Abstract
Protein glycosylation is one of most common protein modifications and is involved in many biological activities. N-linked and O-linked glycosylation not only represents abundant glycan modifications, but also are structurally diverse. Mass spectrometry has emerged as a major method for glycomic analysis. However, glycan extraction from proteins and glycan modification are two critical steps in glycomic analysis of glycans using mass spectrometry. In this protocol, we describe a novel and high-throughput method for isolation and modification of glycans from glycoproteins using a chemoenzymatic approach on solid-phase. Proteins are first immobilized to a solid support and unconjugated molecules are washed away; glycans, while still linked to glycoproteins on the solid support, can be treated enzymatically or chemically on solid-phase for glycan derivatization. Glycans are then released from the solid support for analysis by mass spectrometry. The procedures outlined are robust and useful for high-throughput glycomic analysis from complex biological or clinical samples.
Keywords: Solid-phase, protein immobilization, glycan derivatization, chemoenzymatic processes, MALDI, GIG, mass spectrometry, glycomics
Introduction
Protein glycosylation is one of the most common protein modifications and is involved in many biological pathways including cell-cell signaling, protein stability and solubility, and interactions of ligands and receptors (Ole and Cummings, 2009). Aberrant glycosylation may be associated with a multitude of pathological states such as cancer malignancy (Hakomori, 2002), immune response (Rudd et al., 2001), and neuromuscular disorders (Huizing et al., 2004). Protein glycosylation can be exploited for diagnosis and therapeutic targeting of associated diseases through identification and quantification of glycans independently or in combination with proteins. Potentially new clinical diagnostic procedures, as well as the development of potent pharmaceuticals, can be aided significantly through precise, reproducible, and sensitive glycomic analysis (Mechref and Novotny, 2009). To identify and quantify glycans from complex biological specimens, an efficient method is essential for isolation of glycans which maintains glycan composition and relative abundance intact for analysis.
The method for isolation of glycans depends on the type of glycans conjugated to proteins. N-glycans are conjugated to proteins through asparagine residues in the motif of Asn-X-Ser or Asn-X-Thr, where X is any amino acid except proline. N-glycans can be defined in three categories including oligomannose, hybrid and complex glycans. They can be released from their proteins using peptide-N-glycosidases (PNGases) (Kobata, 1979; Plummer et al., 1984). O-linked glycans are conjugated to both serine and threonine residues. Instead of single core structure in N-glycans, O-glycans consists of eight core structures. There is no specific enzyme comparable to PNGases for removal of O-linked glycans. Chemical methods, e.g., β-elimination, are usually employed for release of O-linked glycans (Carlson, 1968). For the released glycans from proteins, multiple purifications are often required for removal of salts and reagents in order to extract glycans for mass spectrometry analysis. Additionally, modifications of glycans are used to improve glycomic analysis and quantification, but in solution, it is challenging to remove the reagents that are often used for glycan derivatization. This protocol we describe is an improved method for extraction and derivatization of glycan for mass spectrometry analysis.
Basic Protocol
Glycoprotein Immobilization for Glycan Extraction
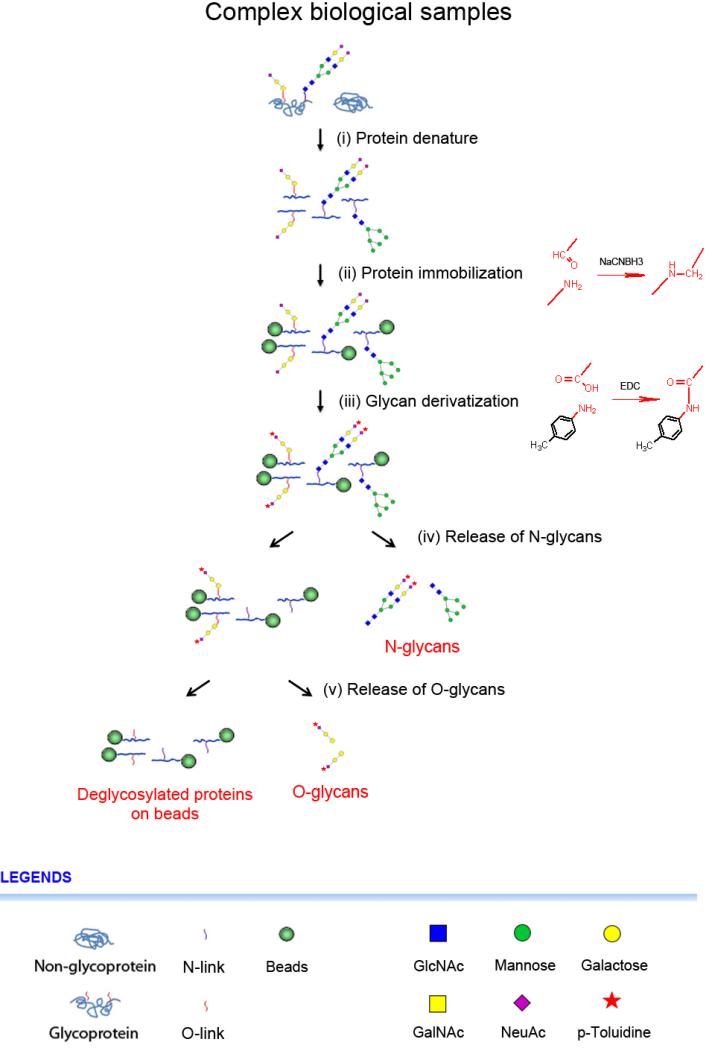
The method, Glycoprotein Immobilization for Glycan extraction (GIG) (Yang et al., 2013a), uses chemoenzymatic solid-phase for capture, modification and release of glycans from glycoproteins directly from complex biological samples. The procedure consists of several key steps including protein conjugation, glycan derivatization, and glycan release as shown in Figure 1. The solid support, or beads, is functionalized with amino-group reactive groups such as aldehydes, which enable to conjugate with N-termini or lysine side chains of proteins in mildly basic condition (e.g., pH 10) and further reduce in the presence of sodium cyanoborohydride in phosphate buffer (pH 7.4) (Figure 1ii). To stabilize sialic acids, glycans on solid-phase are labeled with primary amine via carbodiimide coupling in the presence of N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC; pH 4-6) (Shah et al., 2013) (Figure 1iii). As an example of glycan enzymatic treatment, sialic acids can be optionally removed from glycans using neuraminidase (Yang et al., 2013a). To release N-glycans, the immobilized glycoproteins are digested with PNGase F; after release of N-glycans, O-glycans can be also cleaved using β-elimination in the solution of ammonium hydroxide (Rademaker et al., 1998) or enzymes such as O-glycosidase (Umemoto et al., 1977). The GIG provides a platform to analyze both N-linked and O-linked glycans; the sample loss is largely minimized and glycan treatment is readily performed on solid-phase before release by enzymes or chemicals.
Figure 1.
Glycan extraction from solid-phase using chemoenzymatic immobilization and treatment. (i) Proteins denature, or protein tryptic digestion; (ii) Protein or peptide immobilization (reductive amination); (iii) Glycoprotein glycan derivatization (carbodiimide coupling); (iv) Release of N-glycans; (v) Release of O-glycans (β-elimination). These proteins can be further analyzed using tryptic digestion.
Equipment
Eppendorf Research Plus pipette (2.5 μL, 20μL, 200 μL, and 1 mL; Eppendorf) μFocus MALDI plate (384 circles; 900 μm diameter; Hudson Surface Technology)
Aminolink plus coupling resin (Aldehyde-activated 4% beaded agarose, slurred in water with sodium azide; Thermo Scientific)
15 mL BD Falcon tube (BD Biosciences)
Zeba spin desalting column (2mL; Thermo Scientific)
Sep-Pack C-18 cartridge (3 cc Vac; 200 mg sorbent, 50-105 μm particle size; Waters)
Carbograph extract-clean columns (Carbograph; 3.0 mL; Grace)
Snap-cap spin column (0.5 mL; Thermo Scientific)
230 mm Pasteur pipette, type I (Wheaton)
Corning Costar 96-well cell culture plate (Corning)
Latex dropper bulb (Fisher Scientific)
Barnstead/Thermolyne LabQuake tube shaker (Barnstead International)
Branson Sonifier 250 (Emerson Industrial Automation)
Allegra 6R centrifuge (Beckman Coulter)
Centrifuge 5415 D (Eppendorf AG)
Boekel 13300 oven (Boekel Scientific)
BCA protein assay kit (BCA reagent A:BCA reagent B = 50:1; BSA standard, 2 mg/mL; Thermo Scientific)
μQuant Microplate Spectrophotometer (Biotek Instruments)
Innova 4000 incubator shaker (37°C; New Brunswick Scientific)
Revco Freezers Ultima II (Thermo Scientific)
Lab Dancer Mini Vortexer (VWR International)
Thermo Savant SpeedVac SPD121P Centrifugal Evaporator (Thermo Scientific)
Thermo Savant Refrigerated Vapor Traps (Thermo Scientific)
DPC MicroMix 5 Shaker (Conquer Scientific)
MALDI plate holder (For Shimadzu Axima Resonance; Hudson Surface Technology)
MALDI-QIT-TOF mass spectrometer (Axima Resonance; Shimadzu) equipped with a controllable MSn fragmentation, high resolution precursor ion selection.
Glycoprotein Immobilization for Glycan Extraction
Prepare protein or peptide samples: when sample contains only peptides, dry sample in SpeedVac and re-suspend in pH 10 binding buffer (90 μL; maximum 0.5 mL due to the volume limitation of the snap-cap spin-column); when samples are proteins, we can either denature or digest proteins after being extracted from cells or tissues. Denature proteins using denaturing buffer (10 μL if 90 μL buffer is used; 10x). In this protocol, we used cell or tissue to demonstrate the method.
Note: Use acetonitrile (≤10% in final solution; e.g., 10 μL in 90 μL of binding buffer) to dissolve peptides if binding buffer does not completely dissolve them.
1. Sonication: Centrifuge cells (from 15 cm dish) or tissues (~ 50 mg) at 2000 x g for 2 min, discard the supernatant. Add 1 mL of RIPA lysis buffer (store in 4°C prior to use) to sample. Sonicate sample using Branson Sonifier sonicator tip in RIPA for 30 sec, then cool sample in ice container for 30 sec, repeat 4-6 times. Centrifuge the lysate at 16,000 x g for 10 min at 4°C. Collect the supernatant.
△ CRITICAL STEP Check the sample solution under white light for protein extraction; complete extraction is achieved when sample is transparent and clear.
△ CRITICAL STEP Sonication conditions must be determined empirically for each cell or tissue type, and sonicator (Duty cycle: 20; output control: 1-2; timer: 30 seconds).
□ PAUSE POINT The supernatant can be stored at −80° for at least 12 months.
2. Buffer Exchange: Twist off the desalting column's bottom closure and loosen cap. Place column in a 15 mL conical collection tube (Note: use 2 mL microcentrifuge tube for collection when 0.5 mL desalting column is used). Centrifuge column at 1000 x g for 2min. Discard supernatant and place a mark on the side of the column. Add up to 500 μL of binding buffer or DI water to the column. Centrifuge at 1000 x g for 2 min. Repeat additional 2-3 times. Place column in a new collection tube, remove cap and slowly apply sample to the center of the compact resin. Wait until sample penetrates the resin completely. Centrifuge at 1000 x g for 2 min. Collect supernatant.
△ CRITICAL STEP Keep mark at the same position during all subsequent centrifugation steps. Expect incomplete buffer exchange otherwise.
△ CRITICAL STEP Do not use any buffer containing amines which react with aldehyde beads and compete with protein immobilization.
□ PAUSE POINT The supernatant can be stored at −80° for at least 12 months.
3. Sample Concentration Measurement: Prepare a series of BSA solution for calibration, including 0.0625, 0.125, 0.25, 0.5, 1, 2 mg/mL; Prepare BCA solution by diluting BCA reagent B 1:50 with reagent A. Mix 20 μL BSA or sample with 200 μL BCA solution in a 96-well cell culture plate. Briefly mix in DPC shaker and incubate at 37°C for 30 min. Measure protein concentration using μQuant Microplate Spectrophotometer.
△ CRITICAL STEP Calculate amount of sample used by concentration and volume. Take appropriate amount of beads based on sample amount. Approximately, 1 mL of beads can immobilize 10-20 mg proteins.
□ PAUSE POINT Sample can be stored at −80° for at least 12 months.
4. Protein Denaturation: Dissolve 2 mg protein sample in 180 μL of binding buffer (pH 10) in a 1.5 mL microcentrifuge tube (see Reagent Setup) (The protein amount is based on 2 μL of PNGase F as a ratio of 10:1). Add 20 μL of denaturing buffer (10x), briefly vortex and centrifuge. Close microcentrifuge tube cap and incubate sample in a hot-plate at 100°C for 10 min. Leave the mixture at room temperature for 10 min.
△ CRITICAL STEP Do not use a large volume of sample in this step in order to prevent explosion and loss of sample during heating. Open and close microcentrifuge tube CAP every 3 min.
Note: Peptide standards (1% or 20 μg), AG and/or NT, can be mixed with 2 mg proteins before immobilization. Detection of AG and/or NT in MS indicates non-specific binding.
5. Resin Pre-Conditioning and Immobilization: Resin (or beads) is stored in 0.02% sodium azide at 4°C. Equilibrate resin to room temperature before use. Vortex resin slurry well before transferring resin using a pipette. Cap the bottom of a snap-cap spin column (using a rubber which is provided by Thermo Scientific). Place 100 μL resin slurry in the column (Note: our results show that the binding efficiency is above 95% using 200 μL resin for 4 mg BSA). Remove the bottom cap and centrifuge at 2000 x g for 30 sec. Discard flow-through (Note: Unless specified, centrifugation is 2000 x g for 30 sec). Close the bottom cap (rubber), add 500 μL binding buffer (pH 10) to resin. Close the top cap and vortex briefly. Remove both top and bottom caps. Place spin column in a new microcentrifuge tube. Centrifuge at 2000 x g for 30 sec. Discard flow-through. Repeat washing two additional times. Add 100 μL binding buffer to the resin, cap top and bottom. Briefly vortex to resuspend resin.
△ CRITICAL STEP Resin may aggregate after being conditioned with binding buffer. If aggregation still exists, use 200 μL pipette to break them down.
6. Protein Immobilization – Conjugation: Add 2 mg proteins (after protein denaturation) to the pre-conditioned resin (200 μL). Wash sample tube using 100 μL binding buffer and then transfer to the pre-conditioned resin. Repeat this step once. Vortex mixture of sample-resin for 30 sec. Place the mixture on a tube rotator for end over end mixing. Incubate at room temperature for at least 4 h.
Note: The total amount is 500 μL. The snap-cap spin column can contain up to 600 μL. Make adjustment on initial sample concentration and volume if necessary.
CRITICAL STEP To achieve a high yield on protein immobilization between resin and sample, constant and efficient mixing is critical for the reaction. Resin precipitation may cause incomplete sample conjugation and reduce yield.
7. Protein Immobilization – Reduction: Add 55 μL of 500 mM NaCNBH3 in PBS to mixture of sample-resin. Briefly vortex and incubate at room temperature for at least 4 h on a tube rotator. Remove spin-column caps and centrifuge at 1000 x g for 30 sec and collect flow-through. Wash resin using 500 μL of 1x PBS (during vortex, cap top and bottom of the spin column; remove caps when centrifuge column). Discard flow-through. Repeat this step once. Add 500 μL of 1x reducing buffer to resin. Incubate at room temperature for 4 h with mixing.
Note: This step allows reduction of imine for formation of amine (reductive amination). The total amount CRITICAL STEP Protein immobilization yield can be measured using flow-through and initial samples. More resin is required if yield is less than 90% as measured by BCA assay on the flow-through.
8. Resin Active Site Blocking: Wash resin using 500 μL of 1x Tris-HCl. Discard flow-through and repeat twice. Add 500 μL of 50 mM NaCNBH3 in 1x Tris-HCl. Vortex resin and incubate at room temperature for 30 min with head-to-toe mixing. Wash resin using 500 μL of 1 M NaCl. Discard flow-through and repeat twice. Wash resin using 500 μL DI water. Discard flow-through and repeat twice.
△ CRITICAL STEP It is important to block active aldehyde sites on resin after sample immobilization. The active aldehyde sites can react with any chemicals or enzymes which have primary amines, such as p-toluidine or PNGase F.
□ PAUSE POINT The sample-resin can be stored at 4°C for at least two weeks.
9. Glycoprotein Glycan Derivatization: The immobilization of glycoproteins can be readily performed by use of appropriate chemicals or enzymes (Figure 1iii). The advantages of GIG are demonstrated on sialic acid derivatization and desialylation. Other treatment and modification, such as galactose oxidase, galactosyltransferase and fucosidase may be applied. If sample does not have sialic acid, then directly go to the section of Release of NGlycan. For sialic acid derivatization, see Sialic Acid Derivatization; for removal of sialic acids, see Desialylation.
Sialic Acid Derivatization
Add 833 μL concentrated HCl (36-38%) in 9.167 mL DI water. Vortex gently and weigh 1.071 g of p-toluidine in HCl-DI solution. Mix and heat at 65°C oven for 10 min. If p-toluidine does not dissolve completely, heat at 65°C oven for another 5 min. Add 40 μL of EDC (5.65 M) and 25 μL of the concentrated HCl in 400 μL solution of p-toluidine. Check pH, if pH is not in the range of 4-6, adjust by addition of HCl (decreasing pH) or EDC (increasing pH). Add solution to sample-resin. Briefly vortex and incubate at room temperature for at least 4 h.
! CAUTION: Always add HCl in water slowly. Explosion may occur when water is added to the concentrated HCl!
Note: P-toluidine solution concentration is 1 M. Cut tip of a pipette to facilitate pipetting of EDC if it is viscous.
△ CRITICAL STEP EDC is viscous and need thawing at room temperature for 20 min.
Note: For each sample, use 465 μL solution consisting of 400 μL of p-toluidine, 40 μL of EDC and 25 μL of HCl.
Wash sample-resin using 500 μL of 0.1% formic acid; Centrifuge and discard flow-through; Repeat twice. Wash sample-resin using 500 μL of 10% acetonitrile; Centrifuge and discard flow-through; Repeat twice. Wash sample-resin using 500 μL of 1M NaCl. Centrifuge and discard flow-through. Repeat twice. Wash sample-resin using 500 μL HPLC water; Centrifuge and discard flow-through; Repeat twice.
△ CRITICAL STEP EDC is soluble in strong acid and acetonitrile. EDC-p-toluidine should be removed by steps (vii) and (viii) effectively. Without these washing steps or incomplete washing will result in EDC-p-toluidine contamination in the final product.
□ PAUSE POINT The sample-resin can be stored at 4°C for at least two weeks.
Desialylation
If sialic acid need to be removed, this step is required. Add 40 μL HPLC water per 100 μL resin slurry. Add 4 μL G1 (reaction buffer) to the sample-resin in 40 μL solution. Add 1-2 μL neuraminidase per 1 μg of glycoprotein in sample-resin (please check with manufacturer for actual amount of enzyme required for complete digestion; the ratio given in this protocol is based on New England BioLabs P0720). Vortex sample-resin gently and incubate at 37°C for 1 h. Wash resin using 500 μL of 1 M NaCl; Centrifuge and discard flow-through; Repeat twice. Wash resin using 500 μL HPLC water; Centrifuge and discard flow-through; Repeat twice.
□ PAUSE POINT The sample-resin can be stored at 4°C for at least two weeks.
10. Release of N-Glycan: In contrast from in-solution release of glycans, both N- and O-glycans are able to be released sequentially (Figure 1iv). If N-glycans are released in volatile buffer, e.g., N-glycan Release in Ammonium Bicarbonate, no purification is required. When N-glycans are further modified such as reducing-end fluorescent labeling, use G7 (reaction buffer for PNGase F) in N-glycan Release for Derivatization and purify samples.
N-glycan Release in Ammonium Bicarbonate
Add 38 μL of 10 mM ammonium bicarbonate and 2 μL PNGase F (1-20 μg glycoproteins, detail see product protocol of P0704, New England BioLabs). Gently vortex and incubate at 37°C for 2 h.
Note: 38 μL of buffer used for every 100 μL of resin slurry.
△ CRITICAL STEP Use freshly prepared ammonium bicarbonate. You must check pH. A pH of 7.5-8.5 is required for optimal enzyme activity. Overnight incubation (37°C) is recommended for complex biological samples.
Remove both top and bottom caps. Use a clean microcentrifuge tube (2 mL capacity) to collect flow-through. Centrifuge at 2000 x g for 30 sec and collect flow-through. Add 200 μL of HPLC water, vortex and centrifuge. Collect flow-through. Repeat this step twice. Combine all flow-through. Add 100% formic acid to make 0.1-1% (pH < 3.0). Dry sample in SpeedVac at 37°C. Resuspend sample in 40 μL HPLC water.
△ CRITICAL STEP Use 1% formic acid to adjust sample pH to acid. The positive-ion mode is expected for glycan MS analysis. HPLC water is only used for MS detection.
□ PAUSE POINT The glycan is stable for at least several months when stored at −20°C.
N-glycan Release for Derivatization
Add 34 μL of DI water, 4 μL of G7, and 2 μL PNGase F. Gently vortex and incubate at 37°C for 2 h. Remove both top and bottom caps; Use a clean microcentrifuge tube (2 mL capacity) to collect flow-through. Centrifuge at 2000 x g for 30 sec and collect flow-through. Add 200 μL of DI water, vortex and centrifuge; Collect flow-through; Repeat this step twice. Combine all flow-through. Add 100% formic acid to make 0.1-1% (pH < 3.0). Purify N-glycan using Carbograph cartridge (See Glycan Carbograph Purification).
Note: 200 μL of resin slurry is used.
□ PAUSE POINT Sample is stable for at least several months when stored at −20°C.
11. O-glycan release: After release of N-glycans, use ammonium hydroxide (β-elimination) to cleave O-glycans (Figure 1v). Transfer sample-resin to a 2 mL microcentrifuge tube.
△ CRITICAL STEP Add 500 μL of ammonium hydroxide to sample-resin, vortex and transfer to tube. Repeat twice for complete transfer.
Centrifuge resin at 10,000 x g for 5 min. Discard supernatant. Add 400 μL of ammonium hydroxide, incubate at 65°C for 24 h. Centrifuge and collect supernatant. Wash resin using 500 μL of 0.1% TFA; collect supernatant; Repeat twice. Use concentrated TFA to decrease sample pH to acid.
Note: The amount of TFA is determined by that of ammonium hydroxide added to sample-resin. For 400 μL of ammonium hydroxide, at least 400 μL of TFA is required.
Purify O-glycans using Carbograph cartridge.
□ PAUSE POINT Sample is stable for at least several months when stored at −20°C.
12. Glycan Carbograph Purification: Add 3 mL of acetonitrile (100%) to cartridge. Add 3 mL of 1% TFA, repeat once. Load sample to Carbograph, allow sample solution penetration to cartridge by gravity. Re-load the flow-through to Carbograph for additional time. Wash cartridge with 3 mL of 0.1% TFA, repeat four additional times. Wash cartridge with 3mL of 0.1% TFA in 10% acetonitrile. Elute sample with 400 μL of 0.1% TFA in 80% acetonitrile, repeat once. Dry sample solution in SpeedVac at 37°C. Resuspend sample in 40 μL HPLC water.
Note: Use inert gas or latex dropper bulb to push liquid through cartridge if it is too slow.
□ PAUSE POINT Sample is stable for at least several months when stored at −20°C.
13. Acquisition of MS Spectra: Acquire MS spectra either using MALDI or ESI. For MALDI, we add 1 μL DHB-DMA matrix in μFocus target and then add 1 μL sample. Dry DHB-sample in 37°C in an oven. This setup forms uniform crystals. We set laser power at 100, 2 shots per location and total 100 locations; For ESI, use direct injection for glycan analysis.
Note: DP7 (1 μL at 25 μM) can be added to the sample for normalization of relative abundance of glycans.
Reagents and Solutions
2,5-dihydroxybenzoic acid (DHB; Sigma Aldrich)
Acetic acid (AcOH; ACS reagent, >99.7%; Sigma Aldrich)
Acetonitrile (ACN; LC-MS Chromasolv; Sigma Aldrich)
Albumin from bovine serum (BSA, protein standards; 2 mg/mL; Sigma Aldrich)
Ammonium bicarbonate (NH4HCO3; >99.5%; Sigma Aldrich)
Ammonium hydroxide (NH4OH; 28-30%; Sigma Aldrich)
Angiotensin I human acetate salt hydrate (AG; 1296.48 Da; peptide for internal standard; Sigma-Aldrich)
Fetuin from fetal bovine serum (Fetuin; standard glycoprotein for method validation; Sigma Aldrich)
Formic acid (FA; ACS reagent, >98%; Sigma Aldrich)
Human serum (complex biological sample for glycan extraction; received from Johns Hopkins University)
Hydrochloric acid (HCl; 36.5-38.0%; Sigma Aldrich)
Maltoheptaose (DP7; Oligosaccharide standard for internal standard in MALDI-MS; Sigma-Aldrich)
Mucin from porcine stomach (Type III, bound sialic acid 0.5-1.5%; glycoproteins for O-glycan extraction; Sigma Aldrich)
N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC; >97.0%; 5.65 M; Sigma-Aldrich)
Neuraminidase (α2-3, α2-6, α2-8) (Desialylation of sialic acid; New England Biolabs)
Neurotensin (NT; 1672.92 Da; >90%; peptide for internal standard; Sigma-Aldrich)
N,N-dimethylaniline (DMA; >99.5%; Sigma Aldrich)
P-toluidine (pT; >99.7%; Sigma Aldrich)
Peptide-N-glycosidase F (PNGase F; Enzyme for N-glycan; New England Biolabs)
Phosphate buffered saline (PBS; 10x, pH 7.4; Life Technologies)
Protein denaturing buffer (10x; New England Biolabs) Note: it consists of 400 mM dithiothreitol (DTT) and 5% sodium dodecyl sulfate (SDS)
Protein inhibitor cocktail (Roche)
Reaction buffer for neuraminidase (G1; 10x; New England Biolabs)
Reaction buffer for PNGase F (G7; 10x; New England Biolabs)
Ribonuclease B from bovine pancreas (RNase B; standard glycoprotein for method development; Sigma Aldrich)
RIPA lysis buffer (10x; 0.5 M Tris-HCl, pH 7.4, 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA; Millipore)
Sialylglycopeptide (α2,6) (SGP; >95%; Glycopeptide standard for method development; Fushimi Pharmaceutical; Japan)
Sodium borocyanohydride (NaCNBH3; 98%; Sigma Aldrich)
Sodium chloride solution (NaCl; 5 M; Sigma Aldrich)
Sodium carbonate (Na2CO3, >99.0%; Sigma Aldrich)
Sodium citrate dihydrate (HOC(COONa)(CH2COONa)2, >99.0%; Sigma Aldrich)
Trizma hydrochloride (Tris-HCl; >99.0%; pH 7.6; Sigma Aldrich)
Trifluoroacetic acid (TFA; LC-MS Ultra; Sigma Aldrich)
Water (HPLC grade; Fisher Scientific) (Note: HPLC grade is used for final step of sample preparation prior to MS detection)
DI Water (Generated in lab) (Note: DI water is used for general sample preparation)
RIPA Lysis buffer: Dilute the protein inhibitor cocktail 1:100 with 1x RIPA lysis buffer.
Binding buffer (pH 10): Add 11.76 g sodium citrate and 2.12 g sodium carbonate in water to final volume of 50 mL, measure buffer pH and adjust by either adding sodium citrate or sodium carbonate if necessary.
10x Reducing buffer (pH 7.4): Add 31 mg NaCNBH3 in 1 mL of 1x PBS buffer to make a 500 mM solution (or 10x reducing buffer)
1x Reducing buffer (pH 7.4): Add 31 mg NaCNBH3 in 10 mL of 1x PBS buffer to make a 50 mM solution (or 1x reducing buffer).
Blocking buffer (pH 7.4): Add 31 mg NaCNBH3 to 10 mL of 1x Tris-HCl buffer (or 50 mM solution).
PNGase F (glycerol-free): Add PNGase F in the bead solution (See Release of Glycan for solution used) right before release of N-glycans. According to protocol provided by New
England BioLabs, 1μL of PNGase F is applied for 10 μg glycoprotein..
DHB-DMA matrix solution: Dissolve 100 mg of DHB per mL into 50% ACN (vol) in the presence of 0.1 mM NaCl and 2% DMA.
TFA solution: 0.1% TFA in HPLC water (vol/vol).
Denaturing buffer: See Materials (Protein denaturing buffer)
Commentary
Background Information
Release, modification, and purification of glycans are routinely manipulated in solution combined with chromatography or filtration. Proteins are first digested with PNGase F after denaturation; the supernatant is then collected after passing through cartridge such as C18; the released N-glycans are purified by a chromatographic column such as Carbograph solid-phase extraction cartridge (Morelle and Michalski, 2007). However, complex biological specimens usually consist of many different sialic acids, which are fragile and preferentially lose their moiety during sample preparation and/or mass spectrometry analysis (Sekiya et al., 2005). Without derivatization or stabilization, profiling of glycans from matrix assisted laser desorption/ionization (MALDI) does not necessarily reflect their relative abundance of each N-glycan in the biological sample. Glycan permethylation is typically performed for sialic acid derivatization and improvement of glycan ionization (Morelle and Michalski, 2007). Glycan loss, especially for low abundant species, is expected after the complicated processes. Other than permethylation, chemical reaction for sialic acid derivatization can be performed. Sialic acid can be modified in solution; however, purification of sialic acids after modification, e.g., amidation (Sekiya et al., 2005), in solution is challenging. Further sample loss is expected for purification of the treated glycans after each step, such as glycoside hydrolase enzymes or glycosidases. A high-throughput and efficient method is essential for analysis of complex biological samples.
Critical Parameters and Troubleshooting
To successfully enrich and modify glycans from glycoproteins, it is most critical to effectively conjugate glycoproteins on resin. The two-step conjugation of glycoproteins is essential for improvement of conjugation yield in that over 93% proteins are immobilized to resin during the first step reaction in pH 10 buffer. It is crucial to remove any amine-containing chemicals since they will react with aldehydes on resin and significantly reduce yield. Therefore, reagents such as Tris, and ammonium bicarbonate should not be used in the coupling buffer. The step for buffer exchange can be removed if those interference reagents contain in sample buffer. The solution for sialic acid derivatization must be prepared fresh. The followed washing steps must be performed carefully, since any remained EDC-p-toluidine will severely interfere with ESI or MALDI ionization. In addition, the pore size of the filter used in spin-column must be appropriate. The beads in the resin have diameters ranging from 45-125 μm; thus, any pore size less than 20 μm should be used. Other general precaution should be taken to prevent contamination from other sources such as powders on gloves.
Anticipated Results
N-glycan Modification on GIG
To demonstrate the feasibility of GIG on glycan enrichment, modification and isolation, we first used standard sialylglycopeptide (SGP) which predominantly consists of biantennary sialic acids N-glycans. The SPG contains six amino acids (Lys-Val-Ala-Asn-Lys-Thr) and a biantennary sialic acid linked to the amino acids via Asn residue. At a higher pH condition, e.g., pH 10, N-terminus and lysine residue can conjugate to aldehyde functionalized on beads. When SGP glycan is directly released from beads using PNGase F, several peaks are observed during MALDI-MS detection (Figure 2A). Peaks of 1976.1 and 1663.1 clearly lost one or two sialic acid residues during MALDI ionization; while the intact sialic acid (2289.1) is not the dominant glycan which is supposed to be the only species. Using carbodiimide coupling, the carboxylic acid reacts with p-toluidine to form an amide. The modification is complete and reliably protects fragmentation of sialic acids as shown in Figure 2B.
Figure 2.
Treatment of glycans on solid-phase using GIG, without sialic acid modification (A), sialic acid modification by carbodiimide coupling (C), and desialylation using neuraminidase (C). Loss of sialic acids was observed without modification and fully derivatization was achieved after modification (A) and (B). Removal of sialic acids was complete on resin.
GIG has advantage of glycan enzymatic treatment while glycoproteins are still conjugated on beads. It is ideal for using specific neuraminidases for determination of the linkage of the sialic acid, since they typically have different linkages such as α2-3, α2-6, and α2-8. This can be easily implemented on solid-phase using GIG. To demonstrate the effectiveness of removal of sialic acids on GIG, we use general neuraminidase which can remove all three types of sialic acid linkages. MALDI-MS showed that all sialic acids are effectively removed, only detecting signal peak at 1663.1 Da (Figure 2C). With specific neuraminidases and sialic acid carbodiimide coupling, one could determine the structure of N-glycans using GIG.
Improvement of MS and MS/MS on Modified Sialylated N-glycans
MALDI-MS has been a reliable and robust method to provide a snapshot of the mass profiles as well as the most likely set of glycan structures (Raman et al., 2005). However, the negatively charged sialic acids tend to be lost in MALDI ionization and their stabilization is usually required (Sekiya et al., 2005). The derivatization of sialic acids can be done directly in solution or on solid-phase. The former approach is challenging for glycan purification from the modification reagents, in that reagents have similar properties so that they are difficult to separate using either chromatographic methods such as C18 or Carbograph (Huizing et al., 2004). This issue is effectively mitigated when derivatization is performed on solid-phase, in which reagents are removed by a variety of washing steps without loss of sample.
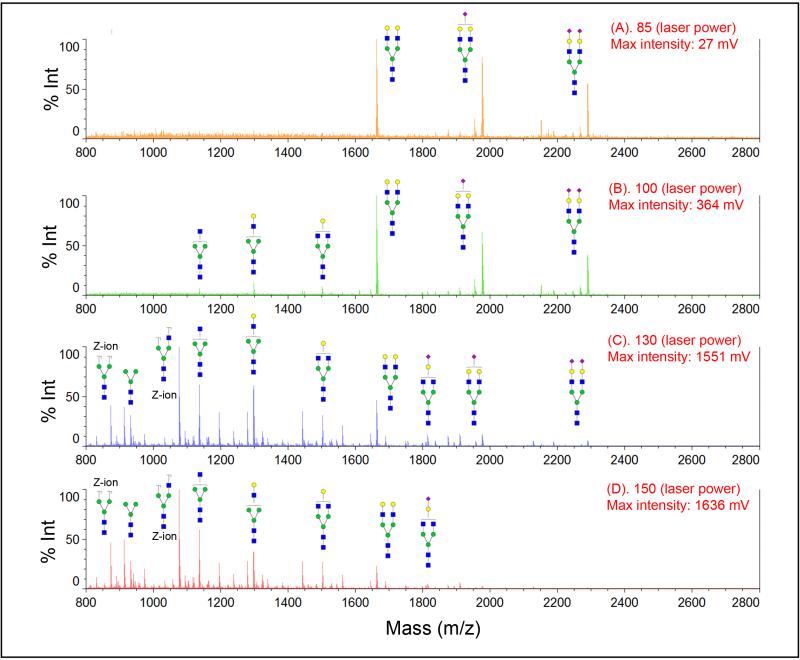
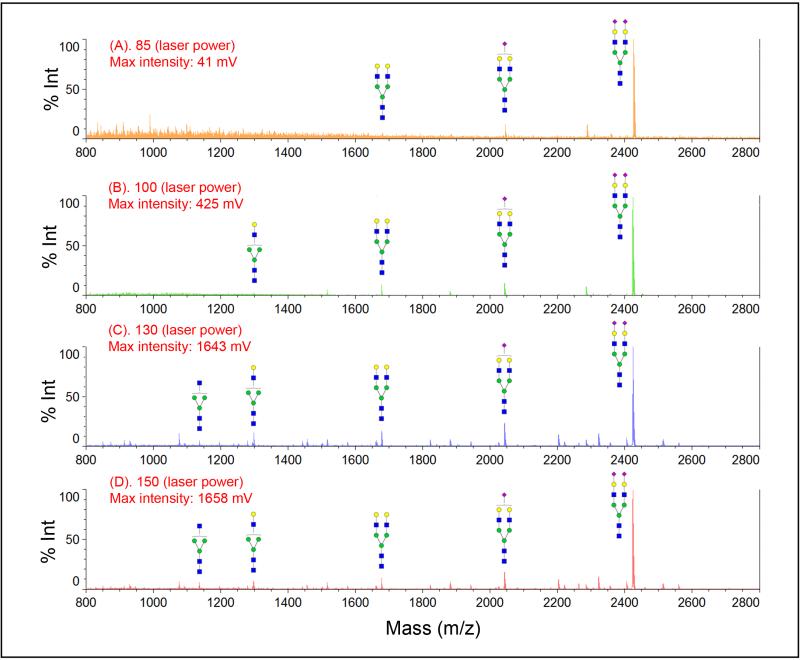
Figure 3 compared the effect of laser power on mass spectra of sialylated N-glycans before and after modification. Even at a lower laser power, e.g., 85 (ranging from 0 to 180 in Shimadzu Resonance Axima), we observed three peaks from SGP N-glycans (Figure 3.1A). The intensity for glycans which lose one and two sialic acids becomes higher than the intact sialic acid glycan at the laser power of 100 (Figure 3.1B). Most sialic acid glycans are fragmented at higher laser energy (Figure 3.1C & D). No intact sialic acid glycan is detected at laser power above 150. For sample whose sialic acid is stabilized by p-toluidine, we detected sialylated biantennary N-glycan across different laser power (85, 100, 130, and 150) as shown in Figure 3.2A-D, although there is small amount of fragmented glycans at a higher laser energy (less than 5%).
Figure 3.
Stabilization of sialic acid with carbodiimide coupling modification in MALDI-MS detection. MS spectra were acquired in different MALDI ionization laser power (A) 85, (B) 100, (C) 130, and (D) 150. Severe loss of sialic acids is observed in native glycan without modification (1), while sialic acids are well protected after carbodiimide coupling stabilization (2). Symbol: solid blue square, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; purple diamond, N-acetyl-neuraminic acid.
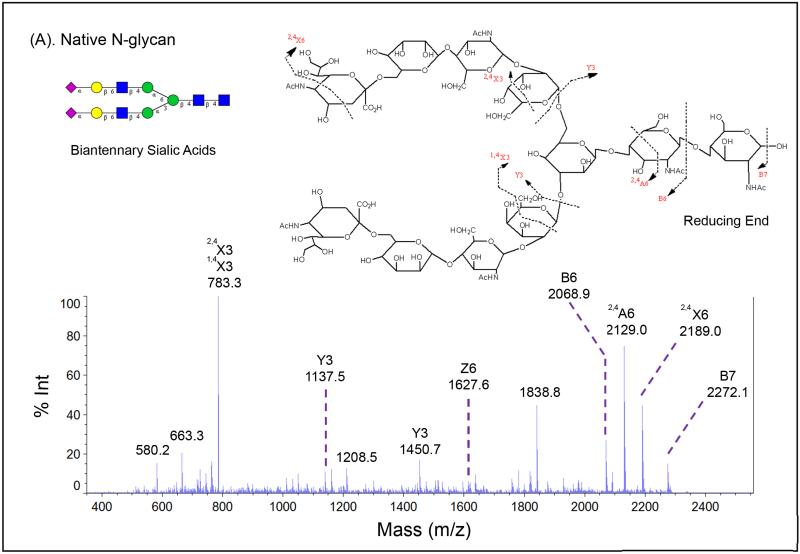
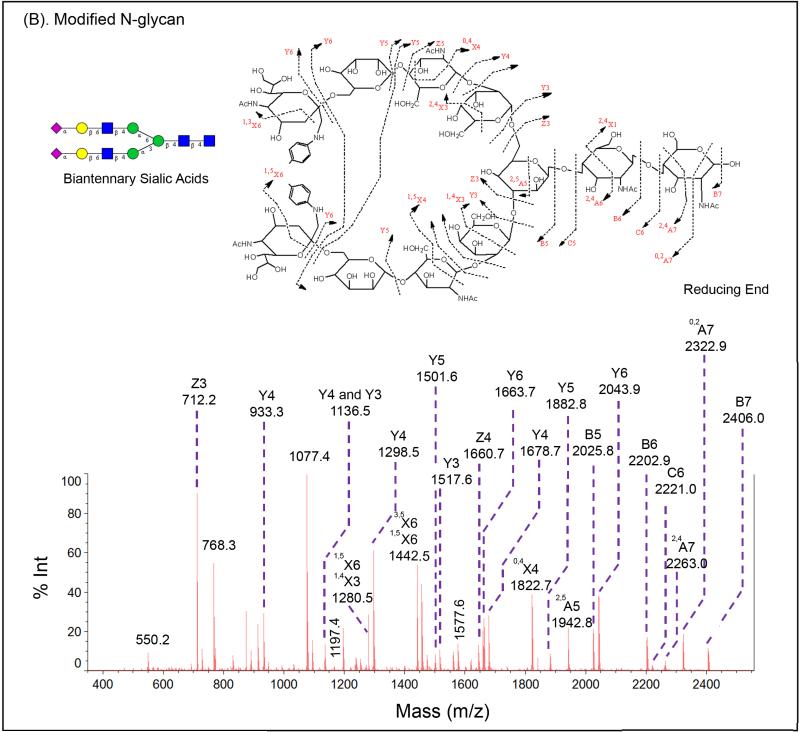
More significant attribute for modified sialic acids may be the improvement of signals for glycan signature ions from its tandem mass spectra. The diverse nature of glycans requires not only specific enzymes but also mass fragmentation to elucidate their linkages and structures. Therefore, it is critical to generate a comprehensive MS/MS spectrum from a glycan precursor. Glycan MS2 fragments may consist of X, Y, and Z ions with reducing terminus, or A, B, and C ions without reducing terminus, as illustrated in Figure 4. We used the SGP N-glycans (sialylated biantennary N-glycan), and one sample was treated with p-toluidine on GIG as a comparison to the control without modification. Their MS1 spectra is shown in Figure 3.1B and Figure 3.2B. MS2 spectra on modified glycans dramatically improve fragment signature ions compare to that on un-modified glycans. The MS2 spectrum of un-modified glycan only exhibits one A ion, two B ions, two X ions, two Y ions, and one Z ion. The overall intensity is low (Figure 4A). For the modified glycan, a much more detailed MS2 spectrum is achieved. The intensity has been significantly improved. Both A and X ions are observed besides B, C, Y, and Z ions (Figure 4B).
Figure 4.
MS2 fragmentation signature ions generated from Shimadzu MALDI-MS on native (A) and modified (B) SGP N-glycans.
Profiling of N-glycans on Complex Biological Sample
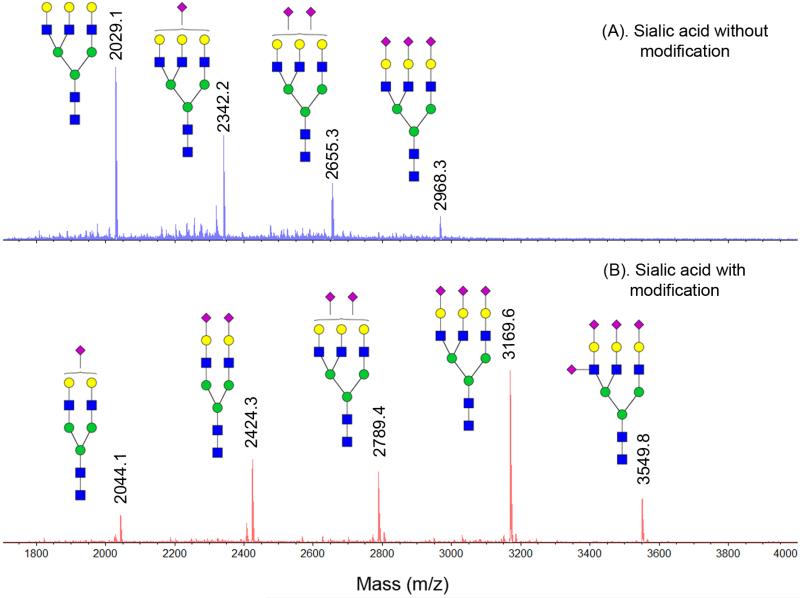
The GIG method isolates N-glycans from glycoproteins. To prove that this method does not create artificial results after sialic acid modification, we first used fetuin from fetal bovine serum. NMR (nuclear magnetic resonance) analysis on fetuin oligosaccharides showed sialylated N-glycans from fetuin, in which triantennary sialic acid is the most abundant among them, followed by the biantennary sialic acid and the tetraantennary structure (Green et al., 1988). In a traditional approach, the glycans are usually isolated from physical chromatography and further modified by permethylation to increase ionization and detectability, as well as sialic acid derivatization. N-glycan profile from fetuin using this approach shows similar data except for relative high abundance of tetraantennary sialic acid than triantennary N-glycan with two sialic acid (Kang et al., 2005). Incomplete permethylation and sample loss during multiple purification steps are potential concerns when the standard method is applied. Comparatively, permanent immobilization of glycoproteins on beads is able to implement high-throughput glycan analysis and significantly minimize sample loss. The N-glycan profile from fetuin using GIG is shown in Figure 5, where Figure 5A is mass spectra without glycan modification and Figure 5B is mass spectra after sialic acid stabilization. Clearly, without stabilization on sialic acids, the most abundant glycans contain less or no sialic acids due to the loss; with stabilization on sialic acids, we observe fetuin N-glycan profile which is consistent with that by traditional approach (Green et al., 1988).
Figure 5.
MALDI-MS profile of sialic acid glycans after carbodiimide coupling using GIG. (A). Sialic acids without modification, showing loss of sialic acids (2029.1 Da and 2322.2 Da); (B). Sialic acids after modification, showing complete labeling. Symbol: solid blue square, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; purple diamond, N-acetylneuraminic acid.
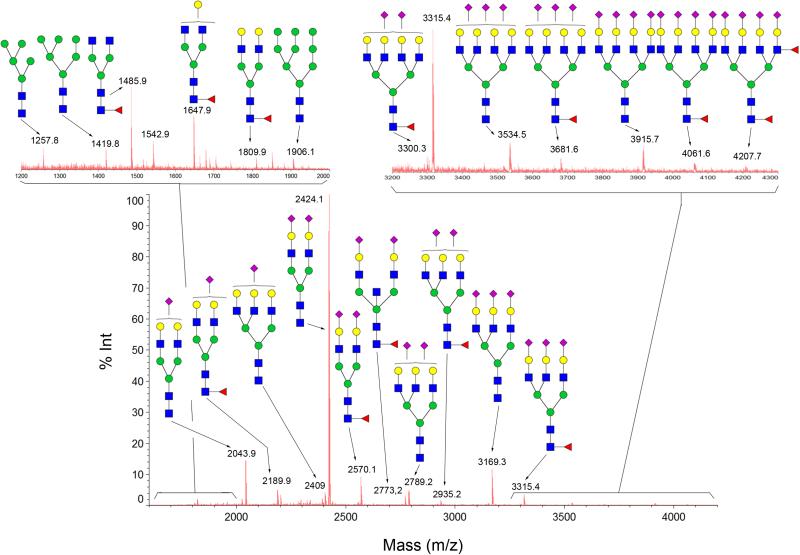
We further applied GIG for profiling of human serum N-glycans as shown in Figure 6. The most abundant species in human glycome is biantennary sialic acid (2424.1 Da), followed by one sialic acid (2043.9 Da), fucosylated biantennary sialic acid (2570.1 Da), and triantennary sialic acid (3169.3 Da). The number of sialic acids and the relative abundance of each sialic acids are consistent with the profile generated by traditional protocol . The glycan above 3000 Da shows good signal to noise ratio (insert in Figure 6). We identified 65 N-glycans from human serum without separation, and up to 150 N-glycans detected after separation by liquid chromatography (chipLC) (Yang et al., 2013b).
Figure 6.
MALDI-MS profile of carbodiimide coupling modified N-glycans using GIG derived from 2 μL of human serum. After conjugation of serum proteins on beads and treated with EDC/p-toluidine, N-glycans released by PNGase F were directly analyzed using MALDI-MS without glycan purification. Only the structures of the major N-glycans are given. Symbol: solid blue square, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; red triangle, fucose; purple diamond, N-acetyl-neuraminic acid.
Time Considerations
Sample preparation using the described protocols requires approximately 24 h, including the sialic acid derivatization. Starting from tissues or cells, it takes 30 min for sonication, 30 min for buffer exchange, 1 h for concentration measurement by BCA, 4 h for conjugation, 4 h for reduction, 30 min for resin blocking, 3 h for sialic acid derivatization, and 2 h for PNGase F digestion. Each washing will take 1 min. In each step, samples may be stored as suggested in the basic protocol. It is recommended that multiple samples from different clinical representatives are prepared in parallel. The proteins on resin may be further analyzed by trypsin digestion (overnight).
Acknowledgments
We thank Dr. Lori Sokoll from Johns Hopkins for providing human serum specimen and Shimdazu for the support of MALDI-TOFMS (AXIMA Resonance). This work was supported by the National Institutes of Health, National Cancer Institute, the Early Detection Research Network (EDRN, U01CA152813), the Clinical Proteomic Tumor Analysis Consortium (CPTAC, U24CA160036), and by National Institutes of Health, National Heart Lung and Blood Institute Programs of Excellence in Glycosciences (PEG, P01HL107153) and the Johns Hopkins Proteomics Center (N01-HV-00240).
Footnotes
Competing Financial Interests
The authors declare no competing financial interest.
Literature Cited
- Carlson DM. Structures and immunochemical properties of oligosaccharides isolated from pig submaxillary mucins. J. Biol. Chem. 1968;243:616–626. [PubMed] [Google Scholar]
- Green ED, Adelt G, Baenziger J, Wilson S, Van Halbeek H. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J. Biol. Chem. 1988;263:18253–18268. [PubMed] [Google Scholar]
- Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Rakocevic G, Sparks SE, Mamali I, Shatunov A, Goldfarb L, Krasnewich D, Gahl WA, Dalakas MC. Hypoglycosylation of α-dystroglycan in patients with hereditary IBM due to GNE mutations. Mol. Genet. Metab. 2004;81:196–202. doi: 10.1016/j.ymgme.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Communi. Mass Spectrom. 2005;19:3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobata A. Use of endo-and exoglycosidases for structural studies of glycoconjugates. Anal. Biochem. 1979;100:1–14. doi: 10.1016/0003-2697(79)90102-7. [DOI] [PubMed] [Google Scholar]
- Mechref Y, Novotny MV. Handbook of Glycomics. 1st ed. Elsevier Inc.; London, UK: 2009. [Google Scholar]
- Morelle W, Michalski J-C. Analysis of protein glycosylation by mass spectrometry. Nat. Protoc. 2007;2:1585–1602. doi: 10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- Ole H, Cummings RD. Essential of Glycobiology. 2nd ed. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- Plummer T, Elder J, Alexander S, Phelan A, Tarentino AL. Demonstration of peptide: N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J. Biol. Chem. 1984;259:10700–10704. [PubMed] [Google Scholar]
- Rademaker GJ, Pergantis SA, Blok-Tip L, Langridge JI, Kleen A, Thomas-Oates JE. Mass spectrometric determination of the sites of O-glycan attachment with low picomolar sensitivity. Anal. Biochem. 1998;257:149–160. doi: 10.1006/abio.1997.2548. [DOI] [PubMed] [Google Scholar]
- Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat. Method. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Wada Y, Tanaka K. Derivatization for stabilizing sialic acids in MALDIMS. Anal. Chem. 2005;77:4962–4968. doi: 10.1021/ac050287o. [DOI] [PubMed] [Google Scholar]
- Shah P, Yang S, Sun S, Aiyetan P, Yarema KJ, Zhang H. Mass spectrometric analysis of sialylated glycans with use of solid-phase labeling of sialic acids. Anal. Chem. 2013;85:3606–3613. doi: 10.1021/ac3033867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto J, Bhavanandan VP, Davidson EA. Purification and properties of an endo-alpha-N-acetyl-D-galactosaminidase from Diplococcus pneumoniae. J. Biol. Chem. 1977;252:8609–8614. [PubMed] [Google Scholar]
- Yang S, Li Y, Shah PK, Zhang H. Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 2013a;85:5555–5561. doi: 10.1021/ac400761e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Toghi Eshghi S, Chiu H, DeVoe DL, Zhang H. Glycomic analysis by glycoprotein immobilization for glycan extraction and liquid chromatography on microfluidic chip. Anal. Chem. 2013b;85:10117–10125. doi: 10.1021/ac4013013. [DOI] [PMC free article] [PubMed] [Google Scholar]