Abstract
The objectives of this study were to detect the presence of Vibrio cholerae in tropical estuaries (Northeastern Brazil) and to search for virulence factors in the environmental isolates. Water and sediment samples were inoculated onto a vibrio-selective medium (TCBS), and colonies with morphological resemblance to V. cholerae were isolated. The cultures were identified phenotypically using a dichotomous key based on biochemical characteristics. The total DNA extracted was amplified by PCR to detect ompW and by multiplex PCR to detect the virulence genes ctx, tcp, zot and rfbO1. The results of the phenotypic and genotypic identification were compared. Nine strains of V. cholerae were identified phenotypically, five of which were confirmed by detection of the species-specific gene ompW. The dichotomous key was efficient at differentiating environmental strains of V. cholerae. Strains of V. cholerae were found in all four estuaries, but none possessed virulence genes.
Keywords: Cholera, Estuaries, Pathogenicity, Genes
Abstract
O objetivo deste estudo foi detectar a presença potencial virulência de Vibrio cholerae isolado de estuários do Nordeste do Brasil. Amostras de água e sedimento foram coletadas e inoculadas sobre meio seletivo para víbrios (TCBS) e colônias com características morfológicas de V. cholerae foram isoladas. A identificação fenotípica seguiu chave dicotômica baseada em caraterísticas bioquímicas. Foram empregadas as técnicas de amplificação da polimerase em cadeia (PCR) utilizando o gene ompW e a de multiplex PCR para detecção de genes de virulência (ctx, tcp, zot e rfbO1). Os resultados da identificação das diferentes abordagens foram comparados. Nove cepas de V. cholerae foram identificadas fenotipicamente e cinco confirmadas através da detecção do gene ompW. A chave dicotômica utilizada foi eficiente para a confirmação da espécie. Os quatro estuários analisados apresentaram estirpes de V. cholerae, e nenhuma das cepas isoladas apresentaram genes de virulência.
INTRODUCTION
The genus Vibrio (family: Vibrionaceae) comprises 104 species6, some of which are pathogenic to humans. One of the best known of these is the Gram-negative species Vibrio cholerae, which is capable of inducing cholera, an acute intestinal infection, when ingested through contaminated water and food16.
In the early 1960s, Colwell demonstrated the importance of aquatic environments to the ecology and epidemiology of V. cholerae 35, contrary to the earlier notion that the organism could only be transmitted by a human source and that water served merely to carry it from host to host.
V. cholerae is widely distributed in estuarine, marine and freshwater environments and has been associated with outbreaks of endemic, epidemic and pandemic proportions11,12,23.
Over 200 serogroups of V. cholerae are known, but only O1 and O139 have been implicated in epidemic cholera12,36, although local sporadic outbreaks of diarrhea associated with non-O1 and non-O139 strains have been documented. Nevertheless, despite the extensive research done over the past years on the ecology, pathogenicity and epidemiological behavior of the species, many questions remain unanswered9,32.
In Brazil, cholera reemerged in 1991, after a century of absence. Between 1992 and 2005, the highest incidences of V. cholerae in samples of water from aquatic ecosystems and foods were registered in the northeastern region4, especially in the state of Pernambuco, making that state one of the most strongly impacted by cholera8. In the state of Ceará, outbreaks were reported between December 1991 and September 199315.
The pathogenesis of cholera is complex and involves the synergy of a number of genes, such as ctx, tcp, zot and rfbO1 10,12. The presence of these genes may be used as an indicator of virulence, although the cholera toxin is considered the most important epidemic marker.
The purpose of the present study was to isolate and test environmental strains of Vibrio cholerae from estuaries in northeastern Brazil, due to the presence of virulence markers.
MATERIAL AND METHODS
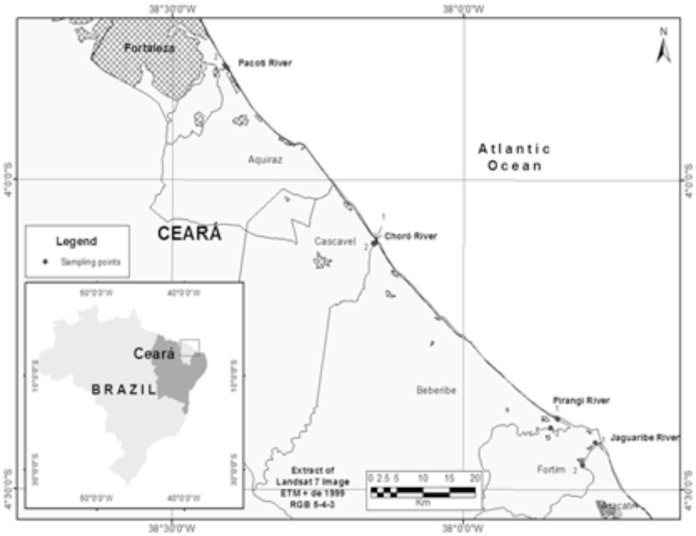
Sampling locations: Sixty-four samples of water (n = 32) and sediments (n = 32) were collected in the estuaries of the rivers Pacoti, Choró, Pirangi and Jaguaribe (east of Fortaleza, Ceará, northeastern Brazil). Sample collections took place on a monthly basis, between January and April 2009. Two points in each river were chosen: one close to and other far away from the river mouth. The coordinates of the sampling locations were registered by GPS (Garmin III Plus) (Fig. 1): Pacoti 1 03°49´16.6˝S and 038°24´11.7˝W, Pacoti 2 03°48´52.4˝S and 038°24´38.1˝W, Choró 1 04°06´07.2˝S and 038°09´01.8˝W, Choró 2 04°06´13.2˝S and 038°09´13.8˝W, Pirangi 1 04°23´11.6˝S and 037°50´18.4˝W, Pirangi 2 04°24´03.8˝S and 037°51´00.3˝W, Jaguaribe 1 04°25´28.7˝S and 037°46´22.5˝W, Jaguaribe 2 04°27´39.9˝S and 037°47´39.5˝W.
Fig. 1 -. Map of the four estuaries Pacoti, Choró, Pirangi and Jaguaribe (Ceará, Brazil) from where water and sediment were sampled between January and April 2009.
Collection of samples: Water samples were collected from a depth of 50 cm and stored in 1-L sterilized amber vials. Core-surface sediment samples were collected using a soil sampler. The samples were transported in isothermal boxes to the Laboratory of Seafood and Environmental Microbiology (LABOMAR/UFC) for immediate analysis. The temperature of the water was registered (thermometer, Incoterm) at the time of sampling.
Isolation of Vibrio cholerae from environmental samples
Water and sediment samples: The sampled water was used to make serial decimal dilutions (from 10-1 to 10-4) in alkaline peptone water (APW) at pH 7.5-8.5. In order to prepare the sediment for analysis, 25-g aliquots were homogenized in 225 mL APW for 30 minutes (10-1). Based on this first dilution, subsequent serial decimal dilutions (from 10-2 to 10-4) were prepared using the same diluent.
One hundred microliters of each dilution was inoculated onto Thiosulfate Citrate Bile salts sucrose (TCBS) agar plates. Inoculated plates were incubated for 18 hours at 37 °C. Colonies with morphological resemblance to V. cholerae (2-3 mm diameter, smooth, yellow, slightly flattened with opaque centers and translucent borders) were reseeded in tryptic soy agar (TSA) for purification19.
Identification of strains of Vibrio cholerae and detection of pathogenic potential
Phenotypic identification: Following purification, the cultures were submitted to biochemical identification using the dichotomous key of NOGUEROLA & BLANCH24. The standard strains V. cholerae O1 Classic 569B and V. cholerae non-O1 IOC 15.177 (supplied by the microbe bank of the Oswaldo Cruz Institute, Rio de Janeiro, Brazil) were used as positive controls. Commercially available antibiotic disks were used to test the susceptibility patterns. Antimicrobial classes used in panel screens included: gentamicin, streptomycin, sulfazotrim, tetracycline, ciprofloxacin, nalidixic acid, penicillin, ampicillin, ceftriaxone, ceftriaxone, aztreonam, cephalothin, chloramphenicol, florfenicol and oxytetracycline. This assay was carried out according to the CLSI3 guidelines.
Genotypic identification
Extraction of chromosomal DNA: Strains of V. cholerae were inoculated in APW + 1% sodium chloride and incubated at 35 °C for 24 hours. Subsequently, 1.0-mL aliquots were submitted to DNA extraction using a commercially available kit (DNeasy Blood & Tissue Kit, Qiagen)12.
Target genes: The identification of V. cholerae was confirmed with the primer for the gene ompW, while virulence was evaluated with the primers for the genes ctxAB (cholera toxin), tcp (toxin co-regulated pilus), rfbO1 (serogroup O1) and zot (zonula occludens toxin)12. The primers were supplied by Croma BioTechnologies (Brazil) (Table 1). The standard strains V. cholerae O1 Classic 569B and V. cholerae non-O1 IOC 15.177 (supplied by the microbe bank of the Oswaldo Cruz Institute, Rio de Janeiro, Brazil) were used as positive controls.
Table 1. Primers and thermocycler conditions used in the molecular study of V. cholerae strains isolated from water and sediments collected in four estuaries in Ceará, Brazil, between January and April 2009.
| Technique | Genes | Primer sequence (5´- 3´) | Thermocycling conditions | Amplicons (bp)f | Source |
|---|---|---|---|---|---|
| PCR | ompW a | F: 5´ -cac caa gaa ggt gac ttt att gtg- 3´ | 304 | ||
| R:5´ - ggt ttg tcg aat tag ctt cac c - 3´. | |||||
| Multiplex PCR | ctxAB b | F:5´ -gcc ggg ttg tgg gaa tgc tcc aag - 3´ | 94°C/10 min. 30 cycles (94°C/1 min., 59°C/1min., 72°C/ 2 min.) 72°C/10 min. | 536 | GOEL et al., (2007) |
| R:5´ - gcc ata cta att gcg gca atc gca tg - 3´ | |||||
| tcp c | F:5´ - cgt tgg cgg tca gtc ttg- 3´ | 805 | |||
| R:5´ - cgg gct ttc ttc ttg ttc g - 3´ | |||||
| rfbO1 d | F:5´ - tct atg tgc tgc gat tgg tg - 3´ | 638 | |||
| R:5´ - ccc cga aaa cct aat gtg ag - 3´; | |||||
| zot e | F:5´- tcg ctt aac gat ggc gcg ttt t - 3´ | 947 | |||
| R:5´- aac ccc gtt tca ctt cta ccc a - 3´ |
ompW a = V. cholerae-specific gene; ctxAB b = cholera toxin; tcp c = toxin coregulated pilus; rfbO1 d = serogroup O1 identification gene; zot e = zonula occluden s toxin; (bp)f = base pair.
PCR: Control strains were used in all amplifications. The total DNA extracted was amplified by PCR to detect ompW (V. cholerae-specific gene) and by multiplex PCR to detect the virulence genes ctxAB (cholera toxin), tcp (toxin co-regulated pilus), rfbO1 (serogroup O1 identification gene) and zot (zonula occludens toxin) using a thermocycler (Techne) (Table 2).
Table 2. Composition and concentrations used in the reactions of the molecular study of V. cholerae strains isolated from water and sediments collected in four estuaries in Ceará, Brazil, between January and April 2009.
| Reagents of the reaction* | PCR | Multiplex PCR | |||
|---|---|---|---|---|---|
| Species-specific gene | Virulence genes | ||||
| ompW | ctxAB | tcp | rfbO1 | zot | |
| Buffer 10X | 20 mM Tris pH 8.4, 50 mM KCl | 20 mM Tris pH 8.4, 50 mM KCl | |||
| dNTP's (2.5 mM) | 0.25 µM | 0.25 µM | |||
| Primer F (10 µM) | 0.4 µM | 0.4 µM | 0.4 µM | 0.4 µM | 0.4 µM |
| Primer R (10 µM) | 0.4 µM | 0.4 µM | 0.4 µM | 0.4 µM | 0.4 µM |
| MgCl2 (50 mM) | 1.5 mM | 1.5 mM | |||
| Taq polymerase (500 U) | 4 U | 4 U | |||
| Sample | 20-35 ng** | 20-35 ng** | |||
| Final reaction volume | 25 µL | 25 µL | |||
= water q.s. was added to each reaction;
= sample concentration varied from 20 to 25 ng.
Visualization of extraction products and amplicons: The DNA extraction products and amplicons were submitted to electrophoresis in 1% agarose gel and Gel Red (GelRed Nucleic Acid Gel stain) and viewed under UV light with a Spectroline transilluminator. The runs lasted 60 minutes each and were performed with 7x14 cm agarose gel at 120V and 500mA. The gels were photo-documented with a digital camera (Kodak EDAS290). A 1000-bp DNA ladder (Sigma) was used as molecular size standard.
RESULTS AND DISCUSSION
Overall, 212 strains of Vibrio spp. were isolated, 98 of which from water samples and 114 from sediment samples. Nine strains were phenotypically identified as V. cholerae, five of which from water samples and four from sediment samples. There was no resistance to the antimicrobial drugs tested in the nine Vibrio cholera strains found.
On PCR, only five percent (55%) of the strains phenotypically identified were confirmed to be V. cholerae (Fig. 2). Phenotypic identification of Vibrio is often ambiguous due to intra-species biochemical variation16, whereas genotypic identification yields a 100% match for V. cholerae using an ompW-specific primer31.
Fig. 2 -. Electrophoretic profile of nine strains identified genotypically as Vibrio cholerae using primers for the species-specific gene OmpW (304 pb) on PCR.
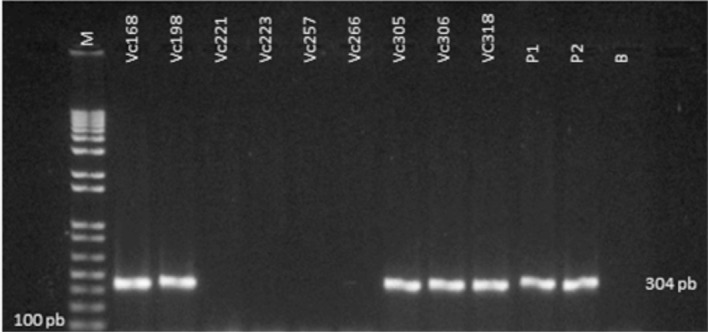
According to GOEL et al. 10,12, OmpW acts as an internal control for V. cholerae, confirming the biochemical identification of suspected strains. This was recently demonstrated by JAIN et al. 17 and by IZUMIYA et al. 16 in studies based on clinical and environmental samples.
In the present study, the greatest number of V. cholerae strains (40%) came from the Pirangi estuary, followed by Jaguaribe, Choró and Pacoti (20% each). The relatively small number of V. cholerae isolated (n = 5) matches the findings of SOUSA et al. 33, who identified only eight strains of V. cholerae among 80 vibrio strains isolated from water and sediment samples collected in the same estuaries.
Figure 3 shows the electrophoretic profile of strains identified as Vibrio cholerae using primers for the virulence genes ctxAB (536 pb), tcp (805 pb), rfbO1 (638 pb) and zot (947 pb) on multiplex PCR. None of the isolates tested positive for these genes.
Fig. 3 -. Electrophoretic profile of strains identified as Vibrio cholerae (Vc) using primers for the virulence genes ctxAB (536 pb), tcp (805 pb), rfbO1 (638 pb) and zot (947 pb) on multiplex PCR.
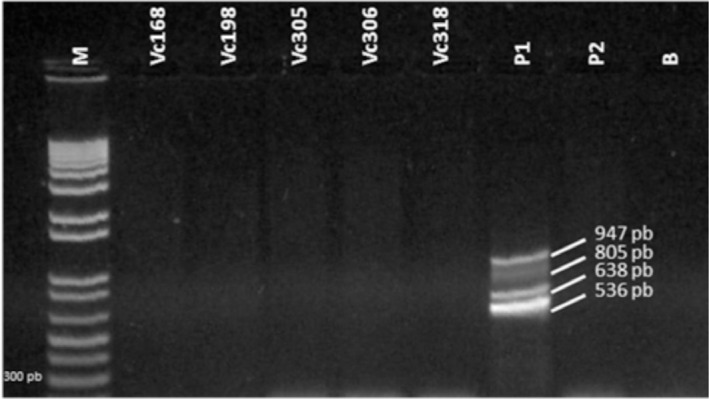
The absence on PCR of virulence genes in these environmental samples is supported by the findings of GONÇALVES et al. 13: none of their 31 strains of V. cholerae isolated from zooplankton, from Baía de São Marcos (São Luis, Maranhão, Brazil), was positive for ctxA, zot or ace.
In an analysis of water collected by LEAL et al. 20 from four rivers in Pernambuco (northeastern Brazil), strains of non-O1 and non-O139 V. cholerae tested negative for ctxA, zot and ace, and only one strain was positive for tcpA. In contrast, THEOPHILO et al. 34 assessed the distribution of virulence markers in Brazilian clinical and environmental strains of non-O1 and non-O139 V. cholerae isolated between 1991 and 2000 and found one of two environmental samples to be positive for ctx, while zot, ace and tcp were detected in clinical and environmental samples.
Nevertheless, according to SINGH et al. 32, environmental strains of V. cholerae do not carry the toxin TCP, which is responsible for one of the factors of pathogenicity in toxigenic strains belonging to serogroups O1 and O139. REEN & BOYD28 have shown the predominant association between ctx and tcp genes and epidemic isolates of V. cholerae O1 and O139 serogroups.
The absence of virulence genes in environmental strains of V. cholerae is explained by DRYSELIUS et al. 5, HEIDELBERG et al. 14 and RASMUSSEN et al. 27. According to these authors, all vibrios possess two chromosomes; in the case of V. cholerae, a large chromosome of 2.96Mb and a small chromosome of 1.07Mb. SCHOONILK & YILDIZ30 have shown that the large chromosome contains most of the genes required for growth and pathogenicity, while the small chromosome encodes a number of essential metabolic components and regulatory pathways. The duplicity of genetic elements can generate false results in DNA extraction-based gene detection studies. Thus, knowledge of the chromosomal location of target genes can help minimize false-negative results by optimizing the extraction and amplification protocols. However, according to NORIEGA-OROZCO et al. 25, genotype studies have shown that V. cholerae, V. vulnificus and V. alginolyticus do not express virulence factors in natural environments.
The strains of non-O1 and non-O139 V. cholerae isolated from the water and sediments collected in four different estuaries for this study presented no virulence markers. However, there is evidence that, even in the absence of these virulence genes, non-O1 and non-O139 V. cholerae can cause diarrhea similar to cholera18, but do not generate epidemics. Many cases of diarrhoeal diseases related to non-O1 and non-O139 V. cholerae were reported2,7,21,26. In addition, environmental strains of V. cholerae may cause outbreaks in the future1,29, possibly triggered by environmental changes, or by lateral or horizontal transference of virulence genes mediated by phages, pathogenicity is lets and/or other mobile genetic elements encoding cholera toxin20,34, as previously reported for marine populations of V. cholerae and V. parahaemolyticus 22.
CONCLUSION
The dichotomous key of NOGUEROLA & BLANCH24 was efficient at differentiating environmental strains of V. cholerae isolated from four estuaries, confirming the importance of aquatic ecosystems in the dissemination, evolution and, in some cases, transmission of this pathogen to humans.
Although these strains possessed no virulence genes capable of causing cholera, environmental strains can evolve into epidemic lineages through contact with toxigenic strains.
ACKNOWLEDGMENTS
The authors would like to thank the government research promotion agencies FUNCAP and CAPES for financial support.
REFERENCES
- 1.Cariri FAMO, Costa APR, Melo CC, Theophilo GND, Hofer E, Melo Neto OP, et al. Characterization of potentially virulent non-O1/non-O139 Vibrio cholerae strains isolated from human patients. Clin Microbiol Infect. 2010;16:62–7. doi: 10.1111/j.1469-0691.2009.02763.x. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, et al. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2009;47:1087–95. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute . Wayne: CLSI; 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. (Document M100-S23) [Google Scholar]
- 4.Colaço W, Silva Filho SV, Rodrigues DP, Hofer E. Vibrio cholerae O1 em amostras de ambientes aquáticos e de alimentos analisados no Estado de Pernambuco, Brasil. Cad Saúde Pública. 1998;14:465–71. doi: 10.1590/s0102-311x1998000300002. [DOI] [PubMed] [Google Scholar]
- 5.Dryselius R, Kurokawa K, Iida T. Vibrionaceae, a versatile bacterial family with evolutionarily conserved variability. Res Microbiol. 2007;158:479–86. doi: 10.1016/j.resmic.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 6.DSMZ Leibniz-Institu Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Bacterial diversity. [[cited 2012 Jan. 24]]; Available from: http://www.dsmz.de/bacterial-diversity.html. [Google Scholar]
- 7.Dutta D, Chowdhury G, Pazhani GP, Guin S, Dutta S, Ghosh S, et al. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg Infect Dis. 2013;19:464–7. doi: 10.3201/eid1903.121156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filizola LRS, Figueirôa ACTA, Araújo MCMD, Cavalcanti VO, Lima CM, Hofer E. Significância de anticorpos vibriocidas circulantes em área pós-epidêmica de diarréia, São Bento do Uma, Estado de Pernambuco. Rev Soc Bras Med Trop. 2007;40:686–9. doi: 10.1590/s0037-86822007000600018. [DOI] [PubMed] [Google Scholar]
- 9.Fraga SG, Pichel M, Costagliola M, Cecilia M, Jurquiza V, Peressutti S, et al. Environment and virulence factors of Vibrio cholerae strains isolated in Argentina. J Appl Microbiol. 2007;103:2448–56. doi: 10.1111/j.1365-2672.2007.03468.x. [DOI] [PubMed] [Google Scholar]
- 10.Goel AK, Jain M, Kumar P, Jiang SC. Molecular characterization of Vibrio cholerae outbreak strains with altered El Tor biotype from southern India. World J Microbiol Biotechnol. 2010;26:281–7. doi: 10.1007/s11274-009-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goel AK, Jiang SC. Genetic determinants of virulence, antibiogram and altered biotype among the Vibrio cholerae O1 isolates from different cholera outbreaks in India. Infect Gent Evol. 2010;10:815–9. doi: 10.1016/j.meegid.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Goel AK, Ponmariappan S, Kambol DV, Singh L. Single multiplex polymerase chain reaction for environmental surveillance of toxigenic-pathogenic O1 and non-O1 Vibrio cholerae . Folia Microbiol. 2007;52:81–5. doi: 10.1007/BF02932143. [DOI] [PubMed] [Google Scholar]
- 13.Gonçalves EGR, Leal NC, Hofer E. Estudo molecular de Vibrio cholerae não-O1 isolado de zooplâncton da Baía de São Marcos/São Luis - MA, Brasil. Rev Soc Bras Med Trop. 2004;37:324–8. doi: 10.1590/s0037-86822004000400007. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg JFJA, Eisen WC, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae . Nature. 2000;406:477–83. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofer E, Reis EMF, Quintaes BR, Rodrigues DP, Feitosa IS, Angelo MRF, et al. Vibrio cholerae resistant to 2,4-diamino-6, 7-diisopropylpteridine (O/129) isolated from patients with enteritis in Ceará, Brazil. J Health Popul Nutr. 2001;19:39–42. [PubMed] [Google Scholar]
- 16.Izumiya H, Matsumoto K, Yahiro S, Lee J, Morita M, Yamamoto S, et al. Multiplex PCR assay for identification of three major pathogenic Vibrio spp., Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus . Mol Cell Probes. 2011;25:174–6. doi: 10.1016/j.mcp.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Jain M, Goel AK, Bhattacharya P, Ghatole M, Kamboj DV. Multidrug resistant Vibrio cholerae O1 El Tor carrying classical ctxB allele involved in a cholera outbreak in South Western India. Acta Trop. 2011;117:152–6. doi: 10.1016/j.actatropica.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Jagadeeshan S, Kumar P, Abraham WP, Thomas S. Multiresistant Vibrio cholerae non-O1/non-O139 from waters in South India: resistance patterns and virulence-associated gene profiles. J Basic Microbiol. 2009;49:538–44. doi: 10.1002/jobm.200900085. [DOI] [PubMed] [Google Scholar]
- 19.Kaysner CA, Depaola A. Bacteriological analytical manual on line. Rockville: U.S. Food and Drug Administration; 2004. Vibrio. Chapter 9. [cited 2012 Jan 13]. Available from: < http://www.cfsan.fda.gov/∼ebam/bam-9.html>. [Google Scholar]
- 20.Leal NC, Figueiroa ACTA, Cavalcanti VO, Silva SC, Leal-Balbino TC, Almeida AMP, et al. Characterization of Vibrio cholerae isolated from the aquatic basins of the State of Pernambuco, Brazil. Trans R Soc Trop Med Hyg. 2008;102:272–6. doi: 10.1016/j.trstmh.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Lukinmaa S, Mattila K, Lehtinen V, Hakkinen M, Koskela M, Siitonen A. Territorial waters of the Baltic Sea as a source of infections caused by Vibrio cholerae non-O1, non-O139: report of 3 hospitalized cases. Diagn Microbiol Infect Dis. 2006;54:1–6. doi: 10.1016/j.diagmicrobio.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Masini L, De Grandis G, Principi F, Mengarelli C, Ottaviani D. Research and characterization of pathogenic vibrios from bathing water along the Conero Riviera (Central Italy) Water Res. 2007;41:4031–40. doi: 10.1016/j.watres.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Nandi B, Nandy R, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38:4145–51. doi: 10.1128/jcm.38.11.4145-4151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguerola I, Blanch AR. Identification of Vibrio spp. with a set of dichotomous keys. J Appl Microbiol. 2008;105:175–85. doi: 10.1111/j.1365-2672.2008.03730.x. [DOI] [PubMed] [Google Scholar]
- 25.Noriega-Orozco L, Acedo-Félix E, Higuera-Ciapara I, Jiménez-Flores R, Cano T. Pathogenic and non pathogenic Vibrio species in aquaculture shrimp ponds. Rev Latinoam Microbiol. 2007;49:60–7. [Google Scholar]
- 26.Ottaviani D, Leoni F, Rocchegiani E, Santarelli S, Masini L, Trani VD, et al. Prevalence and virulence properties of non-O1 non-O139 Vibrio cholerae strains from seafood and clinical samples collected in Italy. Int J Food Microbiol. 2009;132:47–53. doi: 10.1016/j.ijfoodmicro.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–31. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reen FJ, Boyd EF. Molecular typing of epidemic and nonepidemic Vibrio cholerae isolates and differentiation of V. cholerae and V.mimicus isolates by PCR-single-strand conformation polymorphism analysis. J Appl Microbiol. 2005;98:544–55. doi: 10.1111/j.1365-2672.2004.02451.x. [DOI] [PubMed] [Google Scholar]
- 29.Rivera ING, Chun J, Huq A, Sack RB, Colwell RR. Genotypes associated with virulence in environmetal isolates of Vibrio cholerae . Appl Environ Microbiol. 2001;67:2421–9. doi: 10.1128/AEM.67.6.2421-2429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoolnik GK, Yildiz FH. The complete genome sequence of Vibrio cholerae: a tale of two chromosomes and of two lifestyles. Genome Biol. 2000;1:REVIEWS 1016. doi: 10.1186/gb-2000-1-3-reviews1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Chaturvedi AN. Prevalence of virulence genes (ctxA, stn, OmpW and tcpA) among non-O1 Vibrio cholerae isolated from fresh water environment. Int J Hyg Environ Health. 2006;209:521–6. doi: 10.1016/j.ijheh.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Singh DV, Matte MH, Matte GR, Jiang S, Sabeena F, Shukla BN, et al. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol. 2001;67:910–21. doi: 10.1128/AEM.67.2.910-921.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sousa OV, Macrae A, Menezes FGR, Gomes NCM, Vieira RHSF, Mendonça-Hagler LCS. The impact of shrimp farming effluent on bacterial communities in mangrove waters, Ceará, Brazil. Mar Pollut Bull. 2006;52:1725–34. doi: 10.1016/j.marpolbul.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Theophilo GND, Rodrigues DP, Leal NC, Hofer E. Distribution of virulence markers in clinical and environmental Vibrio cholerae non-O1/non-O139 strains isolated in Brazil from 1991 to 2000. Rev Inst Med Trop Sao Paulo. 2006;48:65–70. doi: 10.1590/s0036-46652006000200002. [DOI] [PubMed] [Google Scholar]
- 35.Vezzulli L, Pruzzo C, Huq A, Colwell RR. Environmental reservoirs of Vibrio cholerae and their role in cholera. Environ Microbiol Rep. 2012;2:27–33. doi: 10.1111/j.1758-2229.2009.00128.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Zhang J, Lu X, Liang W, Zhang L, Kan B. O antigen is the receptor of Vibrio cholerae serogroup O1 El Tor typing phage VP4. J Bacteriol. 2013;195:798–806. doi: 10.1128/JB.01770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]