ABSTRACT
Microbial population polymorphisms are commonly observed in natural environments, including long-term infected hosts. However, the underlying processes promoting and stabilizing diversity are difficult to unravel and are not well understood. Here, we use chronic infection of cystic fibrosis airways by the opportunistic pathogen Pseudomonas aeruginosa as a system for investigating bacterial diversification processes during the course of infection. We analyze clonal bacterial isolates sampled during a 32-year period and map temporal and spatial variations in population diversity to different infection sites within the infected host. We show that the ancestral infecting strain diverged into distinct sublineages, each with their own functional and genomic signatures and rates of adaptation, immediately after initial colonization. The sublineages coexisted in the host for decades, suggesting rapid evolution of stable population polymorphisms. Critically, the observed generation and maintenance of population diversity was the result of partitioning of the sublineages into physically separated niches in the CF airway. The results reveal a complex within-host population structure not previously realized and provide evidence that the heterogeneity of the highly structured and complex host environment promotes the evolution and long-term stability of pathogen population diversity during infection.
IMPORTANCE
Within-host pathogen evolution and diversification during the course of chronic infections is of importance in relation to therapeutic intervention strategies, yet our understanding of these processes is limited. Here, we investigate intraclonal population diversity in P. aeruginosa during chronic airway infections in cystic fibrosis patients. We show the evolution of a diverse population structure immediately after initial colonization, with divergence into multiple distinct sublineages that coexisted for decades and occupied distinct niches. Our results suggest that the spatial heterogeneity in CF airways plays a major role in relation to the generation and maintenance of population diversity and emphasize that a single isolate in sputum may not represent the entire pathogen population in the infected individual. A more complete understanding of the evolution of distinct clonal variants and their distribution in different niches could have positive implications for efficient therapy.
INTRODUCTION
Cystic fibrosis (CF) is a life-shortening disease caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Although CF itself is a genetic disease, the associated morbidity and mortality are the result of lifelong, chronic bacterial airway infections and the resulting inflammation. The opportunistic pathogen Pseudomonas aeruginosa is a predominant pathogen in these chronic airway infections. CF patients often acquire intermittent colonization of the airway from early childhood, which eventually proceeds into chronic infection where the same clonal P. aeruginosa lineage can persist for decades despite antibiotic therapy (1). CF airway infections are associated with genetic and phenotypic changes in P. aeruginosa that contribute to its long-term persistence in the airway. It is well established that the phenotypic characteristics of P. aeruginosa isolates from chronically infected CF patients are most often remarkably different from those displayed by the clonal isolates that initiated the infection in the same patients years before. Many phenotypic characteristics of chronically infecting strains are consistently selected in different CF patients, suggesting that adaptation occurs with conserved patterns of evolution (2). This conclusion has been further substantiated by genome sequence analyses, which have led to the discovery of recurrent patterns of mutations in independently evolving P. aeruginosa lineages and, furthermore, provided new insight into the genetic changes underlying the phenotypic changes (3–5).
In parallel with these advances in characterizing the evolutionary pathways in P. aeruginosa, other studies have documented the existence of P. aeruginosa population diversity within individual hosts. For example, a significant level of phenotypic variability within the infecting P. aeruginosa population is commonly observed, and P. aeruginosa clones sampled from the same clinical specimen often display large differences in colony morphology, quorum-sensing regulation, antibiotic resistance, and other phenotypes typically associated with chronic CF infections (6–10). Similarly, a few genome-based studies have provided evidence for genomic diversity among clonal isolates (11) and for clonal sublineage replacements during long-term infections in individual patients (3). Similar results have been obtained for other CF-associated pathogens (12). While these studies add to an emerging picture of underlying and complex population dynamics within the infected host, the specific causes of the observed intraclonal diversity are difficult to unravel and are incompletely understood. Furthermore, since other studies have documented cases with only limited evolution and diversification of P. aeruginosa during chronic CF infections (5, 13) our general understanding of the processes that determine the level of within-host P. aeruginosa population diversification is limited.
Since the CF airway is a highly structured environment with multiple connected compartments, each with different environmental conditions, a possible model is that population diversity is the result of the evolution of sublineages that can efficiently occupy the different niches available in the CF airway. Nevertheless, other processes, such as interactions between variant subpopulations (e.g., the evolution of syntropic relationships) (14) and the evolution of variants with increased mutation rates (i.e., the hypermutator phenotype) (15), have been described for P. aeruginosa CF isolates and could also contribute to population diversification.
Critically, it remains unknown how the observed intraclonal diversity is generated and whether the environmental heterogeneity and structure of the CF airway contribute to diversity generation, how stable these variant populations are as a function of time, and whether the variant populations are geographically segregated in the airway. To address these issues, we analyzed clonal bacterial isolates sampled from a chronically infected individual during a 32-year period. Sampling and storage of P. aeruginosa isolates from chronically infected Danish CF patients has been carried out since 1973, which has resulted in a strain collection that represents 40 years of infection history from a large number of infected individuals. These frozen stocks of P. aeruginosa can easily be revived and analyzed. In our initial examination of the clonal relationship among the stored isolates using low-resolution molecular typing methods, we identified two distinct and dominant P. aeruginosa lineages that were observed repeatedly because of transmission among patients (16). Here, we focus on one of these transmissible lineages, called DK1, and demonstrate rapid diversification of the original infecting strain into three distinct sublineages, each with its own functional and genomic signatures. We show that this functional and genomic diversification remained stable for decades and demonstrate that the sublineages occupy separate niches, such as the sinuses and the lower airway.
RESULTS
Genomic evolution of the DK1 lineage and within-patient diversity.
The DK1 lineage has been isolated from more than 40 CF patients since the start of the sampling program in 1973. We first sought to identify patients who had a long clonal infection history with DK1 and from whom both contemporary DK1 isolates and historical isolates from lung expectorates were available. We found one such patient (P30M0) and sequenced the genomes of 18 isolates sampled between 1980 and 2012 from this patient (Fig. 1). Furthermore, to characterize the evolution of the DK1 lineage from its entry into the CF environment until the present day, we also identified and sequenced the four oldest DK1 isolates in the strain collection. These four isolates were sampled from four different patients in 1973 (Fig. 1).
FIG 1 .
Patient origin and sampling time of genome-sequenced P. aeruginosa DK1 isolates. DK1 P. aeruginosa isolates were sampled between 1973 and 2012 from five different CF patients (P30M0, P33F0, P50F0, P13F1, and P43M1). Bacterial isolates from different patients are specified by symbols as indicated. Multiple symbols indicate that multiple isolates were sampled in the same year from the same patient.
Following sequencing, we identified high-quality single-nucleotide polymorphisms (SNPs) in the nonrepetitive parts of the genomes by mapping sequence reads for each isolate against the DK1-P33F0-1973 genome sequence, as previously described (3), and found a total of 160 SNPs that had appeared since the isolates diverged from their common DK1 ancestor before 1973.
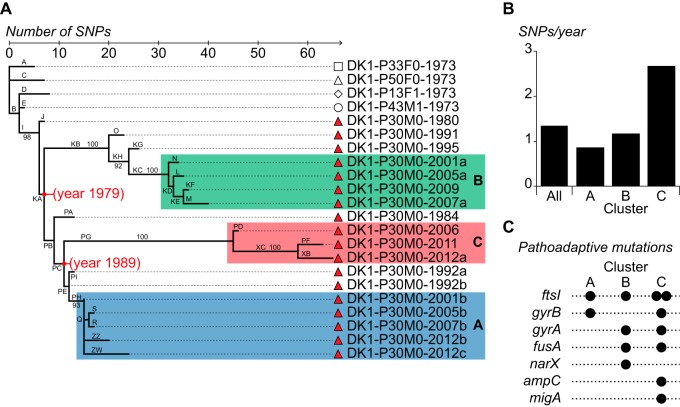
The identification of SNPs enabled us to determine the evolutionary relationship among the DK1 clones (Fig. 2A). Focusing on the isolates from P30M0, we observed that relatively few SNP changes had occurred during 32 years of colonization of the patient (see Table S1 in the supplemental material). For example, only 19 SNP changes separate isolates DK1-P30M0-1980 and DK1-P30M0-2012c, and DK1-P30M0-2012c had only accumulated 24 SNP changes in total since it diverged from the common ancestor of all DK1 isolates (Fig. 2A). Despite this moderate genomic evolution, the phylogenetic tree revealed a divergent population structure of the P30M0 isolates, and the deeply branched topology of the tree provided evidence for three distinct, coexisting sublineages (Fig. 2A, clusters A, B, and C). We performed a Bayesian analysis to estimate the divergence dates and mutation rate (17). Based on the Bayesian phylogenetic reconstruction, we estimated that the infecting population first diverged in 1979 (95% highest posterior density [HPD], 1976 to 1982) (Fig. 2A). This estimate coincides with the first DK1 culture from P30M0, which was sampled in 1980, suggesting that the DK1 population diverged soon after initial colonization. The population further diverged into clusters A and C a decade later (Fig. 2A). Importantly, bacterial isolates from the three clusters were identified in contemporary samples (from 2012 and 2013), demonstrating their coexistence for decades in the same CF patient (see below).
FIG 2 .
Evolutionary trajectory of the DK1 lineage. (A) Maximum-parsimony reconstruction of the phylogeny of DK1 clones isolated from CF patients P30M0, P33F0, P50F0, P13F1, and P43M1. The tree is based on 160 SNPs (identified from genome sequencing) that accumulated in a highly parsimonious fashion (parsimony consistency of 0.99). The bacterial isolates are named according to their clone type, the patient from whom they were isolated, and their year of isolation (e.g., DK1-P30M0-1980). Capital letters indicate branch names, and lengths of branches are proportional to the numbers of SNPs. The specific mutations that have accumulated during each specific branching are listed in Table S1 in the supplemental material. The three phylogenetic clusters in P30M0 are labeled A, B, and C. Median estimates of divergence dates (in calendar years) are given in red for the major nodes. Numbers at nodes indicate bootstrap values of ≥90%. (B) Mutation rates (SNPs per year) for each of the three clusters and for all DK1 isolates. (C) Prevalence of mutations in pathoadaptive genes in the three DK1 sublineages. The genes hit by nonsynonymous SNPs are indicated by filled circles for each of the three DK1 sublineages in P30M0.
Overall, the DK1 lineage accumulated 1.3 SNPs/year, and the relative rates of nonsynonymous and synonymous substitution (dN/dS = 0.56) implied that the long-term evolution of the DK1 lineage was dominated by negative selection (P < 4.72 × 10−10). Nonetheless, isolates from cluster C had higher evolutionary rates than the two other clusters (Fig. 2B). The mean mutation rate for cluster C was estimated to be 2.7 SNPs/year, while the rates for clusters A and B were found to be 0.9 and 1.2 SNPs/year, respectively. The elevated rate in cluster C was not a consequence of horizontally acquired SNPs, as observed in other CF isolates (3), or a hypermutator phenotype caused by mutations in DNA mismatch repair systems (18). Instead, we speculated that the elevated mutation rate was due to selection and fixation of adaptive mutations. In support of this hypothesis, we found a stronger signature of positive selection (i.e., a larger fraction of nonsynonymous mutations) among the SNPs in the C cluster (Fig. 2A, branches PD, PF, PG, XB, and XC) than in the A and B clusters (39/13 versus 37/29; Fisher’s exact test, P = 0.026).
We have previously identified a set of 65 pathoadaptive genes in P. aeruginosa that are involved in CF host adaptation (i.e., genes that undergo adaptive evolution under the pressure of natural selection during chronic CF infections) (3). Although this list of pathoadaptive genes was derived from an unrelated P. aeruginosa lineage (DK2), we reasoned that the prevalence of mutations in these pathoadaptive genes within each of the three sublineages would provide an alternative measurement of the level of positively selected SNPs. Since some of 65 genes might be lineage-specific pathoadaptive genes and since different lineages can follow different evolutionary trajectories, we expected to find mutations in only a subset of these 65 genes. Nevertheless, in agreement with our finding of enhanced positive selection in cluster C, we found that most of the mutations in pathoadaptive genes were found in this cluster, whereas cluster A contained the fewest (Fig. 2C).
Phenotype analysis supports the existence of different sublineages.
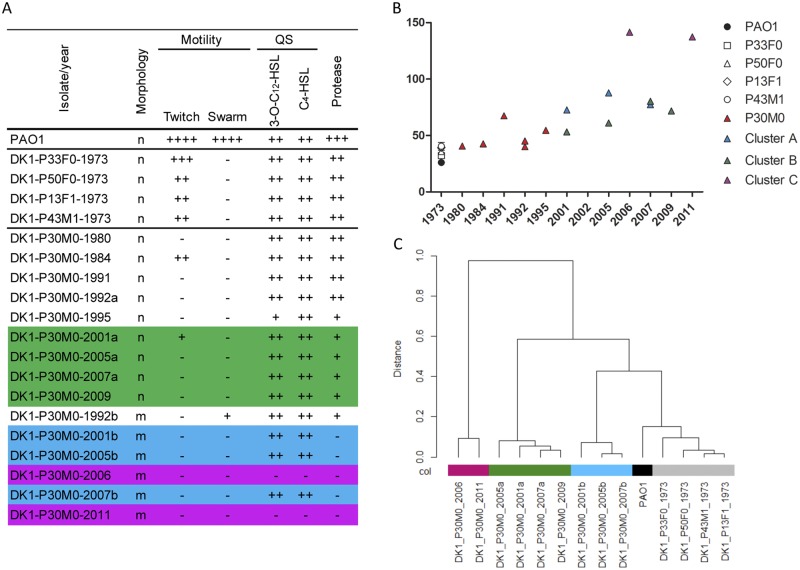
We hypothesized that the existence of three divergent sublineages, each with a different evolutionary trajectory, would result in observable functional diversification. We first analyzed the genotype/phenotype relationship within each cluster by measuring the catabolic performance of the isolates on 125 informative substrates of both carbon and nitrogen sources using Biolog Phenotype MicroArrays. Hierarchical clustering (Ward’s linkage, Euclidian distance) of the data resulted in four principal clusters, each containing isolates with related catabolic profiles (Fig. 3C). It was observed that early isolates (from 1973) clustered together with P. aeruginosa laboratory reference strain PAO1 and that the later isolates from P30M0 grouped in three different clusters identical to the clusters derived from the genome sequence data (Fig. 2A).
FIG 3 .
Phenotypic characterization of DK1 isolates. (A) For morphology, “n” indicates nonmucoid and “m” indicates mucoid cells. Motility is relative to that of strain PAO1. Quorum sensing (QS) was assayed by determining the production of acylated homoserine lactones (HSL), detected by inspection of the bioluminescence of the monitor strain (++, high level; +, low level; −, not detectable). Protease secretion on skim milk plates was determined (+++, high level; +, low level; −, not detectable). (B) Doubling times of historical isolates and P30M0 isolates measured in LB medium. (C) Dendrogram showing the hierarchical cluster analysis (Wards linkage, Euclidean distance) of global catabolic function. Colors represent each cluster; cluster A is blue; cluster B is green; and cluster C is purple.
P. aeruginosa isolates from chronically infected patients often display common phenotypic traits, including conversion into alginate-overproducing mucoid variants (19, 20), reduced growth rates (21), and loss of virulence factor production (4), quorum sensing (22), and motility (23). We measured the variations in these phenotypes for isolates from the three clusters. All isolates had reduced or abolished motility, but lineage-specific differences in other traits were observed (Fig. 3A). For example, cluster B isolates had maintained the production of quorum-sensing molecules and had reduced protease secretion, while isolates from cluster A produced quorum-sensing molecules but no proteases. Cluster C isolates had lost both quorum-sensing-molecule production and protease secretion (Fig. 3A). Furthermore, isolates from this cluster had a significantly longer in vitro doubling time than isolates from clusters A and B (Fig. 3B). All P30M0 isolates contained a nonsense mutation (C349T [a C-to-T change at position 349]) in the anti-sigma factor mucA. Inactivation of MucA leads to activation of the AlgT sigma factor, expression of the alg operon (encoding enzymes for alginate biosynthesis), and overproduction of the alginate exopolysaccharide. As expected from the mucA mutation, isolates from clusters A and C were mucoid. However, isolates from cluster B were nonmucoid, most likely due to missense mutations in algT (see Table S1 in the supplemental material).
Overall, the unique phenotypic profiles in relation to both catabolic capacities and CF-related phenotypes found for each cluster supported the SNP-based genomic evidence for coexisting subpopulations. Furthermore, we note that the isolates from cluster C showed more phenotypic traits typically associated with chronic infections than the other two clusters.
Correlation between genomic evolution and phenotype diversity.
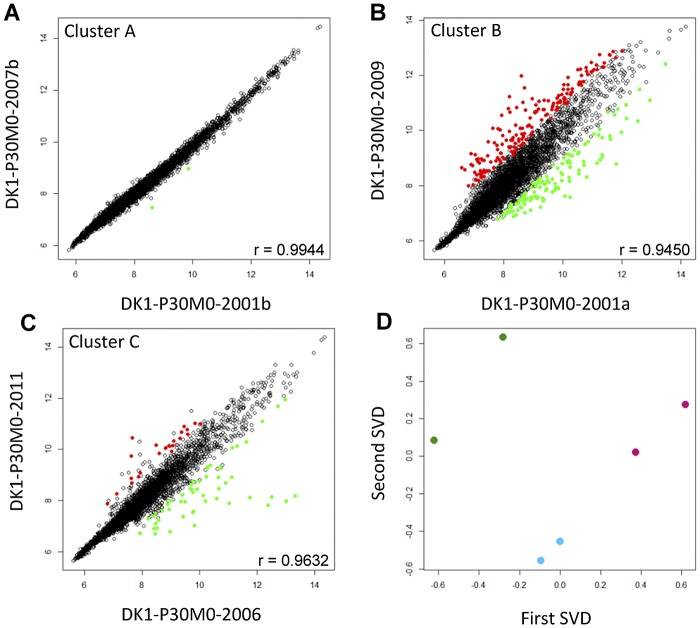
Long-term colonization of CF airways is often associated with significant changes in the global gene expression profiles of the infecting isolates (24, 25), and to examine the level of diversity and its variation as a function of time within each of the three clusters, we measured the whole-genome transcript abundance of two temporally separated isolates from each cluster. The two isolates (DK1-P30M0-2001b and DK1-P30M0-2007b) from cluster A showed very similar gene expression profiles, as indicated by a high Pearson correlation (r = 0.9944) (Fig. 4A) and their close clustering in singular value decomposition (SVD) analysis, whose results are shown in Fig. 4D. In fact, only two genes with significant expression changes (Student’s t test, P < 0.05) were identified in isolate DK1-P30M0-2001b relative to the gene expression profile of DK1-P30M0-2007b. In contrast, temporally separated isolates from both cluster C and B showed more diverse gene expression profiles (r = 0.9632 and r = 0.9450, respectively). Seventy-eight genes with significant expression changes were identified when comparing isolates from cluster C (Fig. 4C), and 298 genes with significant expression changes were identified when comparing the two nonmucoid isolates DK1-P30M0-2001a and DK1-P30M0-2009 from cluster B (Fig. 4B). The differences in the extent of phenotypic diversity developing over time within the three clusters are consistent with a hypothesis that isolates from clusters B and C evolved in different environments than cluster A, with stronger selective pressures mediating higher levels of both genomic evolution and phenotypic diversity.
FIG 4 .
Transcriptome analysis. (A) Comparison of gene expression of the two mucoid isolates DK1-P30M0-2001b and DK1-P30M0-2007b from cluster A. (B) Comparison of gene expression of the two nonmucoid isolates DK1-P30M0-2001a and DK1-P30M0-2009a from cluster B. (C) Comparison of gene expression of the two mucoid isolates DK1-P30M0-2006 and DK1-P30M0-2011 from cluster C. Red circles represent genes with significantly increased expression (fold change of ≥2; P ≤ 0.05), and green circles indicate genes with decreased expression. The r (Pearson correlation) values indicate the strength and direction of the linear relationship between the expression levels of the isolates. (D) Results of single value decomposition (SVD) analysis of the gene expression relationships among isolates from clusters A, B, and C. Each dot represents the mean of duplicates. Blue dots represent isolates from cluster A, green dots represent isolates from cluster B, and isolates from cluster C are shown as purple dots.
Limited parallel evolution among coexisting sublineages.
The long-term coexistence of distinct sublineages enabled us to investigate the extent of parallelism among three evolutionary pathways within the same individual. We examined the genome data for mutations that accumulated independently in the different sublineages (see Table S1 in the supplemental material). As anticipated, such parallel evolution was observed in genes known to be involved in antibiotic resistance and included mutations in DNA gyrases gyrA (clusters B and C), gyrB (clusters A and C), and fusA1 (clusters B and C), encoding elongation factor G (Table S1) (26, 27). Unexpectedly, no other examples of parallel genomic evolution were found, which suggests that the three sublineages exist in separate niches, each with different selective conditions by which evolution is directed. Importantly, this conclusion is in agreement with the observed lineage-specific distribution of mutations in pathoadaptive genes (Fig. 2C).
Mapping coexisting subpopulations to different spatial sites.
Patient P30M0 underwent paranasal sinus surgery in 2012, during which expectorated sputum, bronchoalveolar lavage (BAL) fluid, and sinus material (from both left and right sinuses) were sampled and stored. This provided a unique opportunity to determine the distribution of the three sublineages at different sites in the infected CF airway (i.e., the sinus region versus the lower airway). Sublineage-specific primers were designed on the basis of the genome sequence data, and a cross-sectional analysis of multiple bacterial isolates from each sample was done by PCR screening (see Materials and Methods). We randomly selected sixty-four isolates from each sample (BAL fluid, sputum sample, and left sinus surgery material), with the exception of the right sinus sample, from which we only recovered four isolates. Using PCR screening, we found that all analyzed isolates from the sinus samples belonged to cluster A (64 plus 4 isolates analyzed in total), while all isolates from the BAL fluid and sputum samples belonged to cluster C (2 sets of 64 isolates analyzed in total) (Fig. 5A). These analyses strongly suggest that clusters A and C are spatially segregated in the sinus region and lower airway, respectively. Genome sequencing of one isolate from the BAL fluid sample and two isolates sampled from each side of the sinuses verified that they belonged to cluster C (BAL fluid isolate DK1-P30M0-2012a) and cluster A (sinus isolates DK1-P30M0-2012b and DK1-P30M0-2012c) (Fig. 2A). We also took advantage of the observation that isolates from each of the three clusters exhibited distinct phenotypes. For example, isolates from clusters A and B formed colonies faster than isolates from cluster C (due to differences in growth rates), and isolates from cluster B were nonmucoid while isolates from clusters A and C were mucoid (Fig. 3). Repeated dilutions and plating of the sinus, BAL fluid, and sputum samples combined with visual screening for cluster-defining phenotypes supported the spatial distribution of clusters A and C. Bacterial isolates from cluster B were not found in the samples related to the upper airway (sinus surgery samples) but could be found at an overall frequency of 0.5% in stored sputum samples obtained in 2010, 2011, and 2012 (Fig. 5B). The presence of isolates from cluster B in sputum combined with their absence in sinus samples suggests that isolates of cluster B colonize the lower airway.
FIG 5 .
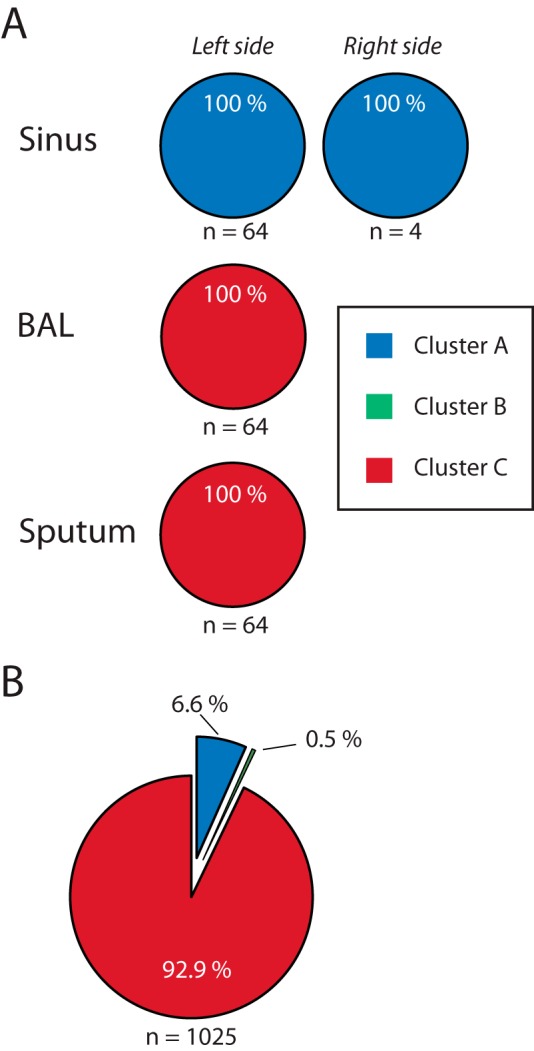
Distribution of the three sublineages at different sites in patient P30M0. (A) Analysis of P. aeruginosa isolates sampled from patient P30M0 during paranasal sinus surgery in 2012. Each randomly chosen isolate was inspected using a cluster-specific PCR method to determine if it was part of either cluster A (blue), B (green), or C (red). All isolates belonged to these three clusters. With the exception of the sample from the right sinus, 64 random isolates from each site were tested. (B) A total of 1,025 isolates from sputum samples collected in 2010, 2011, and 2012 were picked randomly and association with one of the three clusters determined.
Together, these findings strongly suggest that the three coexisting subpopulations evolved in spatially separated niches of the CF airway, with isolates from cluster A colonizing the sinuses of this patient and isolates from clusters B and C colonizing and evolving in the lower airway.
DISCUSSION
Although P. aeruginosa’s population heterogeneity in relation to chronic CF infections has frequently been documented by microbial culturing, our understanding of the temporal and spatial dynamics of this diversity, as well as the underlying diversity-generating processes, is limited. In this study, we combined current genome sequencing technologies, phenotypic profiling, and unique sampling materials which included clonal bacterial isolates sampled during a 32-year period both from sputum and from material obtained from paranasal sinus surgery in a single chronically infected CF patient to gain new insight into bacterial diversification processes in chronic CF infections.
The phylogenomic analysis demonstrated that the initial infecting DK1 strain diversified into three major subpopulations (clusters A, B, and C), each with its individual genetic signature of mutations and limited parallel genomic evolution. Our phenotype profiling of strains from the different clusters supported this result and showed that the accumulation of mutations shaped their phenotypes in such a way that the three clusters were clearly distinguishable with respect to mucoidy, doubling time, metabolic performance, and global gene expression profiles. Although diversity has often been observed in P. aeruginosa populations from the same CF respiratory specimen, we show here that such diversity can have long-term stability, as demonstrated by the decades-long coexistence of the three different sublineages.
Inspection of multiple bacterial isolates from sputum samples and from sinus material after surgery did not reveal the presence of additional major sublineages in the infected patient (Fig. 5). This suggests that the strain collection analyzed here (which consists of multiple single isolates sampled from the patient since 1980) did indeed capture the dominant sublineages in the infected individual. Importantly, our focus on the evolution of the three genetically and phenotypically distinct sublineages does not rule out the existence of an additional layer of population heterogeneity in the patient. For example, it is more than likely that microdiversity exists within each of the three sublineages (i.e., that further cluster-specific diversification has taken place). Indeed, the 14 SNPs that separate cluster A isolates DK1-P30M0-2012b and DK1-P30M0-2012c, sampled from the left and right sinuses, respectively, may illustrate this point. The long-term dynamics of such potential cluster-specific heterogeneity is generally unknown and cannot be investigated with our present single-isolate strain collection and our single-time-point cross-sectional analysis. Future genomic and phenotypic analysis of entire respiratory specimens sampled throughout infection is required to understand the diversification processes at both the lineage and subpopulation level.
From an ecological perspective, our findings indicate population diversification as a result of adaptive radiation processes and niche partitioning. Evolutionary theory and experimental evolution studies—in which microbial populations evolve under defined, artificial conditions in the laboratory—have clearly established a connection between heterogeneous environments (i.e., the introduction of distinct and vacant niches, each with different selective conditions) and the evolution and maintenance of population diversity (28–30). Indeed, this explanation has been used to rationalize the observed population diversity in relation to chronic CF infections, but since most studies on P. aeruginosa population diversity rely on inspection of isolates from sputum or BAL fluid samples which lack spatial resolution, conclusions about niche partitioning have been difficult to reach. In this study, we obtained parallel samples from different compartments in the infected individual, including material sampled from the paranasal sinuses, which enabled us to demonstrate a clear spatial distribution of the sublineages in different niches. We found cluster A to inhabit the paranasal sinuses, whereas we identified clusters B and C in samples from the lung environment (i.e., in sputum samples and in BAL fluid samples). Because of the striking absence of parallel genomic evolution between clusters B and C, which indicates that they populate niches with different selective pressures, we suggest that clusters B and C occupy distinct niches in the lungs of the patient. Furthermore, a distribution of the three sublineages in different niches would result in lineage-specific sets of genes being under positive selection, which was indeed observed (Fig. 2C). However, we note that our findings in relation to clusters B and C cannot exclude the possibility of their coexistence in the same physical lower-airway niche. Although the two sublineages have evolved distinct phenotypes, have followed distinct evolutionary trajectories, and have a distinct set of mutations of pathoadaptive genes, it is not possible to map their spatial distribution within the lower airway of the infected individual, and the precise location of the two clusters therefore remains unknown. Furthermore, laboratory evolution studies with Escherichia coli have shown that coexisting sublineages may evolve in simple, homogenous chemostat cultures and that such population polymorphisms can be maintained through evolution of, e.g., cross-feeding interactions (31, 32). It is therefore a possibility that related niche-partitioning processes (i.e., diversification without geographical isolation) have played a role in the evolution of clusters B and C. However, recent documentations of spatial distributions of microorganisms in CF lungs support our hypothesis of spatial segregation of clusters B and C. For example, spatial heterogeneity of microbial communities in infected CF lungs has been demonstrated recently by culture-independent analysis of CF lung tissue samples (33), and furthermore, microscopy analysis of explanted CF lungs has shown that mucoid and nonmucoid variants of P. aeruginosa are located in different niches, such as the sputum in the conductive zone of the bronchi and the respiratory zones of the alveoli and respiratory bronchioles in the infected CF lung, respectively (34). Importantly, clusters B and C are different with respect to the mucoid phenotype (isolates from cluster C are mucoid as a consequence of mutations in mucA, and isolates from cluster B are nonmucoid as a consequence of mutations in both mucA and algT), and it is possible that a related distribution is present in the patient studied here.
The observation that the faster growing cluster B isolates consistently constituted only a small fraction of the population sampled recently from the lung environment and were even absent from the population in some cases could be related to difficulties in sampling the particular lung niche occupied by cluster B. This is in accordance with observations from CF autopsies and explanted lungs, which showed that cells of the nonmucoid phenotype were actually phagocytosed by polymorphonuclear leukocytes, in contrast to cells of the biofilm-growing mucoid phenotype (34). Alternatively, or complementarily, it is possible that isolates from cluster C are better adapted than isolates from cluster B to the stressful and selective conditions in the lung environment.
Our phylogenomic analysis further showed that cluster B diverged from the population soon after or coinciding with the initial colonization and that the DK1 population further diverged 10 years later. By combining this phylogeny with the spatial location of each of the three clusters, it is now possible to reconstruct the evolutionary history of DK1 in the patient studied here (Fig. 6). We propose that the patient was first colonized in the paranasal sinuses with isolates belonging to cluster A. Following the initial colonization, isolates from the infected sinuses migrated to the lungs in two distinct events (separated by a decade) and evolved into highly successful colonizers of the lungs (i.e., clusters B and C). Based on observations of extensive overlap in colony morphotypes between clonal isolates sampled from the paranasal sinuses and from BAL fluid samples, we have previously demonstrated migration from the sinuses to the lung environment (8). The genomic data presented here provide further evidence for this hypothesis and highlight the dynamic nature of the infecting population, as well as the significance of the paranasal sinuses as functional pathogen reservoirs.
FIG 6 .
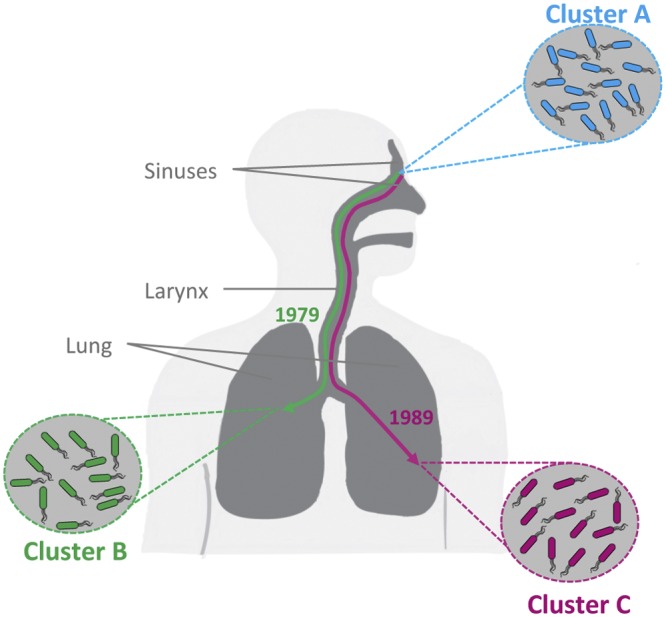
Model of the evolutionary history of DK1 in patient P30M0. Reconstruction of the evolutionary history of DK1 in patient P30M0 on the basis of the phylogenetic relationship among the studied isolates and their distribution in different niches in the infected airway. The patient was first colonized in the paranasal sinuses with isolates belonging to cluster A. Soon after initial colonization, the population diverged and formed a genetically and phenotypically distinct sublineage (cluster B) in the lower airway. A decade later, another sublineage (cluster C) diverged from the population in the paranasal sinuses and migrated to the lower airway. The precise locations of clusters B and C in the lower airway (e.g., left versus right lung or the conductive zone versus the respiratory zone) are not known and for illustrative purposes are shown as being segregated into the left and right lung. However, the differences between clusters B and C in relation to phenotypes such as mucoidity and in the number of pathoadaptive mutations suggest that they inhabit distinct niches with different selective pressures in the lower airway.
Interestingly, the amounts of evolutionary change were different in the three clusters. Isolates from clusters B and C accumulated more mutations in pathoadaptive genes and demonstrated more phenotypic alterations as a function of time than the sinus-associated population in cluster A. We speculate that the observed differences are related to mutation pathways activated by stress responses (i.e., induced when cells are maladapted to their environment) (35, 36) or differences in in vivo growth rates or are due to oxygen radical damage as a result of the inflammatory response, which differs in different niches of the respiratory tract. Although the sinus and lung environments share some physiological properties, such as thick mucus that can function as sites for bacterial growth, there are also significant differences that may explain the different evolutionary trajectories associated with each compartment. For example, the inflammatory response is reduced in the paranasal sinuses, and polymorphonuclear leukocytes, which produce oxygen radicals (37), dominate in the lungs and sputum, whereas they are rare in the paranasal sinuses, where the inflammation is less pronounced due to high production of immunoglobulin A (38). Furthermore, antibiotic penetration and, hence, the achievement of therapeutic levels are hypothesized to be less efficient in the sinuses than in the lungs (38, 39).
We have previously shown a much more pronounced evolutionary response in sinus-associated P. aeruginosa populations in intermittently colonized patients than what we show here for cluster A (8). In addition, studies have shown extensive overlaps in evolved phenotypes like colony morphology and in global gene expression profiles between isolates from sinus and lung samples from the same chronically infected patients (8, 39), which was clearly not observed here. These observational differences suggest that generalizing of conclusions concerning the evolution of P. aeruginosa in the paranasal sinuses should be considered with caution and that our understanding of the factors that influence the evolutionary trajectories of sinus-associated P. aeruginosa populations is incomplete.
Our study provides evidence that the environmental heterogeneity of the CF airway promotes diversification and long-term coexistence of P. aeruginosa sublineages during chronic CF airway infections. It is possible that similar adaptive radiation processes underlie the population diversity observed in other CF-associated pathogens (12). Critically, some P. aeruginosa lineages from chronically infected CF patients exhibit only limited diversity (13), and studies of additional patients are therefore required to understand whether generalities in the microbial diversification processes exist. More broadly, our results show that P. aeruginosa isolates from chronic CF patients may be representatives of an underlying complex and dynamic population structure comprised of distinct clonal variants that are distributed in different niches. Since treatment strategies targeting one population may not be effective against other subpopulations, an incomplete description of the population diversity within the infected individual may lead to inaccurate descriptions of therapy-relevant phenotypes, such as antibiotic resistance profiles in the pathogen population. A more complete understanding of population diversity and its spatial and temporal distribution could therefore have positive implications for efficient therapy and disease outcome.
MATERIALS AND METHODS
Ethics statement.
No patient samples were taken specifically for this study. Isolation and storage of bacterial isolates were part of the established routine at the Department of Clinical Microbiology, Rigshospitalet, Copenhagen, Denmark. The bacterial isolates used in the study included isolates from the bacterial collection available at the Department of Clinical Microbiology, Rigshospitalet, as well as bacteria isolated during functional endoscopic sinus surgery (FESS) on patient P30M0 in 2012. The use of bacterial isolates was approved by the local ethics committee, Region Hovedstaden. All samples were anonymized.
Bacterial strains.
The investigated strains of the P. aeruginosa DK1 clone type consisted of 18 longitudinal isolates from patient P30M0 and four historical isolates from 1973 from patients P33F0, P50F0, P13F1, and P43M1. Patient P30M0 was chosen based on the long history of clonal infection with DK1 and the availability of stored isolates and new lung expectorates. Additional clinical strains from P30M0 were isolated from either sinus or bronchoalveolar lavage (BAL) fluid samples from 2012. The samples from the paranasal sinuses were taken during FESS, as previously described (39). Isolation and identification of P. aeruginosa was done as previously described (40). The isolates were grown in Luria-Bertani (LB) medium and stored at −80°C with 10% glycerol. The P. aeruginosa isolates investigated in this study were confirmed to belong to the DK1 lineage by two independent genotyping methods, pulsed-field gel electrophoresis and SNP genotyping using AT biochips (Clondiag Chip Technologies, Germany), as described previously (41).
Genome sequencing.
Genomic DNA was prepared from P. aeruginosa isolates using the Wizard genomic DNA purification kit (Promega). Genomes were sequenced to a coverage depth of 90-fold (range, 17- to 207-fold) on either an Illumina GAIIx platform generating 75-bp single-end reads (6 isolates) or a HiSeq2000 platform generating either 75-nt single-end reads (7 isolates) or 100-nt paired-end reads (9 isolates). The genome of DK1-P33F0 was de novo assembled into 6,156,016 bp (252 contigs, N50 = 49,984 bp) with the assistance of the genome sequences of P. aeruginosa PAO1, PA14, and LESB58 by using the Columbus module of Velvet, version 1.0.16 (42). The genome assembly of DK1-P33F0 was then used as a reference for the alignment of sequence reads from each of the isolates to identify high-quality SNPs in the nonrepetitive parts of the genome.
Mutation detection was performed as previously described (3). Maximum-parsimony analysis (43) was used to reconstruct the phylogeny of DK1 clones, and the bootstrap support values for branches were calculated using PAUP 4.0 (44). Calculation of the dN/dS ratio was based on the assumption that a random mutation in the total number of possible SNP mutations would cause a synonymous change 25% of the time (13, 45). Bayesian analysis of evolutionary rates was performed using BEAST, version 1.7.2 (17), with a lognormal relaxed molecular clock model and a general time-reversible substitution model. Mutation rates were calculated from a chain length of 50 million steps, sampled every 5,000 steps. The first 5 million steps were discarded as a burn-in. A maximum clade credibility tree was generated using the TreeAnnotator program from the BEAST package, and the effective sample sizes (ESS) of all parameters were >1,000 as calculated by Tracer, version 1.5 (available from http://beast.bio.ed.ac.uk/Tracer), which was also used to calculate the 95% HPD confidence intervals of the divergence times (i.e., an interval within which the modeled parameter resides with 95% probability).
Biolog Phenotype MicroArray.
Phenotype MicroArray (Biolog, Hayward, CA) experiments were performed in duplicate according to the manufacturer’s instructions (46, 47). Bacterial strains were streaked on LB agar plates and incubated at 37°C until colonies appeared on plates (23 to 48 h). Cells were swabbed from the plates and suspended in IF-0 GN Base (inoculation fluid; Biolog) at a density corresponding to 42% transmittance in the Biolog turbidimeter. The cell suspensions were diluted 1:6 in IF-0 minimal medium containing Biolog redox dye mixture D (tetrazolium), and 100-µl aliquots were added to carbon source plates (PM1 microplate). Nitrogen plates (PM3 microplate) with ferric citrate and glucose supplemented to the inoculation mixture were prepared. In total, 190 substrates were available on the two plates. Of these, 125 were found to be informative by showing consistent growth between replicates. OmniLog OL_FM/Kin 1.20.02 software (Biolog) was used to export the OmniLog data, and the average area beneath each kinetic curve was used for analysis. Total catabolic function was calculated as previously described (48). Further data and statistical analyses were performed using R version 2.10.1 (2009; R Development Core Team).
Affymetrix GeneChip.
Strains were grown aerobically at 37°C in LB medium, starting at an optical density at 600 nm (OD600) of 0.01, and harvested at an OD600 of 0.5. Immediately after harvesting, the cells were mixed either with RNAprotect bacterial reagent (Qiagen) if they were nonmucoid cells or with STOP solution (95% ethanol, 5% phenol) if they were mucoid, to avoid clumping. After 10 min of centrifugation at 7,000 × g at 4°C, the supernatant was discarded and the cell pellet was stored at −80°C. RNA extraction, cDNA preparation and labeling, and hybridization were performed as previously described (13). The raw CEL files were obtained by the Affymetrix GeneChip operating system 1.4, and the analysis was performed by using BioConductor tools in the R environment (49). Strains were tested in triplicates.
Motility assays.
The motility of all isolates was tested on ABT agar (50) supplemented with 0.5% Casamino acids and 0.5% glucose as previously described (51). Briefly, ABT plates were inoculated with single colonies on the top, in the middle or on the bottom of the plates to measure swarm, swim, or twitch, respectively. Swim and swarm were measured on 0.3% (wt/vol) and 0.6% (wt/vol) agar, respectively, with incubation for 24 h at 30°C. Twitching motility was determined on 1.5% (wt/vol) agar, with incubation for 48 h at 37°C. The motility zones in triplicates were calculated relative to the motility of the reference strain PAO1.
Detection of quorum-sensing signals.
For detection of quorum-sensing signals, P. aeruginosa strains were cross-streaked against two E. coli monitor strains, MH155 and MH205, as previously described (50). After 48 h of incubation at 37°C, acylated homoserine lactone (AHL) production was detected by inspecting the fluorescence of the monitor strains with a Zeiss Axioplan 2 microscope with a 2.5/0.075× plan Neofluar objective.
Skim milk protease assay.
The production of protease was determined by applying one colony of the isolate grown on LB agar to skim milk plates (LB agar with 10% skim milk). The plates were incubated for 24 h at 37°C, and the clearing zones were measured.
Growth rate measurement and estimation of in situ bacterial generations.
LB broth was inoculated with the isolate and incubated at 37°C overnight. A cell culture volume was transferred into 50 ml of LB broth in a 250-ml shaking flask with baffles, generating an OD600 of 0.01. The flask was incubated at 37°C with shaking at 240 rpm in a New Brunswick Scientific incubator. Growth rates were measured by monitoring the OD600 during growth. The measurements were done in at least duplicates.
Growth rates were calculated from the exponential part of the growth curves and expressed as doubling times in minutes [ln(2)/minutes].
PCR screening.
A multiplex PCR strategy was constructed to determine the different sublineages of P. aeruginosa from patient P30M0. Sublineage-specific primers were designed, including an internal positive control for all lineages (see Table S2 in the supplemental material). PCR amplifications were performed with the following optimized parameters: predenaturation for 5 min at 95°C; 25 cycles of 95°C for 30 s, 66°C for 30 s, and 72°C for 25 s; postextension for 3 min at 72°C; and soaking at 4°C. PCR products were resolved in a 2% agarose gel in 1× Tris-borate-EDTA buffer at 100 V and visualized with ethidium bromide. In total, 64 isolates each (3 × 64) from the sinus, BAL fluid, and sputum samples from 2012 were tested. Furthermore, isolates from sputum samples from 2010, 2011, and 2012 were tested.
SUPPLEMENTAL MATERIAL
SNP changes observed in the P30M0 lineage.
Primers used for multiplex PCR.
ACKNOWLEDGMENTS
We thank S. Damkiær for assistance with Affymetrix Genechips and Biolog and N. Jochumsen for support with the multiplex PCR. We also thank L. Yang (Department of Biological Engineering, Massachusetts Institute of Technology) and the Infection Microbiology Group at DTU for helpful discussions. U. Johansen is thanked for technical assistance.
This work was supported by the Danish Council for Independent Research and the undbeck Foundation. H.K.J. was supported by a clinical research stipend from the Novo Nordisk Foundation. L.J. was supported by a Young Investigator grant from the Villum Foundation.
Footnotes
Citation Markussen T, Marvig RL, Gómez-Lozano M, Aanæs K, Burleigh AE, Høiby N, Johansen HK, Molin S, Jelsbak L. 2014. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 5(5):e01592-14. doi:10.1128/mBio.01592-14.
REFERENCES
- 1. Høiby N. 2011. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 9:32. 10.1186/1741-7015-9-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nguyen D, Singh PK. 2006. Evolving stealth: genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc. Natl. Acad. Sci. U. S. A. 103:8305–8306. 10.1073/pnas.0602526103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marvig RL, Johansen HK, Molin S, Jelsbak L. 2013. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLOS Genet. 9:e1003741. 10.1371/journal.pgen.1003741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tümmler B. 2011. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ. Microbiol. 13:1690–1704. 10.1111/j.1462-2920.2011.02483.x [DOI] [PubMed] [Google Scholar]
- 6. Wilder CN, Allada G, Schuster M. 2009. Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum-sensing populations from cystic fibrosis lung infections. Infect. Immun. 77:5631–5639. 10.1128/IAI.00755-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Høiby N, Yang L, Molin S. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ. Microbiol. 12:1643–1658 [DOI] [PubMed] [Google Scholar]
- 8. Hansen SK, Rau MH, Johansen HK, Ciofu O, Jelsbak L, Yang L, Folkesson A, Jarmer HØ, Aanæs K, von Buchwald C, Høiby N, Molin S. 2012. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J. 6:31–45. 10.1038/ismej.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C. 2011. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183:1674–1679. 10.1164/rccm.201009-1430OC [DOI] [PubMed] [Google Scholar]
- 10. Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, Rabin HR, Surette MG. 2013. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8:e60225. 10.1371/journal.pone.0060225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung JC, Becq J, Fraser L, Schulz-Trieglaff O, Bond NJ, Foweraker J, Bruce KD, Smith GP, Welch M. 2012. Genomic variation among contemporary Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis patients. J. Bacteriol. 194:4857–4866. 10.1128/JB.01050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. 2014. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat. Genet. 46:82–87. 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. 108:7481–7486. 10.1073/pnas.1018249108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin X, Zerr DM, McNutt MA, Berry JE, Burns JL, Kapur RP. 2012. Pseudomonas aeruginosa syntrophy in chronically colonized airways of cystic fibrosis patients. Antimicrob. Agents Chemother. 56:5971–5981. 10.1128/AAC.01371-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. 2010. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 156(Pt 4):1108–1119. 10.1099/mic.0.033993-0 [DOI] [PubMed] [Google Scholar]
- 16. Jelsbak L, Johansen HK, Frost AL, Thøgersen R, Thomsen LE, Ciofu O, Yang L, Haagensen JA, Høiby N, Molin S. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 75:2214–2224. 10.1128/IAI.01282-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1254. 10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- 19. Pedersen SS, Høiby N, Espersen F, Koch C. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6–13. 10.1136/thx.47.1.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 90:8377–8381. 10.1073/pnas.90.18.8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang L, Haagensen JA, Jelsbak L, Johansen HK, Sternberg C, Høiby N, Molin S. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 190:2767–2776. 10.1128/JB.01581-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 8:66–70. 10.1016/j.jcf.2008.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mahenthiralingam E, Campbell ME, Speert DP. 1994. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62:596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thøgersen JC, Mørup M, Damkiaer S, Molin S, Jelsbak L. 2013. Archetypal analysis of diverse Pseudomonas aeruginosa transcriptomes reveals adaptation in cystic fibrosis airways. BMC Bioinformatics 14:279. 10.1186/1471-2105-14-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huse HK, Kwon T, Zlosnik JE, Speert DP, Marcotte EM, Whiteley M. 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. mBio 1(4):e00199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bielecki P, Lukat P, Hüsecken K, Dötsch A, Steinmetz H, Hartmann RW, Müller R, Häussler S. 2012. Mutation in elongation factor G confers resistance to the antibiotic argyrin in the opportunistic pathogen Pseudomonas aeruginosa. Chembiochem 13:2339–2345 [DOI] [PubMed] [Google Scholar]
- 27. Pasca MR, Dalla Valle C, De Jesus Lopes Ribeiro AL, Buroni S, Papaleo MC, Bazzini S, Udine C, Incandela ML, Daffara S, Fani R, Riccardi G, Marone P. 2012. Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb. Drug Resist. 18:23–32. 10.1089/mdr.2011.0019 [DOI] [PubMed] [Google Scholar]
- 28. Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69–72. 10.1038/27900 [DOI] [PubMed] [Google Scholar]
- 29. Hansen SK, Rainey PB, Haagensen JA, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445:533–536. 10.1038/nature05514 [DOI] [PubMed] [Google Scholar]
- 30. Boles BR, Thoendel M, Singh PK. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 101:16630–16635. 10.1073/pnas.0407460101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinnersley MA, Holben WE, Rosenzweig F. 2009. E unibus plurum: genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 5:e1000713. 10.1371/journal.pgen.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herron MD, Doebeli M. 2013. Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol. 11:e1001490. 10.1371/journal.pbio.1001490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. 2012. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 6:471–474. 10.1038/ismej.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Høiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44:547–558. 10.1002/ppul.21011 [DOI] [PubMed] [Google Scholar]
- 35. Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 105:12503–12508. 10.1073/pnas.0801499105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404–1409. 10.1126/science.1082240 [DOI] [PubMed] [Google Scholar]
- 37. Kolpen M, Hansen CR, Bjarnsholt T, Moser C, Christensen LD, van Gennip M, Ciofu O, Mandsberg L, Kharazmi A, Döring G, Givskov M, Høiby N, Jensen PØ. 2010. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65:57–62. 10.1136/thx.2009.114512 [DOI] [PubMed] [Google Scholar]
- 38. Johansen HK, Aanaes K, Pressler T, Nielsen KG, Fisker J, Skov M, Høiby N, von Buchwald C. 2012. Colonisation and infection of the paranasal sinuses in cystic fibrosis patients is accompanied by a reduced PMN response. J. Cyst. Fibros. 11:525–531. 10.1016/j.jcf.2012.04.011 [DOI] [PubMed] [Google Scholar]
- 39. Ciofu O, Johansen HK, Aanaes K, Wassermann T, Alhede M, von Buchwald C, Høiby N. 2013. P. aeruginosa in the paranasal sinuses and transplanted lungs have similar adaptive mutations as isolates from chronically infected CF lungs. J. Cyst. Fibros. 12:729–736. 10.1016/j.jcf.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 40. Høiby N, Frederiksen B. 2000. Microbiology of cystic fibrosis, p 83–107 In Hodson ME, Geddes DM. (ed), Cystic fibrosis, 2nd ed. Arnold, London, United Kingdom [Google Scholar]
- 41. Wiehlmann L, Wagner G, Cramer N, Siebert B, Gudowius P, Morales G, Köhler T, van Delden C, Weinel C, Slickers P, Tümmler B. 2007. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8101–8106. 10.1073/pnas.0609213104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fitch WM. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20:11 [Google Scholar]
- 44. Swofford DL. 2003. PAUP* phylogenetic analysis using parsimony (* and other methods), version 4.0. Sinauer Associates, Sunderland, MA.
- 45. Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496–503. 10.1016/S0169-5347(00)01994-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bochner BR. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309–314. 10.1038/nrg1046 [DOI] [PubMed] [Google Scholar]
- 47. Bochner BR. 2009. Global phenotypic characterization of bacteria. FEMS Microbiol. Rev. 33:191–205. 10.1111/j.1574-6976.2008.00149.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cooper VS, Lenski RE. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407:736–739. 10.1038/35037572 [DOI] [PubMed] [Google Scholar]
- 49. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clark JD, Maaloe O. 1967. DNA replication and the cell cycle in Escherichia coli cells. J. Mol. Biol. 23:99–112 [Google Scholar]
- 51. Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Høiby N, Kjelleberg S, Givskov M. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNP changes observed in the P30M0 lineage.
Primers used for multiplex PCR.