.sinica.edu.tw), rs1347677 is located at intronic enhancer region. A change of “G→T” at rs1347677 may lead to creation of binding sites for transcription factors GATA-1, GATA-2, S8, and CdxA. There were seven additional SNPs (rs297366, rs10219384, rs11023907, rs12274377, rs10832576, rs12798980, rs1837096, and rs2028162) showed significant association signals with hip BMD around the top significant SNP rs1347677. The P values of these SNPs were ranged from 7.32×10−4–1.49×10−6 (Table 2). We characterized the LD for these SNPs using the regional association plot. As shown in Fig. 1b, these SNPs were in high LD with the top significant SNP rs1347677.
Fig. 1.
Regional association plot for SOX6 on chromosome 11. In the left of grey line (a), r2 of pairwise LD is calculated between rs7108738 and other SNPs in the left of grey line. In the right of grey line (b), r2 of pairwise LD is calculated between rs1347677 and other SNPs in the right of grey line. There is no LD between rs7108738 and rs1347677
For the previously reported SNP rs7117858 identified by GWAS [13, 14], significant association was successfully replicated in our study samples, with combined P value of 2.45×10−4. The rs7117858-A was associated with reduced hip BMD value with the beta values of −0.01, −0.0063, and −0.015 in Caucasian sample 1, Caucasian sample 2, and Chinese sample, respectively. The frequency of rs7117858-A detected in our study was consistent with that previously reported. Interestingly, beside the SNP rs7117858, we detected another four SNPs (rs1531903, rs7108738, rs16931831, and rs11827785) around rs7117858 significantly associated with hip BMD. The P values of these four SNPs were 6.55×10−4 to 4.92×10−5 (Table 2). These SNPs had high LD with the SNP rs7117858 (Fig. 1a).
Previous reports [14, 15] found that the variation in SOX6 was skeletal site specific, since the significant signal was only detected from the SNP associated with hip BMD but not with spine BMD. Our study obtained consistent results. No SNP was detected to be significantly associated with spine BMD.
Discussion
SOX6 is a newly identified candidate gene for osteoporosis. A SNP (rs7117858) located at the downstream of the SOX6 gene was reported to be associated with hip BMD by recent GWAS [13, 14] and a replication study [15]. In addition, our group has detected an SNP rs11023787 in the SOX6 to be associated with wrist bone mass by GWAS in the Caucasian sample 2 [22]. Our group has also found two SNPs (rs297325 and rs4756846) associated with BMI and hip BMD by bivariate GWAS in the Caucasian sample 2 [23]. However, most GWAS focused only on those SNPs of top-ranking statistical significance, which may ignore some useful information. In this study, combining all the sample sets in our group, we performed a fine-mapping association study to investigate the relationship between SNPs within and near the SOX6 gene and BMD at both hip and spine. A group of SNPs of the SOX6 gene were identified to be significantly associated with hip BMD both in the Caucasian and Chinese populations. Our results further supported the potential contribution of SOX6 to the variation of BMD and the pathogenesis of osteoporosis.
The SOX6 gene is expressed in various tissues, most abundantly in skeletal muscle. SOX6 is a key transcription factor in chondrogenesis and cartilage formation [7–10]. During embryonic development, trunk, limbs and the majority of the craniofacial skeleton are developed through endochondral bone formation. The process of endochondral bone formation is a complex one that contains multiple stages, including mesenchymal cells differentiating into cartilage cells, pre-hypertrophic chondrocytes, and hypertrophic chondrocytes, as well as mesenchymal cells surrounding hypertrophic chondrocytes differentiating into osteoblasts [24]. A series of molecular signals play important roles in regulating these multiple stages, including genes in the SOX family. For example, SOX6 has been shown to induce chondrocyte hypertrophy and permit formation of prehypertrophic and hypertrophic zones [9]. Sox6−/− mice are born with cartilage defects [24]. Recently, Hsu et al. have found significant differential expression for SOX6 during osteoblast development [13]. Taking into account our association findings and the above lines of evidence, SOX6 might affect BMD or osteoporosis through regulating endochondral bone formation. However, the above mechanism is still speculative and needs extensive functional studies for final validation.
It is necessary to evaluate the association findings in different populations from different ethnicities, since the genomic variation is greater when compared across ethnicities. Our study successfully replicated the association between the previously identified SNP rs7117858 and hip BMD [13–15], which demonstrated the validity of the initial finding. Besides this SNP, we also identified another 19 SNPs of SOX6 significantly associated with hip BMD both in the Caucasian and Chinese populations and the effect directions were perfectly consistent, suggesting that the SOX6 gene might be a common variant for BMD even across different ethnicities.
Our results, together with previous findings [13–15] showed that the association between variations of SOX6 and BMD was skeletal site specific. Significant association signals were only detected for hip BMD but not for spine BMD. Hip fracture and spine fracture occurred most frequently in human; however, the genetic correlation of these two sites was less 0.8 in both females and males [25]. The SOX6 gene might belong to the 20% of genetic determinations of hip BMD that was not shared with spine.
In summary, we performed a fine-mapping association analysis for the SOX6 gene and BMD. We confirmed that the SNP rs7117858 located at the downstream of SOX6 was significantly associated with hip BMD both in Chinese Han and Caucasian populations. Importantly, we detected 15 SNPs located in the introns of SOX6 and 4 more SNPs located at the downstream of SOX6 to be significantly associated with hip BMD. Our findings further highlighted the importance of the SOX6 gene influencing BMD variation and provided more information to the understanding of the genetic architecture of osteoporosis.
Supplementary Table 1
Acknowledgments
This work was supported by grants from NIH (R01 AR050496, R21 AG027110, R01 AG026564, R01 AR057049-01A1 and R21 AA015973) and a SCOR (Specialized Center of Research) grant (P50 AR055081). The study was also funded by grants from National Natural Science Foundation of China (81000363, 31000554), the PhD. Programs Foundation of Ministry of Education of China (20100201120058), the Fundamental Research Funds for the Central Universities, Shanghai Leading Academic Discipline Project (S30501), a grant from Ministry of Education to ShangHai University of Science and Technology, and startup funds from University of Shanghai for Science and Technology and Xi’an Jiaotong University, and the Ministry of Education of China.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00198-011-1626-x) contains supplementary material, which is available to authorized users.
Conflicts of interest None.
Contributor Information
T.-L. Yang, Key Laboratory of Biomedical Information Engineering of Ministry of Education, and Institute of Molecular Genetics, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, People’s Republic of China
Y. Guo, Email: guoyan253@mail.xjtu.edu.cn, Key Laboratory of Biomedical Information Engineering of Ministry of Education, and Institute of Molecular Genetics, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, People’s Republic of China
Y.-J. Liu, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
H. Shen, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
Y.-Z. Liu, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
S.-F. Lei, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
J. Li, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
Q. Tian, School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
H.-W. Deng, Email: hdeng2@tulane.edu, Institute of Bioscience and Biotechnology, School of Science, Beijing Jiaotong University, Beijing 100044, People’s Republic of China. School of Public Health and Tropical Medicine, Tulane University, New Orleans, LA 70112, USA
References
- 1.Ray NF, Chan JK, Thamer M, Melton LJ., 3rd Medical expenditures for the treatment of osteoporotic fractures in the United States in 1995: report from the National Osteoporosis Foundation. J Bone Miner Res. 1997;12:24–35. doi: 10.1359/jbmr.1997.12.1.24. [DOI] [PubMed] [Google Scholar]
- 2.Johnell O, Kanis JA, Oden A, Johansson H, De Laet C, Delmas P, Eisman JA, Fujiwara S, Kroger H, Mellstrom D, Meunier PJ, Melton LJ, 3rd, O’Neill T, Pols H, Reeve J, Silman A, Tenenhouse A. Predictive value of BMD for hip and other fractures. J Bone Miner Res. 2005;20:1185–1194. doi: 10.1359/JBMR.050304. [DOI] [PubMed] [Google Scholar]
- 3.Kanis JA, Oden A, Johnell O, Johansson H, De Laet C, Brown J, Burckhardt P, Cooper C, Christiansen C, Cummings S, Eisman JA, Fujiwara S, Gluer C, Goltzman D, Hans D, Krieg MA, La Croix A, McCloskey E, Mellstrom D, Melton LJ, 3rd, Pols H, Reeve J, Sanders K, Schott AM, Silman A, Torgerson D, van Staa T, Watts NB, Yoshimura N. The use of clinical risk factors enhances the performance of BMD in the prediction of hip and osteoporotic fractures in men and women. Osteoporos Int. 2007;18:1033–1046. doi: 10.1007/s00198-007-0343-y. [DOI] [PubMed] [Google Scholar]
- 4.Arden NK, Spector TD. Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res. 1997;12:2076–2081. doi: 10.1359/jbmr.1997.12.12.2076. [DOI] [PubMed] [Google Scholar]
- 5.Dequeker J, Nijs J, Verstraeten A, Geusens P, Gevers G. Genetic determinants of bone mineral content at the spine and radius: a twin study. Bone. 1987;8:207–209. doi: 10.1016/8756-3282(87)90166-9. [DOI] [PubMed] [Google Scholar]
- 6.Slemenda CW, Christian JC, Williams CJ, Norton JA, Johnston CC., Jr Genetic determinants of bone mass in adult women: a reevaluation of the twin model and the potential importance of gene interaction on heritability estimates. J Bone Miner Res. 1991;6:561–567. doi: 10.1002/jbmr.5650060606. [DOI] [PubMed] [Google Scholar]
- 7.Dy P, Smits P, Silvester A, Penzo-Mendez A, Dumitriu B, Han Y, de la Motte CA, Kingsley DM, Lefebvre V. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev Biol. 2010;341:346–359. doi: 10.1016/j.ydbio.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9(Suppl A):S69–S75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- 9.Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda T, Kawaguchi H, Kamekura S, Ogata N, Mori Y, Nakamura K, Ikegawa S, Chung UI. Distinct roles of Sox5, Sox6, and Sox9 in different stages of chondrogenic differentiation. J Bone Miner Metab. 2005;23:337–340. doi: 10.1007/s00774-005-0610-y. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uusitalo H, Hiltunen A, Ahonen M, Gao TJ, Lefebvre V, Harley V, Kahari VM, Vuorio E. Accelerated up-regulation of L-Sox5, Sox6, and Sox9 by BMP-2 gene transfer during murine fracture healing. J Bone Miner Res. 2001;16:1837–1845. doi: 10.1359/jbmr.2001.16.10.1837. [DOI] [PubMed] [Google Scholar]
- 13.Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang L, Richards JB, Estrada K, Zhou Y, van Nas A, Moffatt MF, Zhai G, Hofman A, van Meurs JB, Pols HA, Price RI, Nilsson O, Pastinen T, Cupples LA, Lusis AJ, Schadt EE, Ferrari S, Uitterlinden AG, Rivadeneira F, Spector TD, Karasik D, Kiel DP. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HA, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FM, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, Spector T, Kiel DP, Stefansson K, Ioannidis JP, Uitterlinden AG. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, Tang NL, Koh JM, Xiao SM, Kwok TC, Kim GS, Chan JC, Cherny S, Lee SH, Kwok A, Ho S, Gretarsdottir S, Kostic JP, Palsson ST, Sigurdsson G, Sham PC, Kim BJ, Kung AW, Kim SY, Woo J, Leung PC, Kong A, Thorsteinsdottir U, Stefansson K. European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS ONE. 2010;5:e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, Pan F, Chen Y, Zhang ZX, Dong SS, Xu XH, Yan H, Liu X, Qiu C, Zhu XZ, Chen T, Li M, Zhang H, Zhang L, Drees BM, Hamilton JJ, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 20.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc. 1995;57:289–300. [Google Scholar]
- 22.Tan L, Liu R, Lei S, Pan R, Yang T, Yan H, Pei Y, Yang F, Zhang F, Pan F, Zhang Y, Hu H, Levy S, Deng H. A genome-wide association analysis implicates SOX6 as a candidate gene for wrist bone mass. Sci China Life Sci. 2011;53:1065–1072. doi: 10.1007/s11427-010-4056-7. [DOI] [PubMed] [Google Scholar]
- 23.Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, Zhang L, Liu XG, Yan H, Wang L, Zhang YP, Levy S, Recker RR, Deng HW. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS ONE. 2009;4:e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13:721–727. doi: 10.1016/s0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 25.Yang TL, Zhao LJ, Liu YJ, Liu JF, Recker RR, Deng HW. Genetic and environmental correlations of bone mineral density at different skeletal sites in females and males. Calcif Tissue Int. 2006;78:212–217. doi: 10.1007/s00223-005-0267-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.