Summary
Availability of diverse genomes makes it possible to predict gene function based on shared evolutionary history. This approach can be challenging, however, for pathways whose components do not exhibit a shared history, but rather, consist of distinct “evolutionary modules.” We introduce a computational algorithm, CLIME (clustering by inferred models of evolution), which inputs a eukaryotic species tree, homology matrix, and pathway (gene set) of interest. CLIME partitions the gene set into disjoint evolutionary modules, simultaneously learning the number of modules and a tree-based evolutionary history that defines each module. CLIME then expands each module by scanning the genome for new components that likely arose under the inferred evolutionary model. Application of CLIME to ∼1000 annotated human pathways, organelles and proteomes of yeast, red algae, and malaria, reveals unanticipated evolutionary modularity and novel, co-evolving components. CLIME is freely available and should become increasingly powerful with the growing wealth of eukaryotic genomes.
Introduction
Biological pathways and complexes represent the fruits of extensive pruning, expansion and mutation that have occurred over evolutionary timescales. For example, mitochondria represent a defining feature of all eukaryotes, yet an estimated one-half of the organelle's ancestral machinery has been lost (Vafai and Mootha, 2012), and the remaining machinery varies significantly across eukaryotic taxa, with many new lineage-specific innovations. Similarly, cilia were likely present in the last common eukaryotic ancestor, though most plants and fungi lost this organelle completely while nematodes have specifically lost motile cilia. Charting the evolutionary history of modern-day pathways and complexes can help to define the taxonomic distribution of pathways and thereby highlight model organisms for experimental studies. Such evolutionary analyses may also teach us about the environmental niches within which they evolved. Importantly, correlated gains and losses can help to predict the function of unstudied genes, and also reveal alternative functions even for genes considered to be well-characterized.
Pioneering work introduced the concept of “phylogenetic profiling” to chart the phylogenetic distribution of genes and relate them to each other (Pellegrini et al., 1999). In this approach, a binary vector of presence and absence of a given gene across sequenced organisms is used to predict function of genes sharing a similar profile, based on the Hamming distance (Hamming, 1950). A number of different computational methods have been developed (Kensche et al., 2008), and have been applied successfully to predict components for prokaryotic protein complexes (Pellegrini et al., 1999), phenotypic traits like pili, thermophily, and respiratory tract tropism (Jim et al., 2004), cilia (Li et al., 2004), mitochondrial complex I (Ogilvie et al., 2005), and small RNA pathways (Tabach et al., 2013).
Although many phylogenetic profiling algorithms are now available, several features limit their utility (Kensche et al., 2008). First, most existing methods compare an input gene to a query gene one at a time – which cannot take advantage of patterns only discernible by analyzing a collection of input genes. Second, most methods do not explicitly model errors in a gene's phylogenetic profile, each of which may be individually noisy due to the inherent challenges of genome assembly, gene annotation, and detection of distant homologs (Trachana et al., 2011). Third, with a few notable exceptions (Barker and Pagel, 2005; Mering et al., 2003; Vert, 2002; Zhou et al., 2006), most existing algorithms do not take into account the phylogenetic tree of the input species, but assume independence across species and hence are highly sensitive to the choice of organisms selected. Available tree-based methods are computationally intensive and not readily scalable to large genomes (Barker et al., 2007; Barker and Pagel, 2005).
Because most existing phylogenetic profiling methods are designed to operate on single genes, they cannot be readily extended to biological pathways, where each member may have different phylogenetic profiles. Our previous experience with mitochondrial complex I illustrates this point (Pagliarini et al., 2008). Human complex I is a macromolecular machine consisting of 44 structural subunits. We observed that these subunits did not share a single, common history of gains and losses across eukaryotic evolution, but clustered into several distinct evolutionary modules. One “ancestral” module consisted of 14 core subunits that were present in bacteria and in humans yet lost independently four times in eukaryotic evolution, whereas other modules consisted of recent animal or vertebrate innovations. By first identifying the “ancestral” module, we could scan the human genome to identify additional genes sharing the same evolutionary history. Five of these genes have since been shown to encode complex I assembly factors that are mutated in inherited complex I deficiencies (Mimaki et al., 2012).
Our previous analysis suggested that biological pathways, as we conceive of them, represent mosaics of gene modules, each sharing a coherent pattern of evolutionary gains and losses. If such modules can be detected accurately, they can then be “expanded” to identify new components. The major challenge in accurate detection is that the number and histories of modules have to be inferred simultaneously.
Here, we introduce a new method that generalizes this approach in a statistically principled manner, using a Bayesian mixture of tree-based hidden Markov models. Our method, called CLIME (clustering by inferred models of evolution), first partitions an input gene set into modules of genes that exhibit coherent evolutionary histories, and then expands each module with new genes sharing the same evolutionary history. CLIME is distinct from existing approaches in that it (i) is a tree-based method for partitioning an input set of related genes, (ii) automatically learns the number of distinct evolutionary modules in the input set, and (iii) leverages information from the entire input gene set to more reliably predict new genes that have arisen with a shared pattern of evolutionary gains and losses.
We systematically applied CLIME to over 1000 human complexes and pathways, two human cellular organelles (cilia and mitochondria), and three entire genomes (red algae, yeast, and the malaria parasite). The results, the software and an online analysis portal, are freely available at www.gene-clime.org.
Results
CLIME: an algorithm for clustering genes based on inferred models of evolution
The CLIME algorithm partitions genes based on inferred models of evolution (Figure 1). CLIME accepts three user-defined inputs: (1) a binary species tree; (2) a phylogenetic profile matrix, X, defining the presence or absence of all genes in a given organism across all species in the tree, and (3) an input gene set G. CLIME partitions the input set G into disjoint evolutionary conserved modules (ECMs), using a Bayesian mixture model to infer simultaneously the number of ECMs, the evolutionary model for each ECM, and gene's membership for each ECM. The algorithm next creates an ECM expansion set, ECM+, that includes other genes in the genome that are likely to have arisen under the ECM's inferred model of evolution compared to a null model.
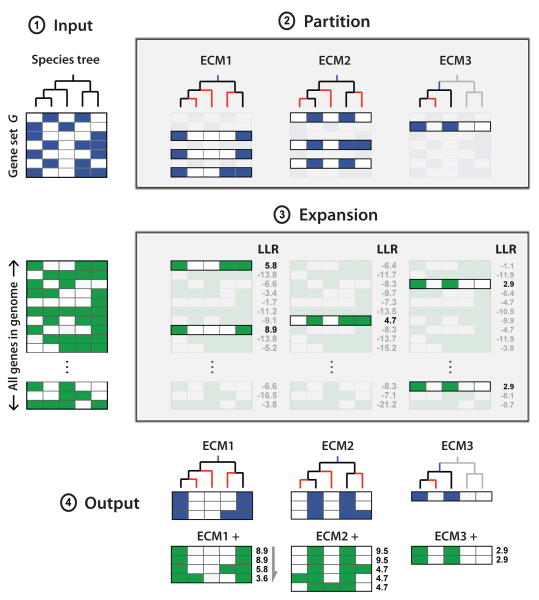
Figure 1. Schematic overview of CLIME.
CLIME partitions an input set of genes into evolutionarily conserved modules (ECMs), and predicts additional genes sharing the same inferred model of evolution. Input: species tree, an input gene set (G), and a phylogenetic matrix (X) for all genes in a reference organism showing presence (green) or absence (white) across all extant species in the tree. For display purposes, a separate blue/white matrix shows the profiles of genes in G, which are a subset of X. Partition: input genes G are partitioned into K distinct ECMs, using a Bayesian mixture of HMMs to simultaneously infer the number of ECMs and the shared evolutionary history of each ECM. Each ECM is modeled by a tree structured HMM with an inferred gain branch (blue) and branch-specific probabilities of gene loss (red). Expansion: each ECM is expanded by identifying genes within the genome that are more likely to have evolved from the ECM's model of evolutionary history compared to a null model of evolution, scored by the log likelihood ratio (LLR). Output: K disjoint ECM clusters and associated ECM+ expansions.
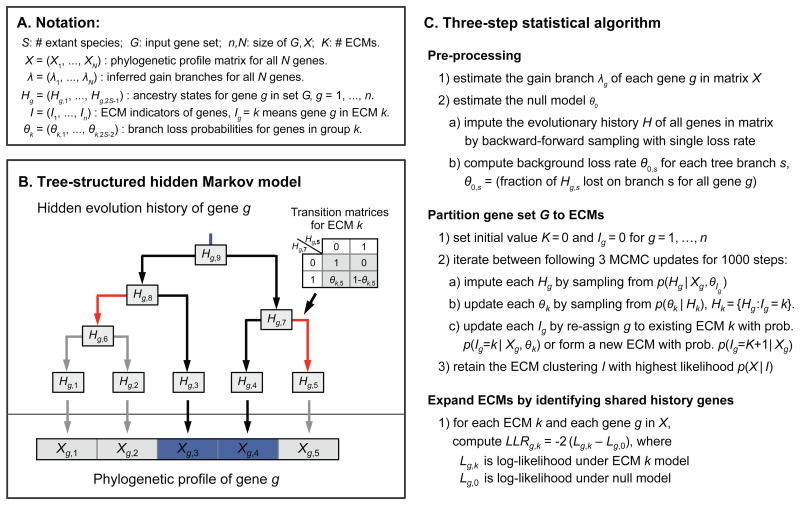
CLIME models the evolution of an individual gene using a tree-based hidden Markov model (HMM), with the assumption that each gene has a single gain event in evolution followed by zero or more loss events on the species tree (Figure 2A,B). CLIME does not consider branch lengths, only the tree topology. For each gene g, the HMM of evolution is based on the presence/absence profile across S living species (Xg the observed states). The HMM contains 2S-1 hidden states (Hg) corresponding to the true presence/absence of that gene in all living and extinct species (Figure 2B). The model includes a user-defined observation error parameter ε (default 0.01) representing the probability that the observed data is an error compared to the true hidden presence/absence (e.g. incomplete genome assembly/annotation). CLIME infers a tree-based HMM to model the evolution of each gene separately, as well as to model the evolution of each ECM. The evolutionary model of each gene g is represented by a single gain branch (λg) and a vector of branch-specific loss probabilities of its ECM (θk) – inferred at the Pre-processing step and Partition step, respectively (Experimental Procedures). Conditional on that gene g is in ECM k, the complete likelihood function for gene g is
Figure 2. The CLIME algorithm.
(A) Notation for random variables in CLIME's statistical model. (B) CLIME's generative tree-structured HMM, including observed states (Xg) and hidden states (Hg) that correspond to the inferred presence/absence of gene g in all living and extinct species in the pre-defined tree. The model is constrained to a single gain branch (blue). Loss events are modeled using branch-specific transition matrices (inset) derived from an ECM or null model (red color indicates branches with high loss probability). This example shows the likely evolutionary scenario that phylogenetic profile of gene g (presence only in species 3 and 4) is generated from ECM k which has high loss rates on two branches (red color), so gene g is likely to be lost on these branches while inherited on other branches. (C) Statistical details for three steps of CLIME.
| (1) |
where Qk,s is the transition matrix for ECM k on branch s (Figure 2A,B), which is the same for all genes in the same ECM and will be inferred from the input data, σ(s) denotes the direct ancestral species of s, T(λg) is the set containing all species in the sub-tree of λg, and
 {·} is the indicator function. The complete likelihood function for CLIME's Bayesian mixture of HMM on phylogenetic profile data is formulated as,
{·} is the indicator function. The complete likelihood function for CLIME's Bayesian mixture of HMM on phylogenetic profile data is formulated as,
| (2) |
where Ig is the ECM assignment indicator for gene g Employing a Dirichlet process prior on the ECM clustering and independent Beta priors on the θ's (i.e., loss probabilities), CLIME uses Markov Chain Monte Carlo (MCMC) sampling (Liu, 2008) of the posterior distribution to simultaneously estimate the optimal partitioning, hidden evolutionary history of genes in G and the probability of gene loss for each ECM on each tree branch. CLIME then scores all genes in the genome for the likelihood of having arisen under an ECM's inferred model of evolution compared with the background null model, using a log-likelihood ratio (LLR). Genes exceeding a threshold (default 0) are included in the expansion ECM+. The CLIME algorithm consists of three main steps (Figure 2C), which are described in Experimental Procedures briefly and in detail in the Supplementary Experimental Procedures.
We have implemented CLIME in C++ software using an algorithm of complexity of O(Sn2) per MCMC iteration, where S is the number of species, and n is the number of genes in G. Using a standard, single computer processor, CLIME can cluster a 100-gene input set in 20 minutes, a 1000-gene input set in less than a day, and (with parallel processing) a 5000-gene input set in under two days (Supplementary Experimental Procedures).
CLIME Inputs: species tree and phylogenetic matrix
CLIME inputs a user-defined species tree and a corresponding phylogenetic matrix. For the current study we used a species tree consisting of 138 diverse, sequenced eukaryotes (Bick et al., 2012) with a prokaryote outgroup. Each gene was deemed to have an ancestral, prokaryotic homologue if it had sequence similarity to at least 20 diverse bacterial/archaeal species. More diverse organisms in the input trees contribute to greater CLIME power, through the increased opportunity for independent loss events (Figure S1).
The user-defined phylogenetic matrix can be constructed using either homology-based or orthology-based methods. Unlike homology matrices, orthology matrices attempt to distinguish between members of multigene families – which is extremely challenging at large evolutionary distances. In the current work, we evaluated seven such methods and found that a simple homology matrix, using a BLASTP expect threshold, performed best (see Experimental Procedures, Figure S2, and Discussion). For the human-centered analyses described below, we created a phylogenetic matrix from 20,834 human genes, where each gene's profile reported whether a homolog was present or absent in each of the 138 eukaryotic species.
Simulation analysis
We used simulation analysis to evaluate CLIME's performance in partitioning and expansion. We varied four simulation parameters: NL, the number of randomly chosen branches having positive probability of gene loss; PL, the probability of gene loss on these branches; NS, the number of singleton genes within each simulated dataset; and ε, the observation error rate in the phylogenetic profile matrix. Higher NL and PL indicated more independent loss events and probability of loss events, hence greater signal; higher Ns and ε introduced more noise.
To evaluate the partitioning ability of CLIME and to compare it to existing phylogenetic profiling methods, we simulated synthetic input gene sets containing 500 genes, comprising a mixture of 50 ECMs, each with 10 genes, that were generated using tree-based as well as tree-independent models of evolution. We compared CLIME to hierarchical clustering based on two existing distance metrics, Hamming distance (Pellegrini et al., 1999) and squared anticorrelation distance (Glazko and Mushegian, 2004), for their ability to recover the simulated ECMs. When phylogenetic profiles were generated from a tree-based model of evolution, as expected CLIME outperformed the other methods in all simulated scenarios (Figure S3A). The simulations showed that CLIME's Dirichlet process mixture model could accurately estimate the correct number of ECMs in data. CLIME was quite accurate at reconstructing modules with at least 6 independent loss events and performed moderately well with 4 loss events (Figure S4). Even when simulations were performed assuming that all 138 species were independent – violating CLIME's fundamental model of evolution – CLIME performed comparably to other methods (Figure S3B). We note that 6 losses from the tree-based model manifests as 20 losses in a tree-independent model – thus these NL values are comparable (Experimental Procedures). Both CLIME and hierarchical clustering could almost perfectly cluster the data from the tree-independent model when there were many simulated absences (NL, exceeding 20), or when there were strongly coherent modules (PL exceeding 0.8).
Next we evaluated CLIME's ability to correctly expand a module. We simulated a scenario in which a genome contained 20,000 genes, 10 of which in actuality form an evolutionary coherent module E* with 10 genes, and 19,990 of which are singleton genes with unrelated evolutionary histories. First, we input a gene set of size 10 consisting of only one member of E*; CLIME correctly partitioned the 10 input genes into 10 singleton ECMs, and in the expansion phase identified 7 of the 9 additional E* members (LLR range 2-16) and 4 false positives (LLR range 2-8) (Figure S5A). In this scenario, CLIME inferred 3 false losses on the tree due to the fact that with only one gene CLIME could not distinguish real loss events from observation errors in the phylogenetic profile. Second we input a gene set with two members of E* and 8 singletons; CLIME properly partitioned the two E* genes together into an ECM and then expanded it with all 8 remaining E* genes (LLR range 4-29), and 2 lower scoring false positives (LLR range 2-3) (Figure S5B). Third, we input a gene set with 5 of the 10 true E* genes; CLIME properly partitioned the 5 genes into an ECM, and expanded it to recover all other 5 simulated ECM genes (LLR range 7-27) and only 1 false positive singleton (LLR = 1.7). In this latter scenario CLIME properly inferred all 5 tree branches with high probability of gene loss (Figure S5C). These simulations demonstrate input sets containing more true E* genes lead to more reliable evolutionary models and hence higher LLR scores in the ECM+ for true versus false positives. Intuitively, these analyses demonstrate how CLIME leverages information from multi-gene inputs to more accurately distinguish between real shared loss events from observation or inferential errors.
Application of CLIME to pathways with well-studied evolutionary histories
Next we applied CLIME to three well-studied gene sets: a macromolecular protein complex (complex I), a single gene (MICU1), and an organelle (cilia) for which there was existing evidence of informative evolutionary histories and for which previous manual phylogenetic profiling methods had been successfully applied to discover novel related proteins (Gabaldón, 2005; Li et al., 2004; Ogilvie et al., 2005; Pagliarini et al., 2008; Perocchi et al., 2010). Analysis of these pathways can help evaluate how faithfully CLIME recovers established evolutionary modules, and also affords opportunity for discovery.
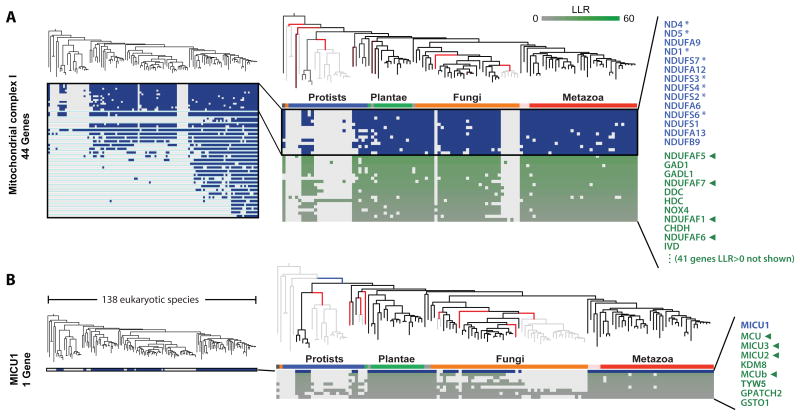
First, we applied CLIME to the 44 human genes encoding complex I of the mitochondrial respiratory chain (Balsa et al., 2012). Since 7 of the complex I genes are encoded by the human mitochondrial genome (mtDNA), we focused this analysis on the subset of 111 species for which mtDNA sequences and annotations were available. CLIME partitioned the 44 complex I genes into 4 non-singleton ECMs (Figure 3A). The ECM with the highest ECM strength (ϕ = 7.6) contained 14 genes, including 8 out of the 14 core essential components conserved to bacteria (Figure 3A). This ECM was nearly identical to the profile identified through extensive manual inspection (Pagliarini et al., 2008). The expansion ECM+ contained 52 predictions with an LLR>0, including five proteins recently shown to assemble complex I (Mimaki et al., 2012). The top predictions are shown in Figure 3A. It has long been known that systematic exposure of insecticides targeting complex I give rise to Parkinson's disease, though the mechanism of selective loss of dopaminergic neurons is unknown. It is notable that two genes, dopamine decarboxylase and glutamate decarboxylase, are also within this ECM+, raising hypotheses about direct links between complex I and the metabolism of two key neurotransmitters.
Figure 3. Application of CLIME to mitochondrial complex I and calcium uniporter.
(A) CLIME partitioning of the 44 subunits of mitochondrial respiratory chain complex I into ECMs (separated by aqua lines). Inset shows ECM1, including the independent loss events (red branches), the phylogenetic profile for the ECM1 genes (blue/white matrix and blue text) and the top genes in ECM1+ (green/white matrix and green text). Tree branch color indicates gene gain (blue), loss (red, brighter hue indicating higher confidence), or inheritance (black), otherwise shown gray. Asterisks indicate core bacterial complex I homologs. Green arrows indicate predictions with recent experimental or human genetic support for functional association with the input set.
(B) CLIME partitioning of the single input gene MICU1, which encodes the first identified protein component of the mitochondrial calcium uniporter complex. The ECM1+ includes four components recently shown to encode uniporter proteins (green arrows).
Next, we analyzed the single gene MICU1. We had previously used simple phylogenetic profiling with three species in combination with RNAi assays to identify MICU1 as the first known component of the mitochondrial calcium uniporter channel (Perocchi et al., 2010). The CLIME expansion, ECM+, contained 8 genes with similar histories, including four genes recently shown to encode additional components of the channel (MCU, MCUb, MICU2, MICU3) (Sancak et al., 2013). Most notably the top-scoring gene (LLR = 10.1), MCU, encodes the pore-forming channel itself (Baughman et al., 2011).
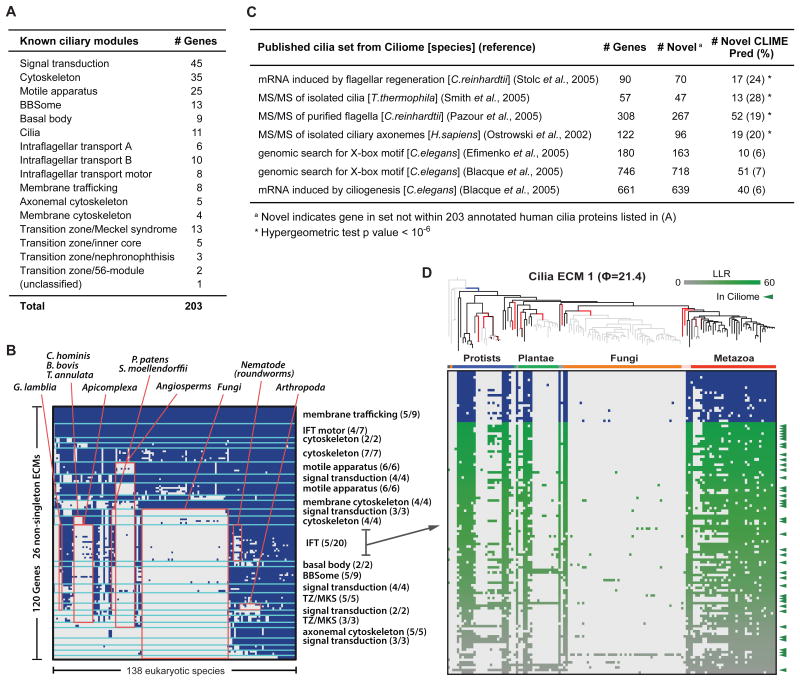
Third, we analyzed a curated set of 203 cilia-localized genes, for which manual annotations into 16 sub-compartments were available (Figure 4A). CLIME automatically partitioned the 203 cilia genes into 26 non-singleton ECMs containing 120 genes. Importantly, many of the ECMs were enriched for specific sub-compartments (cumulative hypergeometric P < 10-4) (Figure 4B), highlighting that in this case functionally related genes have co-evolved and are grouped together by CLIME. Each cilia ECM corresponded to a distinct model of evolution, some with very few loss events (e.g. most of membrane trafficking genes and IFT motor genes didn't show any loss events across 138 species), and others with extensive loss events (e.g. 5 BBSome genes lost 11 times). This evolutionary clustering highlighted particular model organisms for further study, such as arthropods that have specifically lost several transition zone components. CLIME expanded the 25 non-singleton ECMs with 783 additional human genes at an LLR > 0 (excluding ECM12+ that contains a large Zinc finger multigene family). There is a significant overlap between these 783 ECM+ expansion genes and the genes present in the Ciliome database (Inglis et al., 2006), which aggregates data from seven large-scale experimental and computational studies. The expansion list of the top ECM (ϕ = 21.4) contained many highly scoring genes, which are likely to encode novel cilia components (Figure 4D).
Figure 4. Application of CLIME to cilia.
(A) Annotation of 203 human cilia-related genes within 16 sub-compartments.
(B) CLIME partition of 203 cilia genes into modules (separated by aqua lines). Red boxes indicate shared absence in selected clades, labeled above. Sub-compartments with significant enrichment per ECM are labeled at right (parentheses show fraction of ECM genes within sub-compartment).
(C) Overlap between CLIME predictions and seven orthogonal cilia gene sets (Inglis et al., 2006).
(D) The ciliary ECM with highest ECM strength (21.4), including the evolutionary model (gain branch in blue, loss branches in red), the ECM genes (blue text and blue/white matrix) and the ECM+ predictions (green/white matrix). Green tick marks indicate predictions with independent evidence of cilia-related function based on Ciliome database.
Several key points emerge from CLIME's results on complex I, calcium uniporter, and cilia. First, components of pathways do not all share the same evolutionary history but are comprised of distinct sub-modules, each with their own unique history. Second, these sub-modules can correspond to functional subsets such as the cilia motile apparatus. Third, genes that share evolutionary history with ECMs do in fact functionally relate with the original set of genes. Fourth, while the evolutionary signal is boosted from inputs containing more than one gene, the algorithm can be useful even with a single input gene as long as it exhibits a sufficient number of independent loss events. Fifth, the more evolutionary coherent ECMs, reflected by high ECM strength, are more robust and can identify more reliable ECM+ genes.
Exploring the evolutionary modularity of 1025 canonical pathways and complexes
To systematically identify human pathways with informative evolutionary histories, we applied CLIME to over a thousand predefined functional gene sets including physical complexes as well as metabolic and signaling pathways. We hypothesized that a subset of these human pathways will contain modules with highly informative patterns of evolutionary gains and losses that can shed light on the underlying organization of the pathway, can highlight new model organisms for further study, and can predict function of wholly uncharacterized genes for experimental validation.
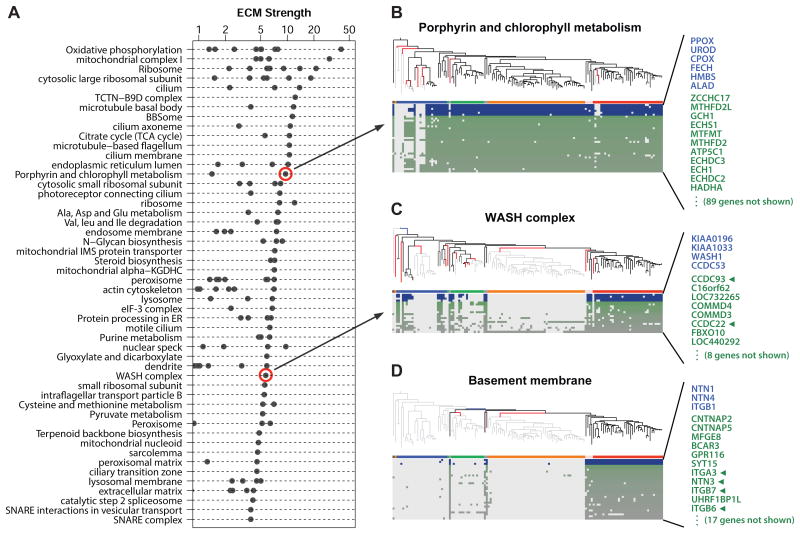
We applied CLIME to 1025 pathways and complexes including 909 cellular components from the Gene Ontology (GO) database (Ashburner et al., 2000) and 116 metabolic and signaling pathways from KEGG (Kanehisa et al., 2012). Overall, we find that 145 canonical cellular components and pathways (14%) show highly informative ECMs, defined as ECM strength > 2 and containing at least 50% nonhomologous genes. Paralogs and other genes with sequence similarity will trivially cluster together since they will share inferred histories, thus they are flagged in CLIME output so that users can optionally filter them out of consideration. We find that approximately half of the identified ECMs contained two or more genes that do not share sequence similarity (Figure S2). The pathways with the highest strength ECMs are shown in Figure 5, and complete results are available (www.gene-clime.org).
Figure 5. CLIME analysis of 1025 human pathways.
(A) Top 50 pathways with highly informative ECMs (strength > 2 and containing at least 50% non-homologous genes), ranked by strength of the top non-homologous ECM. All non-singleton ECMs are shown as separate dots.
(B-D) ECMs for selected pathways. As in Figure 3, the inferred gain/loss events are indicated by blue and red tree branches. Blue/white and green/white matrices show phylogenetic profiles of ECM and ECM+ genes, respectively. Green arrows indicate experimental evidence of functional association with the input gene set.
One KEGG metabolic pathway with a strong evolutionary signature involved six steps of heme biosynthesis (Figure 5B, ECM ϕ = 9.5). While this pathway is highly conserved in most eukaryotes, CLIME highlights a loss event in the nematode lineage – consistent with an experimental study that confirms absence of heme biosynthesis in C. elegans and that proposes pharmacologic targeting of heme transport as potential anti-helminthic therapy (Rao et al., 2005).
One of CLIME's strongest evolutionary signatures and predictions was derived from the small WASH (Wiskott–Aldrich syndrome homologue) protein complex involved in endosome trafficking (Duleh and Welch, 2010) (Figure 5C). Of 9 WASH complex genes, 4 are partitioned into an ECM (ϕ = 5.7) defined by approximately 11 independent loss events. These genes have no apparent bacterial homologs, but were present early in eukaryotic evolution and show absences in four protist clades, five plant clades, all fungi, and one animal species (Schistosoma mansoni). Interestingly, the ECM+ contains 7 genes with LLR > 10 and includes two (CCDC93, CCDC22) recently shown to physically associate with the WASH complex (Harbour et al., 2012). Other ECM+ genes, such as the second-scoring C16orf62 (LLR = 21.2), are completely uncharacterized.
A striking phylogenetic profile was observed for three cell-adhesion genes localized to the basement membrane, which anchors epithelial tissue to connective tissue through adhesion molecules in the extracellular matrix (Figure 5D). These genes (NTN1, NTN4, ITGB1) are present in all animal species as well as three quite distant species (N. gruberi, T. trahens, D. discoideum). CLIME infers an evolutionary model (ϕ = 3.7) for these genes with only two independent loss events in plants and fungi, and suggests that the other instances of profile “presence” calls may be BLASTP errors or horizontal gene transfer. The presence of these cell adhesion molecules in T. trahens and D. discoideum suggests that these may be early innovations in the path to multicellularity. More surprising is their presence in the free-living and single-cell amoeba N. gruberi – however recent evidence suggests that a closely related pathogenic amoeba (N. fowleri) expresses integrins to facilitate invasion within the host extracellular matrix, which might explain their presence (Jamerson et al., 2012). The expanded ECM+ contains 28 genes with LLR > 0, including six members of the integrin complex (half of which do not share sequence similarity to ITGB1) as well as five proteins annotated to reside in the plasma membrane or extracellular matrix (CNTNAP5, CNTNAP2, MFGE8, GPR116, CRIM1), raising hypotheses for shared evolution of proteins required for multicellularity or host invasion.
Application of CLIME to the human mitochondrial proteome
CLIME's evolutionary modeling can be applied not only to individual pathways, but to chart complex evolutionary histories of larger entities – such as the mitochondrion. Standard CLIME analysis of the human mitochondrial proteome (Pagliarini et al., 2008) organized proteins into evolutionary modules that recapitulated many known pathways (e.g., TIM/TOM protein import, fatty acid biosynthesis) and revealed unexpected connections between pathways (e.g. heme and folate biosynthesis) (Figure S6 and Supplementary Experimental Procedures).
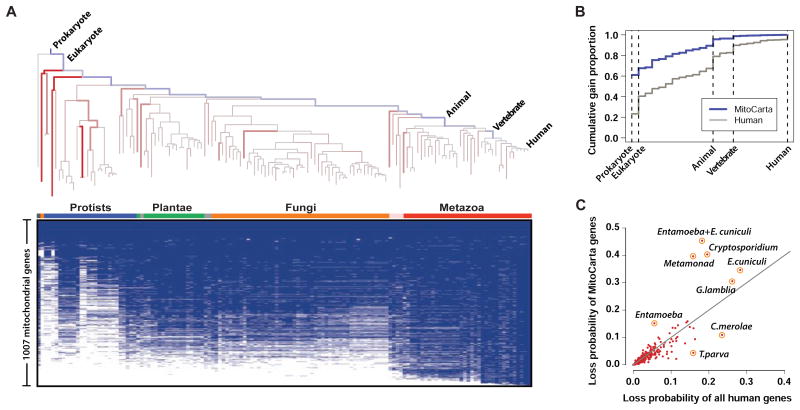
Next we analyzed the gain branches and loss events for each of the 1007 nuclear and mtDNA-encoded mitochondrial genes, inferred during CLIME's preprocessing step, to dissect the complex history of the organelle. We first counted the number of gains observed on each of the 27 potential gain branches between human and the eukaryotic least common ancestor (Figure 6A, blue branches). Mitochondrial genes showed strikingly more ancient evolutionary origins compared to all human genes (Figure 6B), consistent with previous reports (Pagliarini et al., 2008). We next averaged the branch-specific loss probabilities for all mitochondrial genes to highlight the species whose mitochondrial proteomes are greatly reduced relative to rest of their proteomes (Figure 6A, red branches). Analysis of lineages with mitochondrial-specific losses (Figure 6C) highlighted precisely the 7 organisms known to have lost the mitochondrial genome (C. parvum, C. hominis, T. vaginalis, G. lamblia, E. dispar, E. histolytica, E. cuniculi). In contrast, this analysis spotlights the red alga C. merolae that has a greatly reduced proteome in general, without a commensurate reduction in its mitochondrial proteome (Figure 6C). Thus the automated CLIME evolutionary analysis provides insights into the reductive and expansive evolution of this well-studied organelle, defines its gene modules based on evolutionary inference, and highlights specific model organisms for further study.
Figure 6. CLIME analysis of the mitochondrial proteome.
(A) CLIME's estimates of proportions of gene gain (blue branches) and average branch-specific probabilities of gene loss (red branches) on the 138 eukaryotic species tree for the 1007 human mitochondrial genes. Brighter hue indicates higher probability. The presence/absence of the 1007 human mitochondrial genes across 138 species is shown in blue/white matrix.
(B) Cumulative gain proportions of mitochondrial genes versus all human genes (only selected branches labeled).
(C) Average loss probability of mitochondrial genes versus all human genes for each tree branch (only selected branches labeled).
Genome-wide CLIME analysis of malaria, red alga, and yeast
CLIME can also be applied in an unsupervised manner to partition the genes of entire organisms based on evolutionary history. Although it is not currently computationally tractable for CLIME to cluster all ∼20,000 human genes, we have applied it to three diverse species each of whose genomes encode ∼5000 genes (Figure 7). For each species, we created a species-specific phylogenetic matrix generated from homology searches against all 138 eukaryotes (see Methods). From such whole-organism partitioning, we can explore the evolutionary history of features such as the apicoplast or chloroplast, or predict the function of uncharacterized genes. All results are available (www.gene-clime.org), with a few examples highlighted below.
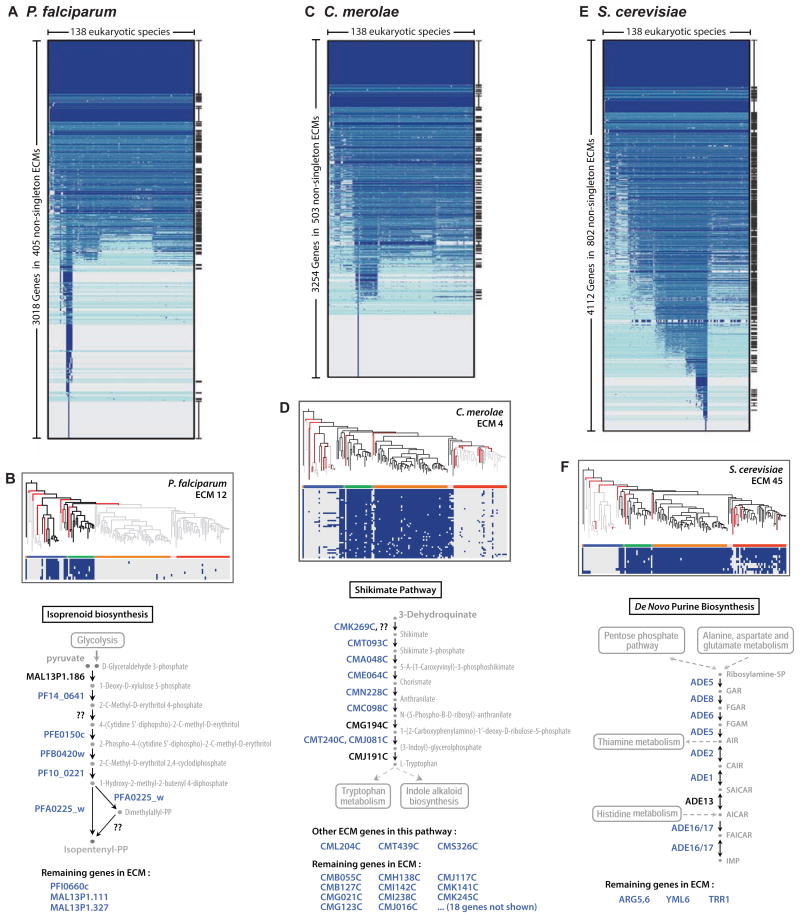
Figure 7. Application of CLIME to the genomes of malaria parasite, red alga, and yeast.
CLIME partitioning of all genes within three model organisms: Plasmodium falciparum (A), Cyanidioschyzon merolae (C), and Saccharomyces cerevisiae (E). ECMs are ordered by mean number of homologs present across taxa, and separated by aqua lines. All ECMs significantly enriched (hypergeometric p-value<10-6) in GO or KEGG gene sets are marked at right. Selected ECMs are shown for the three species (B, D, F), along with schematic pathway diagrams that highlight the location of ECM genes (blue text), genes not in the ECM (black text), and enzymes not known to reside in the species (question marks) based on KEGG. Genes within the ECM but not present in the relevant KEGG pathway are listed below, and may encode novel pathway members.
CLIME analysis of P. falciparum
The malaria parasite P. falciparum is a member of the protozoan phylum Apicomplexa, named for presence of the apicoplast organelle. This non-photosynthetic plastid was derived by secondary endosymbiosis from an alga (Lim and McFadden, 2010), and since it's essential for parasite survival it is an attractive target for drug development. Although the essential apicoplast functions are not well elucidated, it has known roles in the biosynthesis of fatty acids, isoprenoids, heme, and iron-sulfur clusters (Lim and McFadden, 2010). Interestingly, the apicoplast organelle has been lost entirely within one Apicomplexan lineage (Cryptosporidium), but is present in 10 other Apicomplexan genomes analyzed within our 138 eukaryotes.
CLIME analysis partitioned the 5331 P. falciparum genes into 405 non-singleton ECMs (346 of them contain at least two or more non-homologous genes), many of which are significantly enriched for known biosynthetic pathways and cellular compartments annotated in KEGG and GO (Figure 7A). Specifically, 18 distinct ECMs were enriched for apicoplast-localized genes from GO (Figure S7A): some restricted to the Apicomplexan lineage, others sharing homology with plant lineages, and others with broader phylogenetic distribution, consistent with the complex endosymbiotic origin of this organelle. Interestingly, two top apicoplast-enriched ECMs show distinct evolutionary patterns for genes involved in isopentenyl diphosphate biosynthesis (ECM 12, ϕ = 10.2, Figure 7B) and genes involved in fatty acid biosynthesis (ECM 33, ϕ = 8.0) – with the latter module absent in three Apicomplexa (B. bovis, T. annulata, T. parva). These ECMs highlight the ability of CLIME to reconstruct known metabolic pathways, and pinpoints species particularly amenable for dissecting the distinct roles of isoprenoid biosynthesis versus fatty acid biosynthesis in apicoplast function. The results may help to de-orphan the function of uncharacterized genes, such as PFI0660c, MAL13P1.111 and MAL13P1.327, which we predict are involved in isoprenoid biosynthesis.
CLIME analysis of C. merolae
Cyanidioschyzon merolae is a primitive red alga with a highly reduced genome. This organism is not well-studied and many of its genes are uncharacterized – thus unsupervised CLIME clustering has the potential to highlight novel evolutionary modules and identify new members of known pathways.
CLIME analysis partitioned the 5014 C. merolae genes into 503 non-singleton ECMs, 336 of which contained at least two non-homologous genes (Figure 7C). One of the top evolutionary modules contained homologs to the isoprenoid biosynthesis highlighted in the apicoplast analysis above, and it is likely this pathway was present in the plastid ancestor of both the choloroplast and apicoplast. Of interest, ECM 4 (ϕ = 13.9) with 40 genes contained 11 enzymes in the Shikimate pathway involved in the biosynthesis of aromatic amino acids (Figure 7D). This pathway likely resides within the chloroplast, based on homology to A. thaliana genes. It is possible that other genes in ECM 4 may encode some of the missing steps of this pathway (Figure 7D).
CLIME analysis of S. cerevisiae
Lastly, we performed unsupervised CLIME clustering of the 5882 protein-coding genes in yeast – the premiere cellular model organism. CLIME partitioned 4112 genes into 802 non-singleton ECMs (568 of them contain at least two or more non-homologous genes) (Figure 7E). One of the modules (ECM 45, ϕ = 8.5) contains 7 non-paralogous genes (ADE5,7, ADE16, ADE17, ADE6, ADE8, ADE1, ADE2) encoding consecutive enzymatic steps of the de novo purine biosynthesis pathway – that were originally discovered through a classic 1969 genetic screen for mutants accumulating red pigment when grown on adenine-deficient media (Dorfman, 1969). These genes were evidently lost independently six times in evolution (Figure 7F), and highlight several surprising animal species that appear to lack this key pathway (S. mansoni, B. malayi, D. pulex), consistent with one experimental report (Dovey et al., 1984).
The unsupervised clustering of three entire organism genomes recapitulated known functional modules, suggested functions for many uncharacterized genes (Table S1), suggested unexpected links between known pathways, and identified ECMs with entirely uncharacterized genes which raise the hypotheses of potential novel pathways (Table S2).
Online Resources
The website www.gene-clime.org provides access to the CLIME software, source code, pre-computed phylogenetic matrices, results from this report, and a web-based interactive tool for analysis of user-defined gene sets from 10 model organisms: H. sapiens, M. musculus, D. melanogaster, C. elegans, S. cerevisiae, N. crassa, A. thaliana, C. merolae, P. falciparum and T. brucei.
Discussion
We have introduced a new way to partition and expand biological pathways based on inferred models of evolution. A key feature of CLIME is that it explicitly models a pathway as a set of disjoint gene modules, each with its own evolutionary history. The method simultaneously infers the number of modules and evolutionary history that defines each module, and then identifies new members that have arisen under similar models of evolution. The tool is fast and flexible, and reports cluster strength and prediction likelihood using statistical measures that are principled and readily interpretable.
Three key features distinguish CLIME from other algorithms: (i) it operates on an input set of genes, (ii) it models errors in the input phylogenetic profile, (iii) it assumes a tree-based model of gene evolution with a single gain branch and branch-specific loss probabilities. While CLIME can input single genes or entire genomes, best results are obtained from input gene sets with a high prior likelihood of functional relatedness (Figure S5). Leveraging information from multiple genes and modeling profile errors is key, since phylogenetic profiles are often noisy due to incomplete assemblies/annotations, and errors in detecting distant homologs. For instances where the input tree topology is incorrect – or for instances of true horizontal gene transfer or incomplete genome annotations – CLIME will inaccurately model the independent loss events and thus may inflate likelihood scores. However, where the topology is accurate, CLIME's evolutionary model renders it insensitive to overrepresentation of particular clades. Of course, CLIME is not expected to perform well in bacteria, where horizontal gene transfer is rampant and violates CLIME's tree-based model of evolution.
We observed that the chief limitation of CLIME was not due to the algorithm per se but to our homology-based input matrix – which cannot distinguish between members of multigene families. Since CLIME can input any binary phylogenetic matrix, we initially evaluated orthology-based matrices that attempt to resolve multigene families (Figure S2). However ortholog resolution is extremely difficult at large evolutionary timescales, which are the most useful for phylogenetic profiling (since the most power is derived from the most diverse species). Indeed, manual phylogenetic reconstructions of selected multigene families revealed that both best-bidirectional hit (BBH) and orthogroup methods were accurate only within smaller evolutionary time scales (e.g. within fungi/metazoa). In contrast, our homology-based phylogenetic matrix (derived from BLASTP using a simple expect threshold) was more accurate at large timescales but also more limited: “presence” indicates presence of any multigene family member and “absence” indicates absence of the entire family. This homology matrix works well for single gene families (e.g. subunits of complex I) and for multigene families where all members function within the same pathway (e.g. proteins containing the “interflagellar transport domain”). This approach does not work well for genes sharing the same domain that act within fundamentally different pathways (e.g. kinases, G-protein coupled receptors). This limitation could be addressed by using more sophisticated methods to resolve orthology, such as SYNERGY (Wapinski et al., 2007). Alternatively, for pathways with sufficient loss events within a given clade (e.g. opisthokonts), a simple BBH matrix using a smaller tree may be best. Future versions of CLIME may accept a phylogenetic matrix with a probability-based score to account for the uncertainty in resolving homology or orthology.
One of the most important results from the current CLIME analysis is the evolutionary modularity of many pathways and complexes. Application of CLIME to over 1000 human protein complexes, metabolic pathways, and signaling pathways showed that approximately 15% had highly informative evolutionary modules (with strength > 2 and at least 50% non-homologous genes). CLIME analyses have highlighted wholly unanticipated evolutionary modularity within even pathways traditionally considered to be well studied. In less well-studied pathways and organisms, a number of very high scoring modules and predictions have emerged which are ripe for experimental analysis (e.g. WASH complex in human, isoprenoid biosynthesis in red alga and malaria). Excitingly, the power of CLIME will scale with the growing wealth of eukaryotic genome sequences. Inclusion of high quality genomes, especially from more distantly related species or those filling gaps in the tree of life, will increase the opportunity to observe loss events and increase the precision with which CLIME can parse biological pathways.
Experimental Procedures
CLIME Algorithm
Step 0: Pre-processing
For each gene g, the gain branch λg is selected as the branch with the highest likelihood of generating Xg, assuming a branch-independent loss rate (default 0.03, determined based on the genome-wide average observed in our data). CLIME then infers g's evolutionary history by forward-summation-backward-sampling (Liu, 2008) with the same branch-independent loss rate (default 0.03). Next, CLIME uses these models of evolution for all genes in the genome to construct a null model of branch-specific losses (θ0), where the loss rate for each branch s is the fraction of genes lost on branch s.
Step 1: Partitioning
MCMC sampling is used to partition the input gene set G into disjoint ECMs, using a user-defined number of iterations (default 1000). Each MCMC iteration has three updates: (1) for each gene g, we impute the missing evolutionary history, Hg, by sampling from probability distribution P(Hg|Xg,θIg) with forward-summation-backward-sampling (Liu, 2008); (2) for each ECM k, we update branch-specific loss probabilities, θk, by sampling from the conditional distribution P(θk|Hk) where Hk contains evolutionary histories of genes in ECM k; (3) for each gene g, we update the ECM assignment by re-assigning g to an existing ECM k with computed probability P(Ig=k|Xg,θk) or by forming a new ECM with probability P(Ig=K+1 |Xg). To implement, we integrate out θs from the model and run a collapsed Gibbs sampler, which targets the same posterior distribution of partitioning but dramatically improves algorithm efficiency (see Extended Experimental Procedures). CLIME calculates the marginal likelihood of the current ECM partitioning, P(X|I), at the end of each iteration and finally retains the ECM partitioning with the highest marginal likelihood. Once the partitioning is complete, CLIME calculates the ECM strength, ϕk, summarizing how well the evolutionary model of ECM k matches the inferred models of each member gene compared to the null model, using the normalized Bayes Factor (Kass and Raftery, 1995): where Nk is the number of genes in ECM k and P(θ) denotes the prior distribution of loss rates.
Step 2: Expansion
CLIME scores all genes in X for the likelihood of having arisen under an ECM's inferred model of evolution compared with the background null model, using the log-likelihood ratio (LLR) as a measure. Genes with their LLRs exceeding a threshold (default 0) are included in the expansion ECM+, excluding members of input set G.
Simulation analysis
Simulated datasets were constructed as a mixture of 50 non-singleton ECMs (10 genes each) with a certain number of singleton ECMs. 64 simulations were run using a combination of four parameters: (i) the number of singleton ECMs: Ns ∈ {0, 100, 200, 500}; (ii) the number of loss events for each simulated ECM (on randomly selected branches): NL ∈ {4, 6, 8, 10} for tree-based model and NL ∈ {10, 20, 30, 40} for tree-independent model; (iii) the probability of gene loss on each of the NL branches: PL ∈ {0.6, 0.7, 0.8, 0.9}; The simulation observation error rate was set to 0.02. Phylogenetic profiles per ECM for the tree-based simulations were generated from our probabilistic generative model, where loss branches were randomly selected from the 2S-1 branches on the 138-species eukaryotic tree. For the tree-independent simulation, losses were independently selected from the 138 species. Note that the tree-independent simulation is equivalent to tree-based simulation with loss events only happen on the leaf branches of the tree. We chose two different sets of NL for tree-based and tree-independent models of evolution to make their phylogenetic profile matrices equivalent (with comparable absence/presence ratios). For each parameter configuration we simulated 100 datasets, ran CLIME on each dataset, calculated the adjusted rand index (ARI, Hubert and Arabie, 1985) between CLIME's output and the true ECM partitioning, and averaged the ARI across the 100 datasets. For comparison to other distance metrics, we used agglomerative hierarchical clustering with the average method and using 10% singleton genes as cutoff for clusters, as described in (Glazko and Mushegian, 2004). For Figure S5 we used simulation parameters NL = 5, PL = 0.8.
Homology matrix
Protein sequences from 138 eukaryotic organisms corresponding to the published phylogenetic tree (Bick et al., 2012) were downloaded as follows: 132 species from the KEGG Organisms Database, Release 58 (Kanehisa et al., 2006) and 6 species (Thecamonas trahens, Capsaspora owczarzaki, Sphaeroforma arctica, Salpingoeca rosetta, Allomyces macrogynus and Spizellomyces punctatus) from the Origins of Multicellularity Sequencing Project at the Broad Institute (7/9/2012) (Ruiz-Trillo et al., 2007). For each of the ten reference genomes, a species-centric binary phylogenetic matrix Xg,i was constructed to contain 1 if reference gene g shared sequence similarity with any protein in species i (BLASTP, Expect<1e-3) and 0 otherwise. A paralogy matrix was created based on BLASTP (Expect<1e-3). A single “prokaryote” outgroup was added to the eukaryotic tree and to the phylogenetic profile matrix, where Xg,prokaryote =1 if gene g had BLASTP similarity (Expect < 1e-3) to at least 20 out of 502 prokaryotic species in KEGG Organisms Database, Release 58, otherwise 0. These 502 species were selected from 1477 KEGG species, retaining one species per genus (the species with the largest number of annotated proteins).
Comparison of homology matrices
We compared homology matrices using leave-one-out (LOO) cross-validation on the 1025 GO/KEGG pathways (8876 distinct genes; 20594 gene-pathway pairs). For a range of LLR thresholds, sensitivity was calculated as the percent of the genes correctly recovered in any ECM+ derived from the LOO input gene set (from which the gene had been artificially removed) and specificity was calculated as the percent of non-pathway genes correctly absent from all ECM+ derived from the LOO input set. We compared 7 homology matrices: eggNOG (Powell et al., 2012), and six BLASTP-based matrices generated with a combination of thresholds for expect E ∈ {1e-2, 1e-3}, query gene coverage C ∈ {0%, 20%, 30%}, and bi-directionality B ∈ {top-hit, best bidirectional hit} (Figure S2). To assess paralogy effects (Figure S2), we performed LOO cross-validation after removing redundant paralogous genes (BLASTP E<10-3) from each GO/KEGG gene set.
Pathways and enrichment statistics
Metabolic and signaling pathways for 10 model organisms were downloaded from the KEGG Pathway Database, Release 58 (Kanehisa et al., 2006), excluding 3 large terms (‘Human Diseases’, ‘Organismal Systems’, ‘Environmental Response and Signaling’) and excluding all genes that were present in greater than 3 different pathways. Gene ontology terms for cellular compartments were downloaded from the NCBI Gene database (H. sapiens genes, downloaded 12/2012), PlasmoDB version 9.3 (P. falciparum genes), and YeastMine (S. cerevisiae genes, downloaded 11/2011). For unsupervised CLIME clustering, ECMs were tested for enrichment of KEGG or GO categories using the hypergeometric test (P < 10-6).
Supplementary Material
Highlights.
CLIME is a tree-based algorithm that clusters gene sets based on evolutionary history
CLIME predicts new members of a pathway based on shared inferred ancestry
CLIME software and a web-based interface are freely available (www.gene-clime.org)
Acknowledgments
We thank Jagesh Shah for curation of cilia-related genes, and Eran Mick and Daniel Arlow for manuscript feedback. This work was supported in part by grants from the National Institutes of Health (GM0077465 to V.K.M) and National Science Foundations (DMS-1007762 and IIS-1017967 to J.S.L), and the Shenzhen Special Fund for Strategic Emerging Industry grant (ZD201111080127A to J.S.L). V.K.M is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions: V.K.M and J.S.L framed the problem. Y.L, R.G, J.S.L developed the statistical algorithm. Y.L implemented the algorithm and wrote the software. Y.L and S.E.C performed analyses and summarized the results. S.E.C, J.S.L, V.K.M supervised the project. Y.L, S.E.C, J.S.L, V.K.M wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jun S. Liu, Email: jliu@stat.harvard.edu.
Vamsi K. Mootha, Email: vamsi@hms.harvard.edu.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metabolism. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20. doi: 10.1093/bioinformatics/btl558. [DOI] [PubMed] [Google Scholar]
- Barker D, Pagel M. Predicting Functional Gene Links from Phylogenetic-Statistical Analyses of Whole Genomes. PLoS Computational Biology. 2005;1:e3. doi: 10.1371/journal.pcbi.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick AG, Calvo SE, Mootha VK. Evolutionary Diversity of the Mitochondrial Calcium Uniporter. Science. 2012;336:886–886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman BZ. The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics. 1969;61:377–389. doi: 10.1093/genetics/61.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, McKerrow JH, Wang CC. Purine salvage in Schistosoma mansoni schistosomules. Mol Biochem Parasitol. 1984;11:157–167. doi: 10.1016/0166-6851(84)90062-8. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T. Evolution of proteins and proteomes: a phylogenetics approach. Evolutionary Bioinformatics Online. 2005;1:51. [PMC free article] [PubMed] [Google Scholar]
- Glazko G, Mushegian A. Detection of evolutionarily stable fragments of cellular pathways by hierarchical clustering of phyletic patterns. Genome Biology. 2004;5:R32. doi: 10.1186/gb-2004-5-5-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming RW. Error detecting and error correcting codes. Bell System Technical Journal. 1950;29:147–160. [Google Scholar]
- Harbour EM, Breusegem YS, Seaman NJM. Recruitment of the endosomal WASH complex is mediated by the extended“tail”of Fam21 binding to the retromer protein Vps35. Biochemical Journal. 2012;442:209–220. doi: 10.1042/BJ20111761. [DOI] [PubMed] [Google Scholar]
- Hubert L, Arabie P. Comparing partitions. Journal of Classification. 1985;2:193–218. [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. TRENDS in Genetics. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Jamerson M, da Rocha-Azevedo B, Cabral GA, Marciano-Cabral F. Pathogenic Naegleria fowleri and non-pathogenic Naegleria lovaniensis exhibit differential adhesion to, and invasion of, extracellular matrix proteins. Microbiology. 2012;158:791–803. doi: 10.1099/mic.0.055020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jim K, Parmar K, Singh M, Tavazoie S. A cross-genomic approach for systematic mapping of phenotypic traits to genes. Genome Res. 2004;14:109–115. doi: 10.1101/gr.1586704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. Journal of the American Statistical Association. 1995;90:773–795. [Google Scholar]
- Kensche PR, van Noort V, Dutilh BE, Huynen MA. Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. Journal of the Royal Society Interface. 2008;5:151–170. doi: 10.1098/rsif.2007.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS. Monte Carlo strategies in scientific computing. Springer; New York: 2008. [Google Scholar]
- Mering von C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Research. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proceedings of the National Academy of Sciences. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T. eggNOG v3. 0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Research. 2012;40:D284–D289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AU, Carta LK, Lesuisse E, Hamza I. Lack of heme synthesis in a free-living eukaryote. Proceedings of the National Academy of Sciences. 2005;102:4270–4275. doi: 10.1073/pnas.0500877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. TRENDS in Genetics. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493:694–698. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana K, Larsson TA, Powell S, Chen WH, Doerks T, Muller J, Bork P. Orthology prediction methods: a quality assessment using curated protein families. Bioessays. 2011;33:769–780. doi: 10.1002/bies.201100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Vert JP. A tree kernel to analyse phylogenetic profiles. Bioinformatics. 2002;18(Suppl 1):S276–S284. doi: 10.1093/bioinformatics/18.suppl_1.s276. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang R, Li L, Xia X, Sun Z. Inferring functional linkages between proteins from evolutionary scenarios. Journal of Molecular Biology. 2006;359:1150–1159. doi: 10.1016/j.jmb.2006.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.