ABSTRACT
The gut microbiota is essential for human health, but very little is known about how the composition of this ecosystem can influence and respond to bacterial infections. Here we address this by prospectively studying the gut microbiota composition before, during, and after natural Campylobacter infection in exposed poultry abattoir workers. The gut microbiota composition was analyzed with 16S amplicon sequencing of fecal samples from poultry abattoir workers during the peak season of Campylobacter infection in Sweden. The gut microbiota compositions were compared between individuals who became culture positive for Campylobacter and those who remained negative. Individuals who became Campylobacter positive had a significantly higher abundance of Bacteroides (P = 0.007) and Escherichia (P = 0.002) species than those who remained culture negative. Furthermore, this group had a significantly higher abundance of Phascolarctobacterium (P = 0.017) and Streptococcus (P = 0.034) sequences than the Campylobacter-negative group, which had an overrepresentation of Clostridiales (P = 0.017), unclassified Lachnospiraceae (P = 0.008), and Anaerovorax (P = 0.015) sequences. Intraindividual comparisons of the fecal microbiota compositions yielded small differences over time in Campylobacter-negative participants, but significant long-term changes were found in the Campylobacter-positive group (P < 0.005). The results suggest that the abundance of specific genera in the microbiota reduces resistance to Campylobacter colonization in humans and that Campylobacter infection can have long-term effects on the composition of the human fecal microbiota.
IMPORTANCE
Studies using mouse models have made important contributions to our understanding of the role of the gut microbiota in resistance to bacterial enteropathogen colonization. The relative abundances of Escherichia coli and Bacteroides species have been pointed out as important determinants of susceptibility to Gram-negative pathogens in general and Campylobacter infection in particular. In this study, we assessed the role of the human gut microbiota in resistance to Campylobacter colonization by studying abattoir workers that are heavily exposed to these bacteria. Individuals with a certain composition of the gut microbiota became culture positive for Campylobacter. As their microbiotas were characterized by high abundances of Bacteroides spp. and E. coli, well in line with the findings with mouse models, these bacterial species likely play an important role in colonization resistance also in humans.
INTRODUCTION
Our microbes contribute to several important functions for the body. For example, the gut microbiota helps us to digest food and produce vitamins and has an important role in the development of the host physiology and gut immune system (1, 2). The gut microbiota composition has also been associated with disease manifestations, such as atherosclerosis, metabolic disorders, irritable bowel syndrome, and inflammatory bowel disease (3–9). In particular, in Crohn’s disease, patients have been observed to have a disturbed composition and reduced diversity of the intestinal microbiota (10, 11). A recent study proposed that the human gut microbiota can be stratified into 3 major clusters, termed enterotypes, and these enterotypes are defined by a predominance of the genera Bacteroides (enterotype 1), Prevotella (enterotype 2), and Ruminococcus (enterotype 3) (12). In studies of other cohorts, less clear dominance of the genus Ruminococcus and stronger clustering around the genera Bacteroides and Prevotella have been found (5, 13), yet both long-term diet and disease (atherosclerosis) have been linked to these enterotypes (5, 13).
One important function of the microbiota is colonization resistance. However, very little is known about how the human gut microbiota influences susceptibility to infection caused by enteropathogens. In one animal study, transplantation of microbiota from mouse strains refractory to Citrobacter rodentium infection protected susceptible mouse strains from an otherwise lethal infection, suggesting that the protective effect against pathogen colonization and enterocolitis development was dependent on the microbial community composition (14). Another mouse study indicated that a microbiota with a low complexity or the disturbance of the gut microbial community structure with antimicrobials predisposed the subject to the development of Salmonella enterica-induced enterocolitis (15). Stecher et al. (16) in 2010 found that the colonization efficiency of S. enterica correlated with the levels of intestinal Escherichia coli before infection in C57Bl/6 mice and that mice with high levels of lactobacilli were more prone to colonization by a Lactobacillus reuteri strain, suggesting that the presence of closely related species in a host’s microbiota can facilitate colonization by another related bacterial species (16).
Campylobacter is the most common cause of human bacterial enteritis in developed countries, with Campylobacter jejuni and Campylobacter coli causing the vast majority of the infections. Mice are naturally resistant to Campylobacter infection, and challenge leads to only transient colonization. However, mice with a humanized intestinal microbiota could be successfully colonized with C. jejuni, which resulted in intestinal inflammation, further indicating that the constitution of the microbiota might influence resistance to colonization and induction of inflammation (17). Characterization of the microbiotas of mice later infected with C. jejuni revealed that elevated numbers of intestinal E. coli organisms reduced resistance to colonization by C. jejuni (18).
In the present prospective study, we assessed the compositions of the fecal microbiotas of poultry abattoir workers in association with Campylobacter infection. These workers were Campylobacter culture negative at the beginning of the study and were regularly monitored, with fecal samples obtained for Campylobacter culture and analysis of the microbiota composition during the peak season for Campylobacter-positive chicken flocks in Sweden. We found clear differences in the fecal microbiota compositions between participants who later became Campylobacter positive during the study and those who remained negative. Intraindividual stability in the microbiota over time was significantly lower among participants who became Campylobacter positive.
(Part of this study was presented at the 17th International Workshop on Campylobacter, Helicobacter and Related Organisms, Aberdeen, Scotland, 15 to 19 September 2013, and at the 7th Finnish Gut Day, Helsinki, Finland, 24 January 2014.)
RESULTS
Composition of the fecal microbiota.
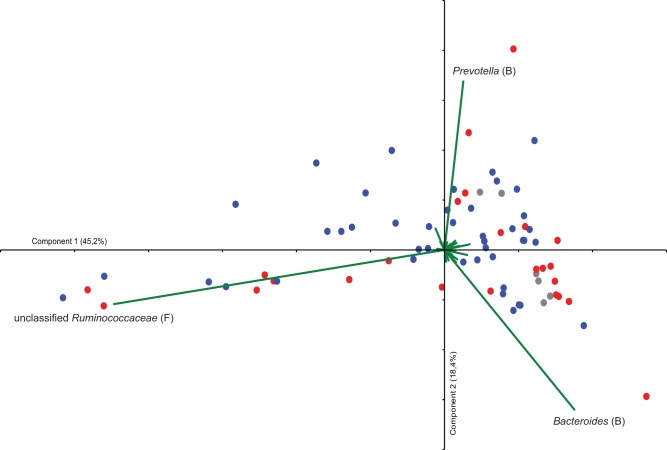
The gut microbiota composition was analyzed in fecal samples collected prospectively from 24 poultry abattoir workers during the peak season of Campylobacter infection in Sweden. 454 pyrosequencing of 16S amplicons from the fecal samples generated a total of 221,238 sequences, with an average of 2,729 sequences per sample (range, 1,409 to 12,785; 90% of the samples had more than 2,110 sequences). Phylogenetic classification revealed that the prevalences of the four major phyla (Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria) varied considerably between the samples. We used principal-component analysis (PCA) in order to identify relationships among samples with regard to their microbial community structure. The PCA identified one individual in the sample set as an outlier, and therefore this particular individual was excluded from the PCA. Analysis of the sequence data, classified to the genus level from the remaining individuals, revealed that primarily three taxonomic groups of bacteria created gradients in the ordination. Interestingly, these taxonomic groups were Bacteroides, Prevotella, and unclassified Ruminococcaceae (Fig. 1), which also have been reported to represent main contributors to the different enterotypes in the human gut (12). The samples did, however, not cluster as distinct enterotypes (Fig. 1).
FIG 1 .
Bi-plot from a principal-component analysis (PCA) of 454 pyrosequencing data classified to the taxon level. Each sample obtained from the poultry abattoir workers is represented by a colored circle, and the variables are shown as green vectors. Eigenvalues for components 1 and 2 are shown in parentheses. The colors of samples represent different groups, as follows: red, individuals who became Campylobacter positive; blue, individuals who remained Campylobacter negative; and gray, individuals with earlier reported Campylobacter infection. Abbreviations represent the phylogenetic origins of the different taxa, as follows: B, Bacteroidetes, and F, Firmicutes.
The composition of the microbiota correlates with a positive Campylobacter culture.
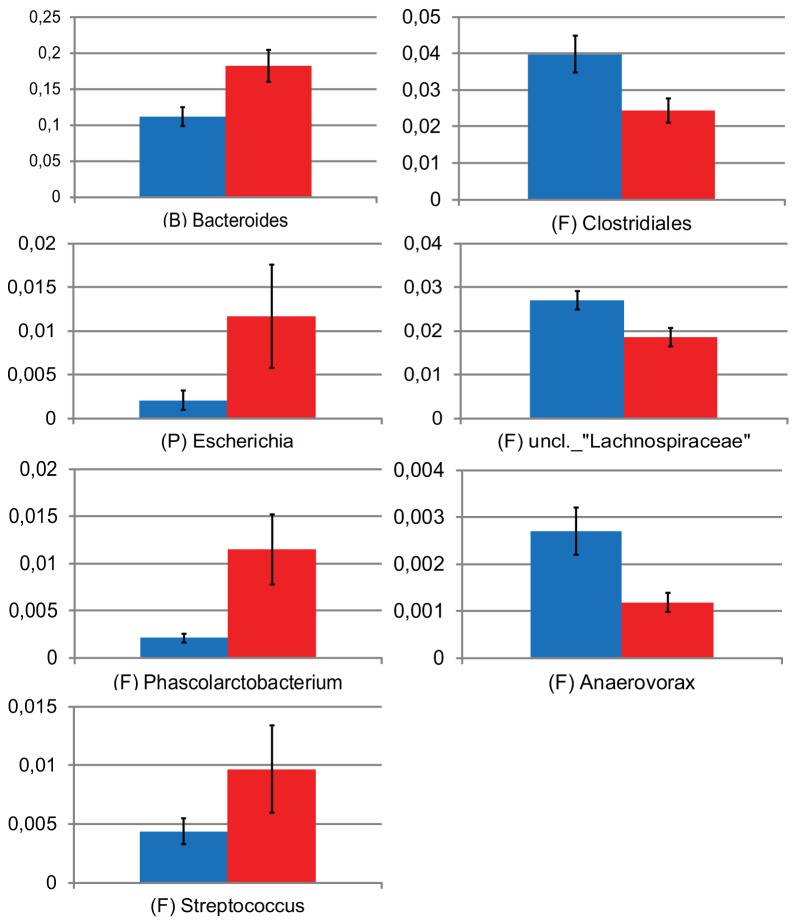
In this study, Campylobacter-positive samples were obtained from seven participants, all but one with asymptomatic infection, whereas 17 remained culture negative. We analyzed differences in the compositions of the microbiotas between individuals who became Campylobacter positive and those who remained negative. There were no significant differences in taxon diversity between the two groups (data not shown). However, coloring of samples in the PCA plot showed that most of the samples from Campylobacter-positive individuals (red) clustered with only little overlap with the samples from Campylobacter-negative individuals. A nonparametric multivariate analysis of variance (np-MANOVA) showed that this clustering pattern was significant, with differences between those who became positive and those who remained culture negative (P = 0.017). In addition, samples from individuals with an earlier-reported Campylobacter infection (gray) clustered primarily together with those from Campylobacter-positive individuals. Interestingly, in the PCA plot, it seemed like the clustering of the samples of the individuals who became culture positive was linked with the abundance of Bacteroides species (Fig. 1). Therefore, we further investigated specifically which microbial taxa differed between the Campylobacter-positive and -negative individuals. As indicated in the PCA, the abundance of Bacteroides sequences was higher in Campylobacter-positive individuals (P = 0.007, Mann-Whitney test) (Fig. 2). In addition, this particular group was associated with higher abundances of Escherichia (P = 0.002), Phascolarctobacterium (P = 0.017), and Streptococcus (P = 0.034) sequences than was the Campylobacter-negative group (Mann-Whitney’s test). Participants who remained Campylobacter negative (blue) had higher abundances of sequences classified as Clostridiales (P = 0.017), unclassified Lachnospiraceae (P = 0.008), and Anaerovorax (P = 0.015, Mann-Whitney test) (Fig. 2). Sequences classified as Campylobacter spp. were found only in low abundances (below 0.2%) and only in three samples of the same number of individuals who became Campylobacter culture positive in the present study.
FIG 2 .
Relative abundances of taxa that differ significantly between individuals who became Campylobacter positive (red) and those who remained negative (blue) (Mann-Whitney’s test). Error bars show standard errors of the means. Abbreviations represent the phylogenetic origin of the different taxa as follows: B, Bacteroidetes; F, Firmicutes; and P, Proteobacteria. uncl., unclassified.
Stability of the gut microbiota composition over time.
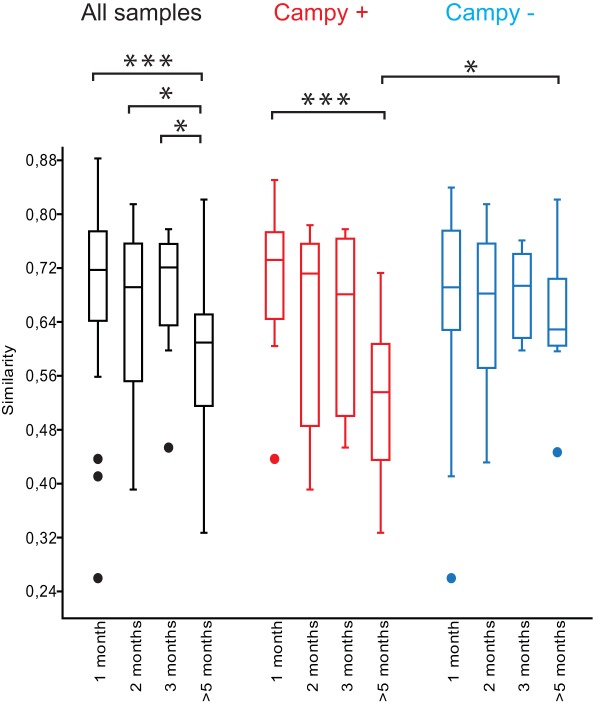
The stability of a participant’s gut microbiota over time was assessed by calculating Bray, Curtis similarities from samples collected from the same individual at intervals of 1, 2, 3, or >5 months. Comparisons of intraindividual samples separated by >5 months generally yielded larger differences than short-term intervals. When Campylobacter-positive and -negative participants were analyzed together, the similarity index for samples separated by >5 months was significantly lower than those separated by 1 (P < 0.001), 2 (P < 0.05), or 3 (P < 0.05, Kruskal-Wallis) months (Fig. 3). However, when the Campylobacter-positive participants were compared to -negative participants that had not reported previous Campylobacter infection, the similarity index for intraindividual samples separated by >5 months was significantly lower for the group that became Campylobacter positive (P < 0.005, Kruskal-Wallis). In fact, most of the drop in the similarity index was explained by this group, as the similarity index for samples separated by >5 months within the control group did not significantly differ from comparisons over shorter time intervals (P = 0.25, Kruskal-Wallis). Furthermore, in the Campylobacter-positive group, the follow-up sample obtained in February accounted for the decreased similarity index, as the comparisons over shorter time periods yielded only small differences (Fig. 3). We analyzed whether this drop could be explained by changes in the abundances of certain taxonomic groups of bacteria in the last sample obtained in February. However, we could not find any specific bacterial taxa that had consistently increased or decreased among all Campylobacter-positive individuals in the long-term follow-up sample.
FIG 3 .
Box plot showing the distribution of similarity scores for intraindividual comparisons of community profiles from samples separated 1, 2, 3, or >5 months in time. Box plots in black show the distribution of similarity scores from all samples, whereas box plots in red and blue show distributions of similarity scores from individuals who became Campylobacter positive (red; Campy +) and those who remained negative (blue; Campy −). Significance was tested using the Kruskal-Wallis test with the Mann-Whitney test as the post hoc test. *, P value below 0.05; ***, P value below 0.005. The horizontal line in the box plot represents the median value, and the box is drawn from the 25th to 75th percentiles. Whiskers show minimum and maximum values, and circles represent outliers.
DISCUSSION
The concept that the microbiota can protect the host from colonization by enteric pathogens, i.e., lead to colonization resistance, has been known for a long time (19), but few studies have been successful in generating evidence to define key species or functions involved. The recent advances in sequencing technology have made such analyzes possible (20), and a few animal studies have indicated a role of the microbiota in colonization resistance (16–18, 21, 22). We aimed to address the role of the human fecal microbiota in colonization resistance and prospectively followed staff members at chicken abattoirs with analyses of their fecal microbiota compositions before, during, and after the summer peak of Campylobacter colonization of chicken flocks in Sweden. We identified significant differences in the microbiota compositions between individuals who became Campylobacter positive during the study and those who remained negative. This is as far as we know the first microbiota study of resistance to Campylobacter colonization in humans.
We found that the main human enterotypes, namely, Bacteroides, Prevotella, and unclassified Ruminococcaceae, created gradients in our material, indicating that the compositions of the fecal microbiotas among the abattoir workers in this study did not differ dramatically from those of other cohorts studied (5, 12). However, we found significant differences in the abundances of certain bacterial taxa between the individuals who became culture positive for Campylobacter and those who remained negative. Importantly, these differences not merely were due to the presence or absence of Campylobacter in the samples but were evident in all consecutive fecal samples of the individuals, both before and after the Campylobacter-positive sample, indicating a general difference in the fecal microbial community structures between individuals who became positive for Campylobacter and those who remained negative. The taxa Bacteroides and Escherichia were present in significantly higher proportions in the positive group (Fig. 2). A recent study linked the abundance of E. coli with a gastrointestinal dysfunction in cystic fibrosis patients (23). Interestingly, recent studies on resistance to Campylobacter colonization in experimentally infected mice showed that colonization with C. jejuni was associated with significantly elevated numbers of both intestinal E. coli and Bacteroides/Prevotella organisms (17, 18). In addition, oral supplement of E. coli to mice increased the susceptibility to Campylobacter infection, and furthermore, high abundances of Proteobacteria in the fecal microbiota have previously been reported to facilitate colonization by bacterial pathogens of the same phylum (16, 18). Whether the correlation between certain bacterial genera and susceptibility to Campylobacter infection is due to a direct inhibitory effect of the commensals or due to their effects on the intraluminal milieu remains to be studied. We also found higher proportions of Phascolarctobacterium and Streptococcus sequences in individuals who became positive for Campylobacter, and these bacterial taxa have previously not been associated with colonization resistance.
The gut microbiota is generally considered to be stable over time in healthy adults (24). This is in agreement with our finding that the intraindividual stabilities of the fecal microbiotas were similar in samples separated in time by 1, 2, or 3 months. In addition, samples from individuals who remained Campylobacter culture negative also had a stable microbiota when samples separated more than 5 months in time were compared. However, among the individuals who became positive for Campylobacter during the study, the similarity index was significantly reduced when samples separated by >5 months were compared, indicating that the microbiota had changed in composition. The fact that the last follow-up sample accounted for most of this change suggests that Campylobacter positivity had caused long-term alterations in the fecal microbiota compositions of these individuals. Possible reasons for such a change might be related either to bacterial competition or to the immune responses of the Campylobacter-positive participants. The fact that we did not find consistent changes in the abundances of certain bacterial taxa in all Campylobacter-positive participants suggests that induction of such changes is complex and might have different outcomes in different individuals in terms of which bacterial species are affected.
Microbial diversity has also been linked with colonization resistance. Stecher et al. in 2010 studied the impact of microbial diversity on the resistance to colonization in mice and found that a microbiota with a high complexity had better resistance to Salmonella infection than both a low-complexity microbiota and that after antibiotic treatment (16). Our results showed no difference in levels of fecal microbial diversity between individuals who became Campylobacter positive and those who remained negative. It is, however, not possible to rule out the importance of diversity. In our study, all but one of the culture-positive individuals were asymptomatic, and all of them had a microbial diversity similar to that of healthy noninfected individuals.
The risk of acquiring Campylobacter infection at a chicken abattoir is high (25), especially during the summer peak in June to September, when approximately 25 to 40% of the slaughtered chicken flocks are Campylobacter positive in Sweden (26). This suggests that all staff members must be exposed to Campylobacter at several occasions during the seasonal peak. Therefore, the chicken abattoir workers make an interesting cohort for studying resistance to Campylobacter infection. Although the number of participants in the present study was low, the findings were clear enough to reach statistical significance. With the background evidence from mouse studies, we find it reasonable to assume that the higher abundances of both Bacteroides and Escherichia species were associated with an increased susceptibility to the establishment of Campylobacter infection, as shown by positive fecal cultures in the abattoir workers. Furthermore, we identified significant alterations in the compositions of the gut microbiotas after 5 months in the individuals who became Campylobacter positive. Whether the microbiota composition, alone or together with the immune status of the individuals, also plays a significant role in the eradication of Campylobacter from the intestine remains to be studied.
In conclusion, elevated proportions of Bacteroides and Escherichia species in the gut microbiota might predispose humans to Campylobacter infection, and Campylobacter infection can cause long-term alterations in the composition of the human gut microbiota. To the best of our knowledge, this is the first microbiota study presenting evidence of resistance to Campylobacter colonization/infection in humans.
MATERIALS AND METHODS
Participants and sample collection.
In this prospective study, a total of 31 abattoir workers (18 women and 13 men) at 3 different poultry abattoirs in Sweden were included. Fecal samples were collected once a month during the summer months June to September of 2010, with an additional follow-up sample at the end of February 2011. On each sampling occasion, one fecal sample for Campylobacter culture and one sample for analysis of microbiota were collected by the participants and sent to the Clinical Microbiology Laboratory at Uppsala University Hospital. Fecal samples were cultured immediately upon arrival for Campylobacter by routine methods, and samples for microbiota analysis were stored at −70°C for subsequent analysis. Information on age, gender, length of employment at the abattoir, previously documented Campylobacter infections, and use of antimicrobial therapy, as well as gastrointestinal disorders and symptoms, was obtained by questionnaires filled out by the participants. All fecal samples were Campylobacter culture negative at the beginning of the study. During the study, 7 participants became culture positive for Campylobacter. One additional abattoir worker was enrolled because he fell ill and was diagnosed with Campylobacter infection at the county hospital of the region where he lived. None of the other infected participants reported any symptoms of Campylobacter infection. Of the 31 participants, 1 Campylobacter-positive individual was excluded due to too little fecal material for microbiota analysis and, for another Campylobacter-positive individual who was treated with antimicrobials, the follow-up samples after antimicrobial therapy were excluded from analysis. Of the culture-negative participants, 2 were excluded because they had gastrointestinal disorders (1 had celiac disease, and 1 reported symptoms related to irritable bowel syndrome). Two were excluded because early antimicrobial therapy was initiated, and the last follow-up sample from one individual was excluded due to antimicrobial therapy in connection with that sample. Furthermore, 2 were excluded because of too little fecal material for microbiota analysis. In total, samples from 7 Campylobacter-positive participants and 17 Campylobacter-negative participants were analyzed for fecal microbiota composition. Five participants (1 Campylobacter positive) had worked at the abattoir ≤1 month from the beginning of the study and had not reported any previously documented Campylobacter infection. Six participants (2 of whom were Campylobacter positive in the present study) had reported previously documented Campylobacter infection. None of the individual characteristics collected in the questionnaires was associated with Campylobacter positivity (see Table S1 in the supplemental material).
The study was approved by the regional board of the ethical committee at Uppsala University. All participants gave their written informed consent.
Bacterial isolation and typing.
Fecal samples were cultured for Campylobacter by routine methods on modified charcoal cefoperazone deoxycholate (mCCDA) agar plates (Oxoid, Basingstoke, England), and isolates were originally identified by colony appearance, oxidase testing, and microscopy. Five isolates were further subtyped by PCR analysis (27), 4 of them to C. jejuni and 1 to C. lari. Two isolates were not available for subtyping.
Preparation of samples for 454 pyrosequencing.
DNA was extracted from 250-mg fecal samples using the Mo Bio PowerSoil DNA kit (Solana Beach, CA, USA) according to the manufacturer’s instructions. The only modification from the kit protocol was that the bead-beating step was performed twice for 45 s at level 5 on a FastPrep-24 (MP Biomedicals, Solon, OH). The bacterial primers Bakt_341F (CCTACGGGNGGCWGGAG) and Bakt_805R (GACTACHVGGGTATCTAATCC) were used to amplify the bacterial 16S rRNA genes from the isolated DNA. The primers were complemented with 454 adapters and sample-specific barcodes (28). PCRs were prepared in triplicate and amplified under conditions that have been described elsewhere (29). PCR products were confirmed using agarose gel electrophoresis, and the triplicate products were pooled and purified using an Agencourt AMPure system (Beckham Coulter Genomics). Purified PCR products were quantified using a Qubit fluorometer (Invitrogen) and mixed into equimolar amounts. The mixed pool of PCR products was sequenced from the reverse primer direction using the Roche/454 GS Titanium technology platform (Branford, CT, USA).
Taxonomic analysis.
The sequence processing was performed mainly as described previously (28). In brief, sequences were checked for quality, and those that were <400 bp in length excluding the primer sequence, contained incorrect primer sequences, or contained any ambiguous base were discarded. Remaining sequences were then subjected to complete linkage clustering using the pyrosequencing pipeline at the Ribosomal Database Project (RDP) (30), with a conservative 5% dissimilarity to define operational taxonomic units (OTUs). The most abundant sequence from each OTU was selected as a representative sequence and was taxonomically classified by BLAST searching against a local BLAST database comprised of 600,316 bacterial 16S rRNA gene sequences longer than 1,200 bp with good Pintail scores from RDP v.10.7. The OTU inherited the taxonomy (down to the genus level) of the best-scoring RDP hit that fulfilled the criteria of ≥95% identity over an alignment of length ≥380 bp. Assignment of sequences to samples was based on the 5-bp barcode.
Statistical analyses.
Samples that contained fewer than 1,000 sequences (in total, five samples) were excluded from the analysis. The statistical analyses were performed in the multivariate statistical software Past (31). Principal-component analysis (PCA) based on abundance data from sequences classified to the genus level was performed to find clustering patterns among the subjects. Validation of the clustering pattern was confirmed with an np-MANOVA using Bray, Curtis distances and 10,000 permutations. Calculations of the individual stability of the microbiotas were performed with Bray, Curtis metrics. Differences in the relative abundances of specific bacterial groups or between similarity indexes were tested with Mann-Whitney’s test and the Kruskal-Wallis test, respectively. Simpson’s index of diversity was used to assess the microbial diversity in the samples (32).
Nucleotide sequence accession number.
The sequences have been deposited in the sequence read archive (SRA) at the NCBI under the accession number SRP045781.
SUPPLEMENTAL MATERIAL
Characteristics of the participants according to Campylobacter-positive or -negative stool culture.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council (grant 521-2011-3527), the Swedish Research Council FORMAS (grant 221-2012-1442), and the Söderbergs Foundation.
The skilled technical assistance of Annika Roos and Maria Nygård is gratefully acknowledged.
Footnotes
Citation Dicksved J, Ellström P, Engstrand L, Rautelin H. 2014. Susceptibility to Campylobacter infection is associated with the species composition of the human fecal microbiota. mBio 5(5):e01212-14. doi:10.1128/mBio.01212-14.
REFERENCES
- 1. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 2. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. 2013. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- 3. Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. 2011. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 141:1782–1791. 10.1053/j.gastro.2011.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. 2012. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143:913–916.e7. 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- 5. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. 2012. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 3:1245. 10.1038/ncomms2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knights D, Lassen KG, Xavier RJ. 2013. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 62:1505–1510. 10.1136/gutjnl-2012-303954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. 2006. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55:205–211. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54. 10.1053/gast.2002.30294 [DOI] [PubMed] [Google Scholar]
- 9. Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 10. Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK. 2008. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2:716–727. 10.1038/ismej.2008.37 [DOI] [PubMed] [Google Scholar]
- 11. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Järnerot G, Tysk C, Jansson JK, Engstrand L. 2010. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139:1844–1854.e1. 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- 12. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium. Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. 2011. Altering host resistance to infections through microbial transplantation. PLoS One 6:e26988. 10.1371/journal.pone.0026988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. 2011. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One 6:e20338. 10.1371/journal.pone.0020338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt WD. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 6:e1000711. 10.1371/journal.ppat.1000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kühl AA, Dashti JI, Zautner AE, Muñoz M, Loddenkemper C, Groß U, Göbel UB, Heimesaat MM. 2011. Novel murine infection models provide deep insights into the “menage a trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953. 10.1371/journal.pone.0020953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haag LM, Fischer A, Otto B, Plickert R, Kuhl AA, Gobel UB, Bereswill S, Heimesaat MM. 2012. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One 7:e35988. 10.1371/journal.pone.0035988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (Lond.) 69:405–411. 10.1017/S0022172400021653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyren P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8:e1002995. 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. 2014. Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63:116–214. 10.1136/gutjnl-2012-304119 [DOI] [PubMed] [Google Scholar]
- 23. Hoffman LR, Pope CE, Hayden HS, Heltshe S, Levy R, McNamara S, Jacobs MA, Rohmer L, Radey M, Ramsey BW, Brittnacher MJ, Borenstein E, Miller SI. 2014. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin. Infect. Dis. 58:396–399. 10.1093/cid/cit715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zoetendal EG, Akkermans AD, de Vos WM. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Perio MA, Niemeier RT, Levine SJ, Gruszynski K, Gibbins JD. 2013. Campylobacter infection in poultry-processing workers, Virginia, 2008-2011. Emerg. Infect. Dis. 19:286–288. 10.3201/eid1902.121147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jore S, Viljugrein H, Brun E, Heier BT, Borck B, Ethelberg S, Hakkinen M, Kuusi M, Reiersen J, Hansson I, Engvall EO, Lofdahl M, Wagenaar JA, van Pelt W, Hofshagen M. 2010. Trends in campylobacter incidence in broilers and humans in six European countries, 1997–2007. Prev. Vet. Med. 93:33–41. 10.1016/j.prevetmed.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 27. Ellström P, Hansson I, Söderström C, Olsson Engvall E, Rautelin H. A prospective follow-up study on transmission of Campylobacter from poultry to abattoir workers. Foodborne Pathog Dis., in press. 10.1089/fpd.2014.1753 [DOI] [PMC free article] [PubMed]
- 28. Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5:1571–1579. 10.1038/ismej.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ye Y, Carlsson G, Wondimu B, Fahlen A, Karlsson-Sjöberg J, Andersson M, Engstrand L, Yucel-Lindberg T, Modeer T, Putsep K. 2011. Mutations in the ELANE gene are associated with development of periodontitis in patients with severe congenital neutropenia. J. Clin. Immunol. 31:936–945. 10.1007/s10875-011-9572-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hammer O, Harper DAT, Ryan DT. 2001. Paleontological statistics software package for education and data analysis. Paleontol. Electronica 4:1–9 [Google Scholar]
- 32. Begon M, Harper JL, Townsend CR. 2006. Ecology: from individuals to ecosystems, 4th ed, p 471–472 Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the participants according to Campylobacter-positive or -negative stool culture.