The genus Dendrobium, one of the largest genera in Orchidaceae, exhibits enormous vegetative diversification. Such a situation has hindered the establishment of consistent classification systems. To clarify phylogenetic relationships in Dendrobium section Dendrobium and allied groups, we performed molecular phylogenetic analyses of 210 taxa. The results showed that many sections are not monophyletic. Most of the morphological characters that have been believed to reflect phylogenetic relationships are, in fact, the results of convergence. Consequently, recircumscription of the infrageneric classification is required.
Keywords: Dendrobium, evolution, ITS, matK, Orchidaceae, phylogeny, systematics, taxonomy.
Abstract
It is always difficult to construct coherent classification systems for plant lineages having diverse morphological characters. The genus Dendrobium, one of the largest genera in the Orchidaceae, includes ∼1100 species, and enormous morphological diversification has hindered the establishment of consistent classification systems covering all major groups of this genus. Given the particular importance of species in Dendrobium section Dendrobium and allied groups as floriculture and crude drug genetic resources, there is an urgent need to establish a stable classification system. To clarify phylogenetic relationships in Dendrobium section Dendrobium and allied groups, we analysed the macromolecular characters of the group. Phylogenetic analyses of 210 taxa of Dendrobium were conducted on DNA sequences of internal transcribed spacer (ITS) regions of 18S–26S nuclear ribosomal DNA and the maturase-coding gene (matK) located in an intron of the plastid gene trnK using maximum parsimony and Bayesian methods. The parsimony and Bayesian analyses revealed 13 distinct clades in the group comprising section Dendrobium and its allied groups. Results also showed paraphyly or polyphyly of sections Amblyanthus, Aporum, Breviflores, Calcarifera, Crumenata, Dendrobium, Densiflora, Distichophyllae, Dolichocentrum, Holochrysa, Oxyglossum and Pedilonum. On the other hand, the monophyly of section Stachyobium was well supported. It was found that many of the morphological characters that have been believed to reflect phylogenetic relationships are, in fact, the result of convergence. As such, many of the sections that have been recognized up to this point were found to not be monophyletic, so recircumscription of sections is required.
Introduction
It is always difficult to construct coherent classification systems for plant lineages having diverse morphological characters. Dendrobium Sw. (Orchidaceae) represents such difficult groups and so far has been established in many alternative systems (e.g. Lindley 1830; Bentham and Hooker 1883; Kränzlin 1910; Schlechter 1912; Brieger 1981; Clements 2003; Wood 2006; Schuiteman 2011). This genus is one of the largest orchid genera, with around 1100 species (Wood 2006). The distribution range extends from Sri Lanka and India throughout tropical Asia and Oceania, north to Japan, east to Tahiti and south to New Zealand. Enormous diversification of the vegetative organs in accordance with habitat shifts and lack of accessory structures of the pollinarium, a cardinal character in orchid classification, has hindered establishment of consistent classification systems covering all major groups of this genus. Previous studies of systematics based on the morphological characteristics of the group were reviewed by Wood (2006). Given the limits to what can be understood of affinities using morphological characters, Yukawa et al. (1993) analysed the molecular phylogenetics of the subtribe Dendrobiinae (Lindley 1830), which includes the genus Dendrobium and putatively related genera Cadetia, Diplocaulobium, Flickingeria, Epigeneium and Pseuderia based on chloroplast DNA restriction site variation. This analysis resulted in presentation of the first probable phylogenetic relationship between members of this genus. Yukawa et al. (1993) demonstrated that Dendrobium is not monophyletic and comprises two major clades (Asian and Australasian clades: sensu Clements 2003). The Asian clade is predominantly diversified west of Weber's line, and the Australasian clade, containing genera Cadetia, Diplocaulobium and Flickingeria, is distributed mostly in Australasia and the Pacific Islands. Subsequent studies on representative members of subtribe Dendrobiinae (e.g. Yukawa et al. 1996, 1999, 2000; Yukawa 2001; Clements 2003, 2006; Schuiteman 2011) incorporated additional macromolecular markers and taxa in their analyses. In addition to providing further support for the above-mentioned phylogeny, these studies identified other infrageneric monophyletic groups.
Dendrobium section Dendrobium is one of the largest sections in the genus Dendrobium, comprising ∼60 species (Wood 2006) distributed across almost the entire geographical range of the genus, with the exception of Micronesia and Melanesia. A number of species are considered important as crude drug sources and are highly sought-after genetic resources with potential value in medicine (Takamiya et al. 2011, 2013). Yukawa et al. (1993) demonstrated that section Dendrobium is nested within the Asian clade. Wongsawad et al. (2001, 2005) analysed sequences for the maturase-coding gene (matK) located in the plastid genome and the internal transcribed spacer regions (ITS) of the nuclear ribosomal DNA of 78 Asian clade species including 35 of section Dendrobium (Wongsawad et al. 2001) and 93 Asian clade species including 42 members of section Dendrobium (Wongsawad et al. 2005). Based on these analyses, Wongsawad et al. demonstrated that section Dendrobium is not monophyletic; that its core clade includes species of sections Breviflores, Densiflora, Holochrysa and Stuposa; and that sections Amblyanthus, Breviflores, Densiflora, Formosae, Holochrysa, Oxyglossum and Pedilonum are not monophyletic. These relationships were confirmed by Xiang et al. (2013). However, given that these studies did not include several species of section Dendrobium and only included a small number of species in sections Aporum, Calcarifera, Calyptrochilus, Crumenata, Distichophyllae, Oxyglossum, Pedilonum, Platycaulon, Stachyobium and Stuposa, which are likely to be closely related to section Dendrobium, our understanding of the relationships between section Dendrobium and other groups within the Asian clade remains incomplete. In this study, we conducted comprehensive phylogenetic analyses of representative species in the Asian clade using the ITS and matK regions to clarify the relationships and the taxonomic position of section Dendrobium.
Methods
Plant materials
The samples for analysis consisted of 210 Asian clade species (214 samples), including 56 species belonging to section Dendrobium. As an outgroup, we chose 10 species of the Australasian clade, based on the results of Yukawa (2001). Plant materials were collected from the living collection of Tsukuba Botanical Garden, National Museum of Nature and Science. All voucher specimens were deposited at the Herbarium, National Museum of Nature and Science (TNS). Voucher information and GenBank accession numbers for all ITS and matK sequences used in this study are listed in Appendix 1. We adopted an infrageneric classification based on that proposed by Schlechter (1912) and modified by Wood (2006).
DNA extraction and sequencing
Genomic DNA was isolated from fresh leaves or flowers by a DNeasy Plant Mini Kit (Qiagen, Hamburg, Germany) following the manufacturer's instructions, and was used as a template for PCR amplification. ITS1-5.8S-ITS2 regions were amplified and sequenced as described by Sun et al. (1994) and Hidayat et al. (2005). The amplification reactions included GC buffer I or II and LA Taq DNA polymerase (Takara Bio, Shiga, Japan). The PCR profile consisted of an initial 2-min premelt at 94 °C and 30 cycles of 50 s at 94 °C (denaturation), 1 min at 60 °C (annealing) and 30 s at 72 °C (extension), followed by a final 7-min extension at 72 °C. matK regions were amplified and sequenced as described by Hidayat et al. (2005). The amplification reactions included Ex Taq buffer and Ex Taq DNA polymerase (Takara Bio). The PCR profile consisted of an initial 5-min premelt at 94 °C and 30 cycles of 30 s at 94 °C (denaturation), 30 s at 53 °C (annealing) and 3 min at 72 °C (extension), followed by a final 7-min extension at 72 °C.
The PCR products were cleaned using a Montage PCR centrifugal filter device (Millipore, Billerica, MA, USA) and then sequenced in both forward and reverse directions using primers described by Hidayat et al. (2005). Sequences were obtained with an ABI PRISM 377 sequencer (Applied Biosystems, Foster City, CA, USA) following the manufacturer's instructions for auto-cycle sequencing reactions.
Molecular phylogenetic analysis
Except for ITS and matK sequences of Dendrobium ovatum obtained from GenBank (accession ID for ITS: HM054721; matK: HM055325), all sequence data were obtained by our own analyses. Two hundred and twenty-four DNA sequences were aligned with ClustalW software. A phylogenetic analysis based on the maximum parsimony (MP) was performed using PAUP* version 4.0b10 (Swofford 2002) for three data sets: ITS, matK and a combination of the two. Gaps were treated as missing data. All characters were equally weighted and unordered (Fitch 1971). Each data set was analysed by a heuristic search method with tree bisection–reconnection branch swapping and the MULTREES option on 100 replications of random addition sequence with the stepwise addition option, and each replicate after 1 × 106 rearrangements was assessed. Bootstrap support values were obtained from 1000 replicates using ‘fast’ stepwise addition. Although the fast stepwise addition analyses are expected to provide a lower support value than obtained when comprehensive branch-swapping is performed (DeBry and Olmstead 2000; Mort et al. 2000; Barkman et al. 2004), we used this option because bootstrap analyses with a full heuristic search method were not computationally feasible with a large data set. Bootstrap percentages (BPs) of 50–74 were defined as weak, 75–84 as moderate and 85–100 as strong, as in Chase et al. (2000) and Kim et al. (2010). The number of steps, consistency index (CI) and retention index (RI) were calculated with one of the most parsimonious trees in each analysis using the TREE SCORES command in PAUP*. To test congruence among data partitions, the incongruence length difference test (Farris et al. 1994, 1995), also designated the partition homogeneity test in PAUP*, was employed to measure character conflicts under a parsimony framework among data sets using 100 heuristic search replications.
The same data sets were analysed by Bayesian analysis using MrModeltest ver. 2.3 (Nylander 2004) to determine the sequence evolution model that best described the data. The GTR + I + G model was chosen for ITS, matK and the combined data by hierarchical likelihood ratio tests. The chosen model was used to perform a Bayesian analysis using the program MrBayes ver. 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). For analysis, two simultaneous runs of four chains each were carried out with the Markov chain Monte Carlo algorithm for 8 000 000 generations, sampling every 100 generations. Data from the first 20 000 generations were discarded as the ‘burn-in’ period. The 50 % majority rule consensus phylogeny and posterior probability (PP) of nodes were calculated from the remaining samples. Clades with PP ≥95 were regarded as strongly supported (Martínez-Azorín et al. 2011).
Ancestral state reconstruction of morphological characters
Among various morphological characters, stem shape and leaf number were examined with ancestral state reconstruction analysis due to their complex pattern of character states even among closely related taxa. Character states for each taxon were obtained from the literature (Seidenfaden 1985; Wood 2006) and observation by the authors. A parsimony reconstruction was performed with the ‘Unordered’ model in Mesquite 2.75 (Maddison and Maddison 2011). The Bayesian tree based on the combined sequence data (ITS and matK) was used as the standard tree topology (see Fig. 1).
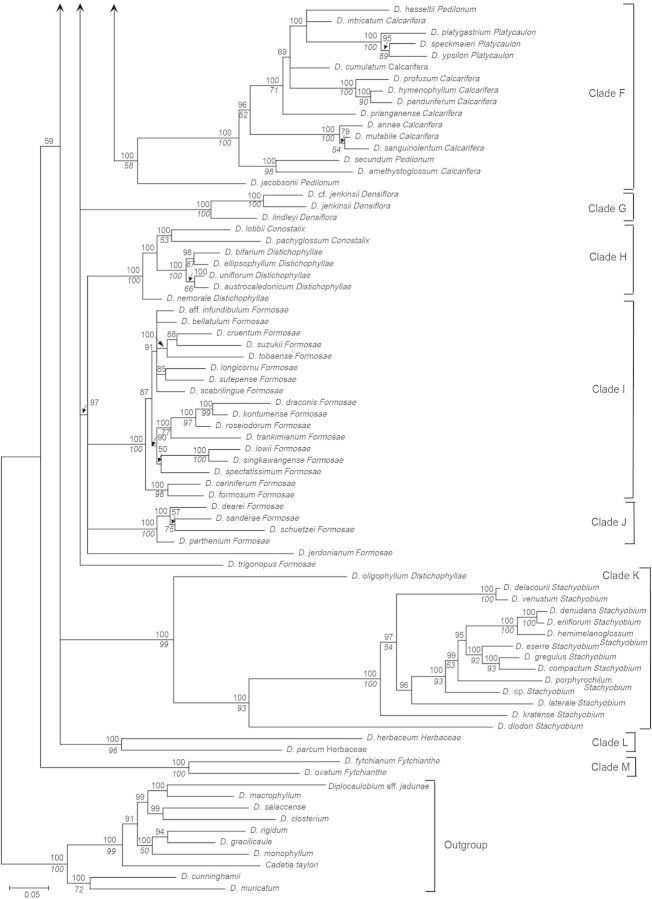
Figure 1.
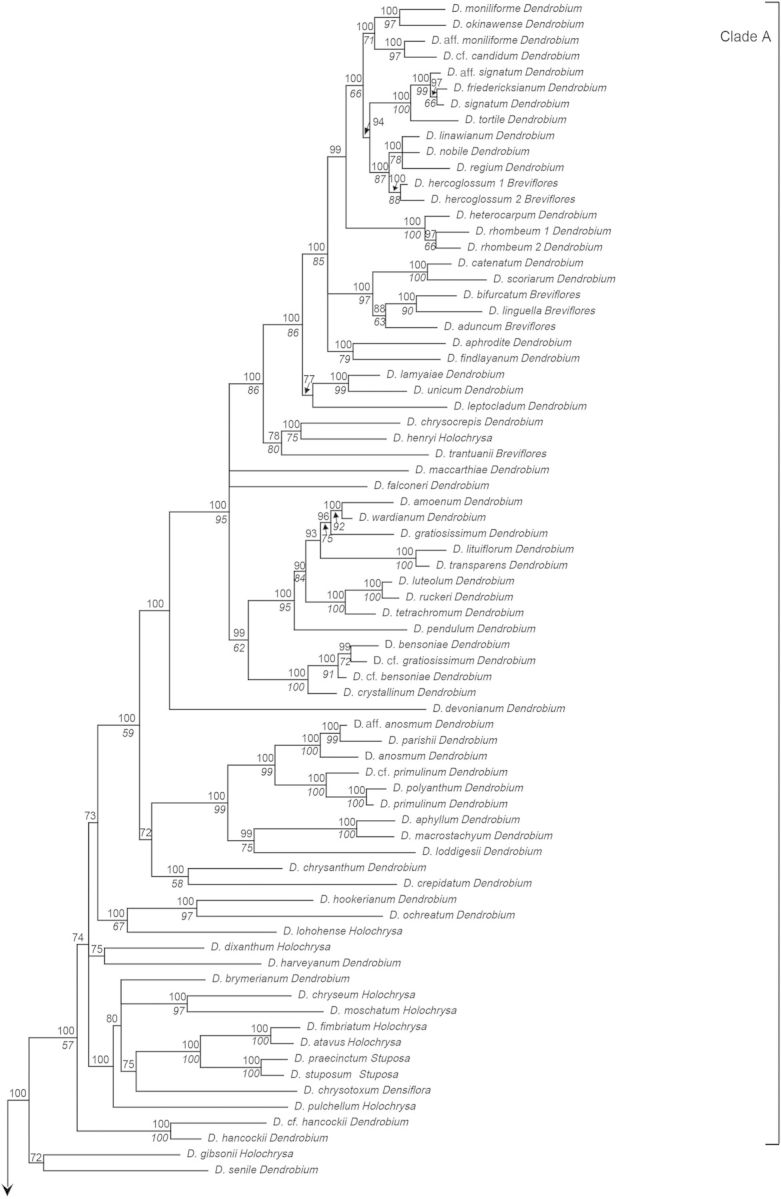
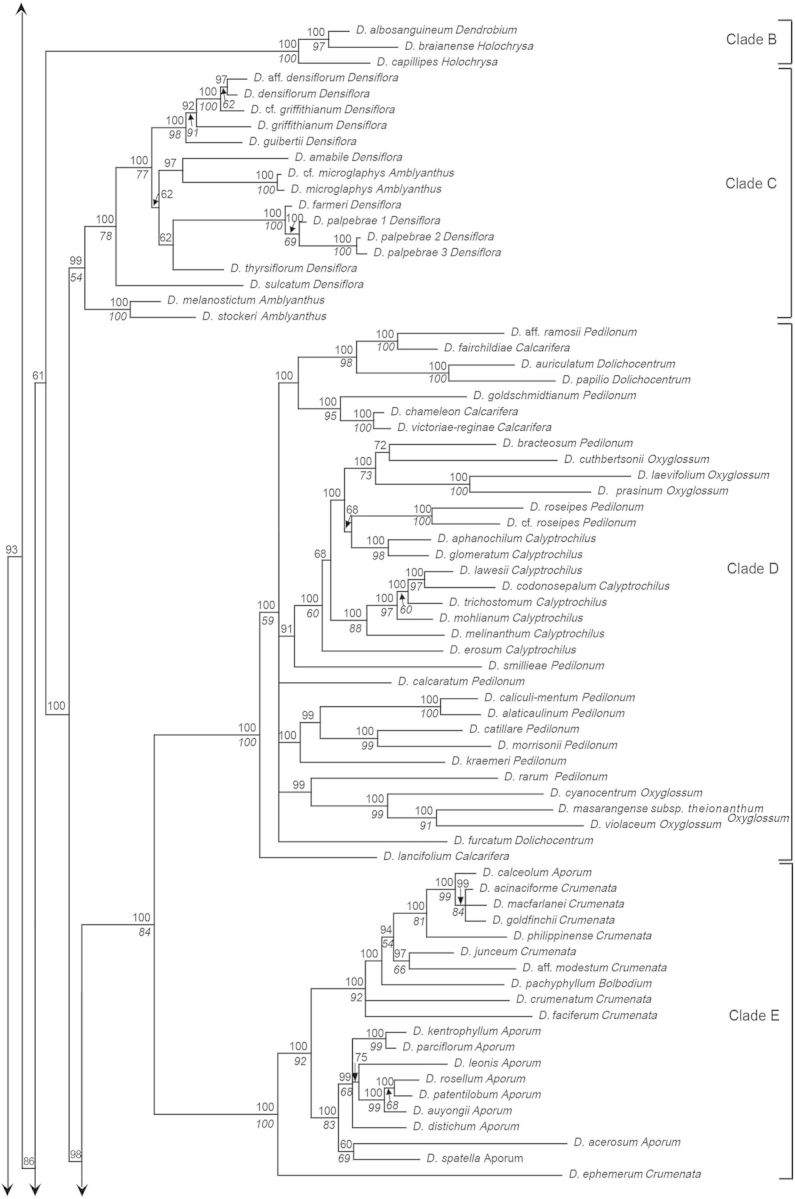
Consensus phylogram of 210 taxa of Dendrobium section Dendrobium and allied groups obtained from 96 596 Bayesian trees from the combined ITS and matK sequence data set. Values below and above branches indicate percentage bootstrap values from maximum parsimony analysis and Bayesian posterior probabilities, respectively.
Results
Parsimony analyses of ITS, matK and combined sequences of the two
Table 1 shows statistics from ITS, matK and combined sequences. We compared topologies between strict consensus trees of ITS and matK data sets [see Supporting Information]. Visual inspection of the topologies between the two regions did not show significant incongruence, although an incongruence length difference test did not support congruence (P value = 0.01). Inconsistent topologies where bootstrap support is weak can be ignored as a soft incongruence (Johnson and Soltis 1998). Moreover, strongly supported clades by matK were generally strongly supported by ITS except for six inconsistent regions [see Supporting Information]. Since analyses of combined data sets provide more resolution and internal support for relationships than individual data sets (Soltis et al. 1998), we combined ITS and matK data for further analysis.
Table 1.
Characteristics of DNA datasets used in this study.
| Characteristics | ITS | matK | Combined data |
|---|---|---|---|
| Number of taxa | 224 | 224 | 224 |
| Total number of characters | 906 | 1630 | 2536 |
| Number of constant characters | 303 | 1092 | 1395 |
| Number of parsimony uninformative characters | 102 | 279 | 381 |
| Number of parsimony informative characters | 501 | 259 | 760 |
| Tree length | 5522 | 1092 | 6692 |
| Consistency index (CI) | 0.226 | 0.6016 | 0.2847 |
| Retention index (RI) | 0.715 | 0.7915 | 0.7198 |
| Number of trees | 140 | 4638 | 103 |
| Bayesian model of evolution | GTR + I + G | GTR + I + G | GTR + I + G |
In the majority of cases, BPs for trees based on the combined data set were higher than those obtained from analysis based on ITS or matK alone. For example, six clades supported by BP at <85 % in both genetic regions when based on either individual data set had BP >85 % in the combined case.
Bayesian analyses of ITS, matK and combined sequences of the two
Comparing the Bayesian trees and strict consensus tree of the most parsimonious trees constructed from ITS and matK sequences, we found that in terms of the differences in results based on the sequence used, the clades of strict consensus trees that were strongly supported by bootstrap value in both ITS and matK trees were also strongly supported by the PP value [see Supporting Information]. However, when we compared the strict consensus tree of the most parsimonious trees and the Bayesian trees with the ITS sequence, we found phylogenetic relationships in four parts where they were not strongly supported by PP value, even if they were by BP value. Specifically, these were the clade comprising Dendrobium amoenum, D. gratiosissimum, D. wardianum, D. lituiflorum, D. transparens, D. luteolum, D. ruckeri and D. tetrachromum (BP86, PP90); the clade comprising D. aff. densiflorum, D. densiflorum, D. cf. griffithianum and D. griffithianum (BP87, PP84); the clade comprising D. farmeri and D. palpebrae (BP87, PP75); and the clade comprising D. speckmaieri and D. ypsilon (BP89, PP92). That said, the PP values for these regions were still fairly high. Furthermore, as in the case for MP analysis, Bayesian analysis revealed that there are no inconsistencies between ITS and matK data sets, so the two regions were combined for subsequent analysis.
The combined tree obtained from a majority-rule consensus of 96 596 trees produced by two runs of the Markov chain Monte Carlo algorithm is presented in Fig. 1. With the exception of one clade consisting of D. aff. densiflorum, D. densiflorum, D. cf. griffithianum and D. griffithianum (BP91, PP92), all of the clades that were strongly supported in bootstrap tests based on MP analysis were also strongly supported by PP values in Bayesian analysis. In this study, major clades whose BP values from MP analysis were >50 % and those whose PP values from Bayesian analysis were >95 %—i.e. clades at least weakly supported by bootstrap values and strongly supported by PP values—were assigned letters A through M. Clade A comprises the majority of sections Dendrobium and Holochrysa, as well as sections Breviflores and Stuposa, and Dendrobium chrysotoxum in section Densiflora. Clade B comprises Dendrobium albosanguineum in section Dendrobium and Dendrobium braianense and D. capillipes in section Holochrysa. Clade C comprises the majority of section Densiflora and section Amblyanthus. Clade D consists of some members of sections Pedilonum and Calcarifera along with sections Dolichocentrum, Oxyglossum and Calyptrochilus. Clade E comprises sections Aporum, Crumenata and Bolbodium. Clade F comprises section Platycaulon and some members of sections Pedilonum and Calcarifera. Clade G comprises D. cf. jenkinsii, D. jenkinsii and D. lindleyi in section Densiflora. Clade H comprises section Conostalix and the majority of section Distichophyllae. Clade I comprises the majority of section Formosae. Clade J comprises D. dearei, D. parthenium, D. sanderae and D. schuetzei in section Formosae. Clade K comprises section Stachyobium and Dendrobium oligophyllum in section Distichophyllae. Clade L comprises section Herbaceae. Clade M comprises section Fytchianthe. No clear relationship to other clades was indicated for Dendrobium gibsonii in section Holochrysa, D. senile in section Dendrobium, or D. jerdonianum and D. trigonopus in section Formosae.
Ancestral state reconstruction of morphological characters
In the analysis of stem shape morphology, the terete, non-succulent stem was supported as the plesiomorphic state in the Asian clade (Fig. 2A). The other stem character states, namely, pseudobulbous, heteroblastic, succulent stem; clavate, non-heteroblastic, succulent stem; basally bulged stem; entirely flattened stem; and thin, wiry stem, were inferred to evolve twice, more than 14 times, twice, three times, and more than seven times, respectively. As for the evolution of leaf number, a character state bearing more than five leaves was suggested to be plesiomorphic in the Asian clade (Fig. 2B). The other character states of leaf number, namely, between three and five; two; and one, were inferred to evolve more than 10 times, once, and once, respectively.
Figure 2.
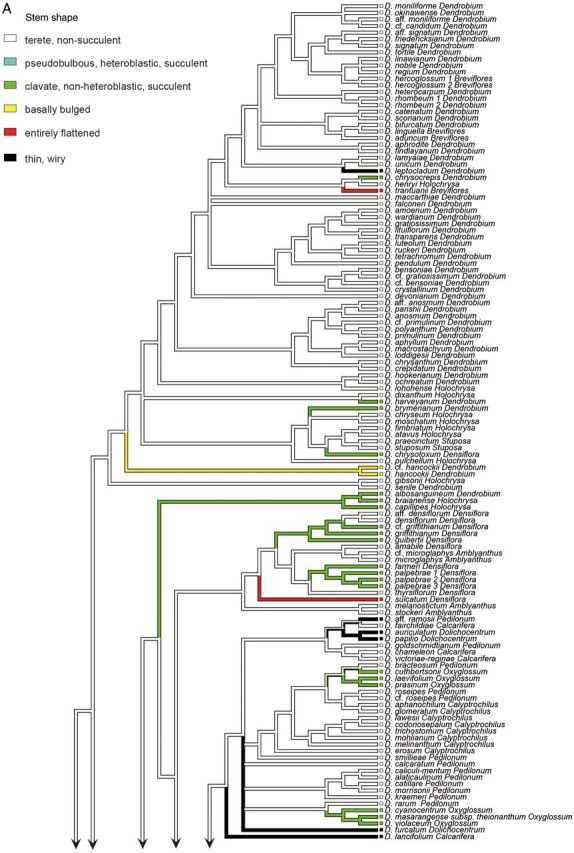
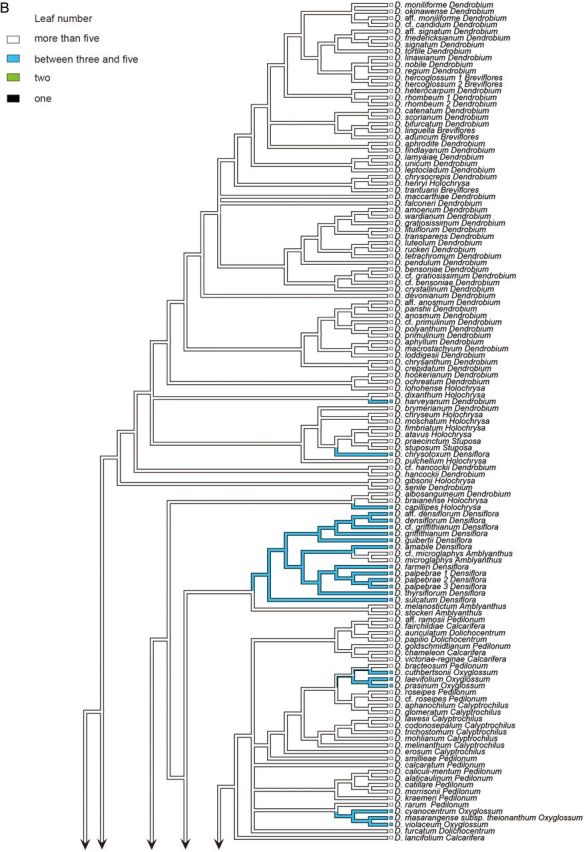
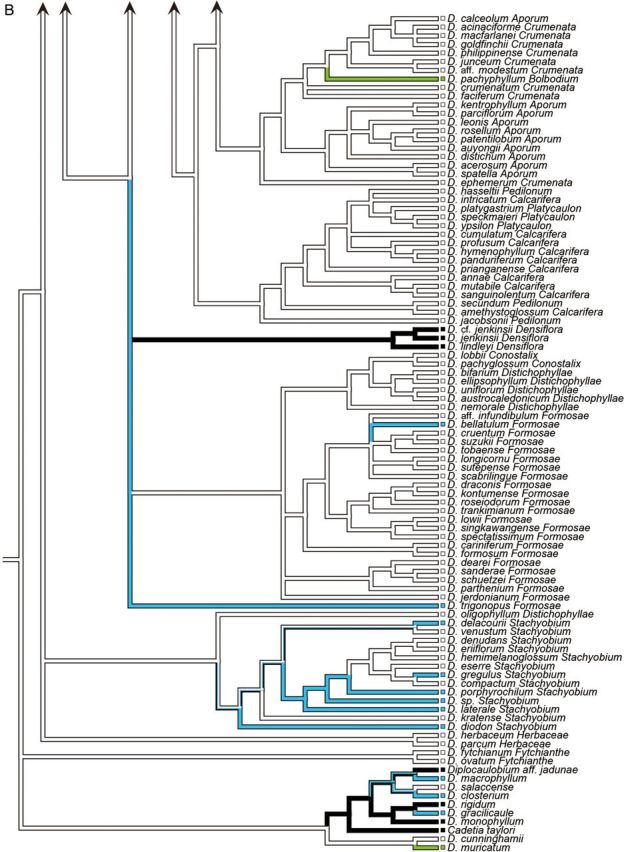
Ancestral state reconstruction of morphological characters ((A) stem shape; (B) leaf number) of 210 taxa of Dendrobium section Dendrobium and allied groups. All analyses were based on the parsimony reconstruction.
Discussion
Phylogenetic relationships at the section level
Section Dendrobium is divided into three subclades. The first is the core clade, which includes the type species Dendrobium moniliforme (Clade A). The second is D. albosanguineum, included in Clade B, and the final subclade is D. senile, with a phylogenetic position that is unresolved. The possibility is suggested (PP72) that D. senile and D. gibsonii in section Holochrysa may form a clade. Furthermore, while there is a support of PP100 suggesting that these two species are sister groups to Clade A, due to the weak bootstrap support in MP analysis (BP < 50), they were not included in Clade A for the purposes of this study. In addition, Clade A also contains all species in section Breviflores, D. chrysotoxum in section Densiflora, all species in section Stuposa, and the majority of species in section Holochrysa, thus rendering section Dendrobium polyphyletic and paraphyletic. These results support the findings of Wongsawad et al. (2001, 2005) and Xiang et al. (2013).
Five species in section Breviflores (Dendrobium aduncum, D. bifurcatum, D. hercoglossum, D. linguella and D. trantuanii) are divided into three subclades within Clade A, making them a polyphyletic group. These results are consistent with the findings of Wongsawad et al. (2005) and Xiang et al. (2013). Two species in section Stuposa (Dendrobium stuposum and D. praecinctum) represent a monophyletic group within Clade A (BP100, PP100), supporting the findings of Wongsawad et al. (2005). Section Holochrysa was found to not be monophyletic, and its eight species (Dendrobium atavus, D. chryseum, D. dixanthum, D. fimbriatum, D. henryi, D. lohohense, D. moschatum and D. pulchellum) in Clade A are divided into six lineages. Further, a subclade comprising D. braianense and D. capillipes is placed within Clade B and the phylogenetic position of D. gibsonii is unresolved. These results are consistent with the analyses by Yukawa (2001), Wongsawad et al. (2001, 2005) and Xiang et al. (2013).
Section Densiflora was also found to not be monophyletic but rather to consist of three groups comprising the majority of Clade C, including the type species D. densiflorum, D. chrysotoxum belonging to Clade A, and Clade G, which is made up of D. jenkinsii, D. cf. jenkinsii and D. lindleyi. The analyses by Yukawa (2001), Wongsawad et al. (2001, 2005) and Xiang et al. (2013) also demonstrated the polyphyly of section Densiflora. Section Amblyanthus was found to nest within Clade C. Four species of section Amblyanthus were found to be divided into two subclades within Clade C, and were polyphyletic, supporting the results of Yukawa (2001) and Wongsawad et al. (2005).
Section Pedilonum was found to not be monophyletic, its members forming eight polyphyletic groups within Clade D and three polyphyletic groups within Clade F. The type species of section Pedilonum, D. secundum, was included in Clade F. Our results were consistent with analyses by Yukawa (2001), Wongsawad et al. (2001, 2005) and Clements (2003, 2006), which also revealed section Pedilonum to not be monophyletic.
Section Calcarifera represents another non-monophyletic group, which is divided into three subclades within Clade D and six subclades within Clade F. Consistent with our results, Clements (2003, 2006) also found that the section was not monophyletic and that its members formed two distinct groups. It is clear that the type species Dendrobium pedicellatum, which was not investigated in this study, belongs to Clade F because a morphologically related species, Dendrobium mutabile, is placed in this clade.
Clade D was shown to also include sections Dolichocentrum, Oxyglossum and Calyptrochilus. Section Dolichocentrum was found to be polyphyletic, with its three species splitting into a lineage represented by Dendrobium furcatum and a subclade (BP100, PP100) made up of Dendrobium auriculatum and D. papilio.
Section Oxyglossum was also found to not be monophyletic. Its six constituent species were divided into two groups, one comprising Dendrobium cyanocentrum, D. masarangense subsp. theionanthum and D. violaceum (BP99, PP100) and the other comprising D. cuthbertsonii, D. laevifolium and D. prasinum, along with D. bracteosum, a member of section Pedilonum (BP73, PP100). Consistent with our results, Clements (2003, 2006) also found that section Oxyglossum was not monophyletic and that its members split into two groups.
Our results suggested that section Calyptrochilus forms a polyphyletic group made up of three subclades: the first comprising Dendrobium aphanochilum and D. glomeratum (BP98, PP100); the second comprising D. lawesii, D. codonosepalum, D. trichostomum, D. mohlianum and D. melinanthum (BP88, PP100); and the third comprising D. erosum.
Section Aporum was found to be polyphyletic, nesting within Clade E along with sections Crumenata and Bolbodium. The members form two subclades: the first comprising nine species (BP83, PP100) and the second comprising Dendrobium calceolum. The monophyly of the first subclade was suggested by Yukawa (2001) and Wongsawad et al. (2001, 2005).
Section Crumenata was also found to nest within Clade E and to not be monophyletic. Its species are divided into two subclades: the first including section Bolbodium and parts of section Aporum (BP92, PP100) and the second representing the earliest diverging clade within Clade E, comprising Dendrobium ephemerum. Analyses by Yukawa (2001) and Clements (2003, 2006) also suggested the first of these subclades to be paraphyletic.
Section Distichophyllae was found to not be monophyletic, with its members splitting into a core clade (Clade H) containing the type species of the section, Dendrobium uniflorum, and the earliest diverging clade within Clade K represented by D. oligophyllum, which is likely a sister group to section Stachyobium. Further, in Clade H, two species of section Conostalix, Dendrobium lobbii and D. pachyglossum, were nested within members of section Distichophyllae. Section Conostalix is probably monophyletic (BP53, PP100).
Members of section Formosae were scattered into four lineages: Clade I, Clade J and two unplaced species, D. jerdonianum and D. trigonopus. Clade I, which contains the type species of the section, D. formosum, comprises only species of section Formosae (BP100, PP100). Clade J similarly comprises only species of section Formosae (BP100, PP100). While the possibility that these four groups represent a monophyletic lineage cannot be ruled out, it is likely that they are polyphyletic. Analyses by Wongsawad et al. (2005) and Clements (2006) also identified two clades within Section Formosae corresponding to Clades I and J.
There is a strong possibility that section Stachyobium forms a monophyletic group within Clade K (BP93, PP100). Consistent with our results, monophyly of section Stachyobium was also suggested by analyses by Yukawa (2001), Wongsawad et al. (2001, 2005), Clements (2006) and Xiang et al. (2013). Furthermore, it is evident that section Stachyobium is a sister group to D. oligophyllum in section Distichophyllae (BP99, PP100).
Section Herbaceae comprises only two species, Dendrobium herbaceum and D. parcum. We found that this section is clearly monophyletic because the two species constitute Clade L (BP96, PP100). Similarly, it is highly probable that section Fytchianthe represents a monophyletic lineage because Dendrobium fytchianum and D. ovatum, members of this section, constitute Clade M (BP100, PP100). While not strongly supported in statistical terms (BP < 50, PP59), Clade M may represent the earliest divergent clade within the Asian clade. Similarly supported by our results, analyses by Wongsawad et al. (2001, 2005) also indicate the possibility that section Fytchianthe is the earliest divergent clade within the Asian clade. Our analyses further indicate that sections Herbaceae and/or Stachyobium may represent the second-earliest divergent clade within the Asian clade after section Fytchianthe.
Evolution of morphological characters
Vegetative stems
The vast majority of species belonging to Clades A, D, F, H, I, J, L and M, and several species in Clades C and K have terete, non-succulent vegetative stems. Results of ancestral state reconstruction analysis showed that this character state represents a plesiomorphy of the Asian clade (Fig. 2A).
Species with a pseudobulbous, heteroblastic, succulent stem in which from one to a few internodes on the upper portions of vegetative stems thicken to become pseudobulbs, account for all species in section Bolbodium and some members of sections Densiflora. Among species analysed in this study, the heteroblastic stem is exhibited by Dendrobium pachyphyllum (section Bolbodium) in Clade E, D. lindleyi, D. jenkinsii and D. cf. jenkinsii (all in section Densiflora) in Clade G. Results of ancestral state reconstruction analysis showed that this character state likely evolved twice in the Asian clade (Fig. 2A).
Dendrobium brymerianum, D. chrysocrepis, D. harveyanum (both in section Dendrobium) and D. chrysotoxum (section Densiflora) in Clade A, D. albosanguineum (section Dendrobium) and D. braianense and D. capillipes (both in section Holochrysa) in Clade B, D. cf. griffithianum, D. farmeri, D. griffithianum, D. guibertii and D. palpebrae (all in section Densiflora) in Clade C, all species of section Oxyglossum in Clade D, D. bellatulum (section Formosae) in Clade I, D. delacourii, D. diodon, D. gregulus, D. porphyrochilum, D. laterale and Dendrobium sp. (all in section Stachyobium) in Clade K and D. trigonopus (section Formosae), an unplaced taxon, have a clavate, non-heteroblastic, succulent stem. Results of ancestral state reconstruction analysis indicated that this character state evolved more than 14 times in the Asian clade (Fig. 2A).
All species of section Crumenata, and Dendrobium hancockii and D. cf. hancockii (both in section Dendrobium) exhibit a basally bulged stem characterized by particular thickening of several internodes positioned on lower portions of vegetative stems. Results of ancestral state reconstruction analysis showed that this character state represents an apomorphy, resulting from two independent evolutionary events within the Asian clade (Fig. 2A).
The stems of the majority of members of section Calcarifera have elliptical cross-sections, while the stems of all members of section Platycaulon are entirely flattened. A few species belonging to sections Breviflores and Densiflora also have flattened stems. Among species analysed in this study, in addition to species of section Platycaulon (Clade F), D. trantuanii (section Breviflores) in Clade A and D. sulcatum (section Densiflora) in Clade C also had flattened stems. Results of ancestral state reconstruction analysis showed that this character state evolved three times in the Asian clade (Fig. 2A).
A further apomorphy of the vegetative stem is thin, wiry stems, observed in all species of sections Aporum and Dolichocentrum and a few species of sections Dendrobium, Calcarifera, Conostalix and Pedilonum. Among species investigated in this study, in addition to species of sections Aporum and Dolichocentrum, thin and wiry stems were shared by Dendrobium leptocladum (section Dendrobium) in Clade A, Dendrobium lancifolium (section Calcarifera) and Dendrobium aff. ramosii (section Pedilonum) in Clade D and D. lobbii (section Conostalix) in Clade H. Results of ancestral state reconstruction analysis suggested that this character state evolved more than seven times in the Asian clade (Fig. 2A). Species belonging to section Aporum have succulent leaves, while other species with thin and wiry stems have fleshy roots. This suggests that, in species with thin and wiry stems, the components responsible for water storage have shifted from the stems to the leaves or roots.
Shoot architecture
The majority of species in the genus Dendrobium exhibit an architecture ubiquitously observed in perennial herbs in which vegetative shoots emerge in a repeated sympodial branching pattern from nodes on basal portions of the vegetative stem and produce roots also at the stem base. However, all species of section Herbaceae and a few species of sections Dendrobium, Calcarifera, Dolichocentrum and Crumenata produce vegetative shoots in a sympodial branching pattern from nodes on the upper parts of the stem, forming ramified stems, whereby roots do not grow from basal portions of new shoots. Among the species examined in this study, in addition to species of section Herbaceae, this architecture was observed in Dendrobiuim falconeri and D. hancockii (both in section Dendrobium) in Clade A; D. chameleon (section Calcarifera), D. furcatum (section Dolichocentrum) and D. victoria-reginae (section Calcarifera) in Clade D; and D. junceum (section Crumenata) in Clade E. This character state is not present in Clade M, which likely represents the earliest divergent clade within the Asian clade. In addition, given that this character state is only observed in a small number of species in a few lineages of both Asian and Australasian clades (Wood 2006), it is likely that ramified stems represent an apomorphy for the Asian clade, resulting from at least four distinct evolutionary events.
Although reproductive shoots are produced by axillary branching from vegetative shoots in most Dendrobium species, all species of sections Stachyobium, Herbaceae and Fytchianthe have terminal buds of vegetative shoots that develop into reproductive shoots. In this study, such a terminal inflorescence was observed in all species belonging to Clades M, L and K, which likely represent the earliest divergent clades within the Asian clade. Dressler (1981) proposed that terminal inflorescence is historically primary, being found in the most primitive orchid. Given that members of the genus Epigeneium, which represents the earliest divergence within subtribe Dendrobiinae, also exhibit a terminal inflorescence, terminal inflorescences may represent a plesiomorphy for the Asian clade.
Roots
Given that almost all Asian clade species have roots with a smooth, shiny white surface and that the majority of the sister Australasian clade species also have smooth roots, it appears that smooth roots represent a plesiomorphy for the Asian clade. In contrast, a number of species belonging to Clade F (Dendrobium annae, D. cumulatum, D. hymenophyllum, D. intricatum, D. mutabile, D. panduriferum, D. profusum and D. sanguinolentum in section Calcarifera and D. platygastrium, D. speckmaieri and D. ypsilon in section Platycaulon) have verrucose roots. All members of section Brevisaccata in the Australasian clade and some species of the genus Epigeneium also exhibit this feature. Such roots certainly evolved three times in the subtribe Dendrobiinae. However, this rough surface feature has no known function, except for increasing the absorptive surface of the root (Wood 2006).
Leaves
While almost all species belonging to the Asian clade have conduplicate leaves with both adaxial and abaxial surfaces, all species in section Aporum and some species in section Crumenata have laterally flattened or terete leaves in which the adaxial surface has been reduced, leaving a unifacial leaf consisting only of the abaxial surface. This feature is accompanied by thickening of the mesophyll, and these character states represent a xerophytic adaptation. In fact, the species with unifacial leaves generally grow as epiphytes in dry habitats of tropical Asia (Yukawa and Uehara 1996). Among the species analysed in this study, all species in section Aporum and section Crumenata species Dendrobium acinaciforme, D. macfarlanei, D. goldfinchii, D. philippinense, D. junceum and D. aff. modestum in Clade E exhibit a unifacial leaf. Since evolution of a unifacial leaf occurred only in Clade E in the Asian clade, it represents an apomorphy resulting from one or a few evolutionary events.
Members of the genus Dendrobium exhibit two shedding patterns: a deciduous pattern, in which leaf life-span is shorter than 1 year and stems lose all leaves temporarily, and an evergreen pattern, in which leaf life-span is longer than 1 year and stems retain their leaves at all times. While the vast majority of species are evergreen, all species in sections Herbaceae, Fytchianthe and Stachyobium and some members in sections Dendrobium and Holochrysa are deciduous. Among species investigated in this study, in addition to species of sections Herbaceae, Fytchianthe and Stachyobium, section Dendrobium species D. aphyllum, D. crystallinum, D. pendulum, D. wardianum, D. amoenum, D. anosmum, D. bensoniae, D. crepidatum, D. devonianum, D. gratiosissimum, D. lituiflorum, D. parishii, D. primulinum, D. transparens, D. unicum and D. dixanthum (section Holochrysa) in Clade A, along with D. albosanguineum (section Dendrobium), D. capillipes (section Holochrysa) in Clade B and D. senile (section Dendrobium), with unresolved phylogenetic position, are all deciduous. Furthermore, while members of the Australasian clade as well as Epigeneium and Bulbophyllum, allied genera to Dendrobium, have evergreen leaves, given that species of sections Fytchianthe, Herbaceae and Stachyobium, which likely are the earliest divergent clades within the Asian clade, are deciduous, deciduousness may represent a plesiomorphy for the Asian clade.
Results of ancestral state reconstruction analysis showed that a stem with more than five leaves represents a plesiomorphy of the Asian clade and that reduction of leaf number likely evolved more than 12 times (Fig. 2B). As shown in Fig. 2A and B, the species with less than six leaves on each stem usually exhibit pseudobulbous, heteroblastic, succulent or clavate, non-heteroblastic, succulent stems except for D. aff. densiflorum, D. amabile, D. densiflorum, D. sulcatum and D. thyrsiflorum (all in section Densiflora) in Clade C. Furthermore, leaves of these species are either succulent or deciduous. Yukawa and Uehara (1996) and Wood (2006) suggested that these combinations of character states show adaptation to a xeric environment.
Schlechter (1912), in his infrageneric classification system of Dendrobium, emphasized the presence or absence of leaf sheaths as a cardinal diagnostic character at the subgenus level. In species with the above-mentioned succulent stems producing a small number of leaves in the upper part, leaf sheaths do not develop. However, in species that produce non-succulent stems, leaf sheaths do not develop in the uppermost leaves, but do develop in lower leaves. Consequently, reduction of the leaf sheath in the uppermost leaves is a common character state both in the succulent and non-succulent stem species, and it is not appropriate to use the presence or absence of leaf sheaths as a character for classification at least in the Asian clade.
Anatomical characters of vegetative organs
Yukawa and Uehara (1996) demonstrated that modifications in size and number of parenchymatous cells of vegetative stems substantially contributed to vegetative diversification in Dendrobium. This observation implies that a simple structural adjustment can result in a major modification of growth habit in this group. They also found that several anatomical characters associated with xeromorphy, such as a thick outer wall of epidermal cells on the stem surface and a thick sclerenchymatous cap on vascular bundles in the stem evolved in members of sections Aporum and Crumenata. In other words, these character states are likely to have evolved in the common ancestor of Clade E. Acquisition of xeromorphic anatomical characters in this clade may have facilitated evolution of unique vegetative characters suited to dry environments such as a succulent, laterally flattened leaf and a wiry or basally bulged stem.
Namba and Lin (1981a, b), Singh (1986) and Morris et al. (1996) investigated the anatomical characters of roots of Dendrobium. Morris et al. demonstrated that root characteristics including the number of layers in the velamina are not useful characters to use in determining sectional relationships.
Reproductive organs
All species of section Densiflora and most members of section Dendrobium have velvety lips with many papillae on the surface. Among species analysed in this study, all Clade A species with the exception of D. unicum and D. lamyaiae in section Dendrobium, all Clade B, C and G species, and D. gibsonii exhibit this character state. When Dendrobium flowers are stained with neutral red, which is taken up by osmophores, or odour-producing cells (Stern et al. 1986), the papillae of the above-listed species are strongly stained (Yukawa 1993). Consequently, it is likely that the papillae on the lip surface accumulate substances responsible for fragrance (Müller 1935). Observation of the near-ultraviolet (near-UV) reflectance of the flowers using near-UV reflectance photography reveals that the parts with papillae absorb near-UV, while other areas of the perianth lobes reflect near-UV, resulting in distinct high-contrast patterns (Yukawa 1993; Indsto and Weston 2000). Given that this combination of characteristics is typical of bee-pollinated flowers, we demonstrate that the dense packing of papillae on lip surfaces represents a character state that evolved in adaptation to bee pollination. The only Dendrobium species producing lips with dense papillae for which pollinators have been observed is D. anosmum, which two species of Apis are known to pollinate (Burkill 1919). Since lip surfaces with dense papillae are not observed in species of the Australasian clade or in Clades K, L and M, which likely represent the earliest divergent clades within the Asian clade, this character state was probably acquired after the divergence of the Asian clade. However, it is unclear how many evolutionary events have resulted in the character state, because phylogenetic relationships of Clades A, B, C and G have not been resolved.
Meanwhile, specialization has occurred in terms of flowering behaviour. Reproductive shoots of Dendrobium generally branch sympodially from vegetative shoots and disappear immediately after flowering. However, in all species in Clade E, namely, members of sections Aporum, Bolbodium and Crumenata, the reproductive shoots remain alive even after flowering and continue to flower repeatedly. There is a high probability that the repeated flowering behaviour was acquired by a common ancestor of Clade E. Further, all species of section Bolbodium and some species of sections Crumenata and Aporum in this clade exhibit gregarious flowering in which, after the flower has differentiated, its growth is suspended at the bud stage and all flowers begin to grow and then bloom at once when the temperature decreases to a certain level or when the difference between daytime and night-time temperatures narrows to a certain point (Seifriz 1923; Coster 1925; Gerlach 1992). This behaviour is observed, for example, in D. pachyphyllum (section Bolbodium), D. crumenatum (section Crumenata), D. ephemerum (section Crumenata), D. acerosum (section Aporum) and D. spatella (section Aporum). It is possible that acquisition of repeated flowering in the common ancestor of Clade E provided further character evolution that increased plant attractiveness to pollinators, because it enabled an individual plant to have numerous buds at once. Acquisition of repeated flowering thus may trigger the evolution of gregarious flowering.
Reappraisal of infrageneric classification
There is no single criterion, but rather, several options for deciding what level of monophyletic unit warrants designation as a genus. In order to fulfil the phylogenetic consistency and conservation of a widely accepted concept of the genus Dendrobium, Yukawa et al. (1993, 1996, 1999, 2000), Yukawa (2001), Burke et al. (2008), Adams (2011) and Schuiteman (2011) have recommended expanding the genus Dendrobium to include the genera Cadetia, Diplocaulobium and Flickingeria. In contrast, Clements (2003, 2006) subdivided Dendrobium and established many new genera to recover the monophyly of the genus. The advantages and disadvantages of these two approaches were discussed by Yukawa et al. (1999) and Schuiteman and Adams (2011), who concluded that the former approach has greater merit. Here, we follow the former approach and examine the infrageneric classification system of Dendrobium proposed by Wood (2006).
We demonstrated that a number of sections used by Wood (2006) are not monophyletic. In order to restore the monophyly of these groups, a revised system at the section level is necessary. Clades A through M defined in this study were all strongly supported, indicating that they are stable units whose monophyly will continue to be supported even if the sampling intensity and gene regions analysed were increased. Furthermore, given that considerable numbers of clades roughly correspond to traditionally used sections, it would seem reasonable to assign, for the most part, Clades A through M to the section level.
We found that in Clade A, section Dendrobium is a paraphyletic group comprising all species in sections Breviflores and Stuposa, most species in section Holochrysa and D. chrysotoxum in section Densiflora. If we recognize these heterogeneous elements at the section level, section Dendrobium also must be subdivided. As sections Breviflores, Stuposa, Holochrysa and D. chrysotoxum in Densiflora do not deviate from a broad concept of section Dendrobium, it would be appropriate to redefine section Dendrobium as the range of Clade A and to include sections Breviflores and Stuposa, and most species in section Holochrysa, and D. chrysotoxum of section Densiflora in section Dendrobium. Schuiteman (2011) also suggested that section Breviflores is polyphyletic and should be included in section Dendrobium. Further, Xiang et al. (2013) proposed to subsume these three sections and D. chrysotoxum into section Dendrobium. Our analyses indicated that D. gibsonii (section Holochrysa) and D. senile (section Dendrobium) may form a monophyletic group with Clade A (BP < 50, PP100). At present, it is appropriate to include these two species in section Dendrobium.
Clade B can be characterized by the following combination of characters: clavate, non-heteroblastic, succulent stems; long flowering stems produced from the upper part of vegetative stems; lip with numerous papillae on the adaxial surface; tapered anther cap; and leaf sheath with brownish margins. Among these characters, leaf sheath with brownish margins represents synapomorphy of this clade. While Clade B itself is strongly supported (BP100, PP100), its relationship to other clades remains to be resolved. Therefore, it would seem appropriate to treat Clade B as an independent section. Clade B is comprised of members of sections Dendrobium and Holochrysa, both of whose type species belong to Clade A. Given that no name corresponding to the three species constituting Clade B currently exists, a new section name will have to be assigned.
Clade C comprises sections Densiflora and Amblyanthus, both of which are not monophyletic in the clade. Dendrobium microglaphys of section Amblyanthus occupies a nested position among members of section Densiflora and lacks scales outside the perianth lobes that characterize section Amblyanthus; instead, it has reduced leaf sheaths and pendulous flowering stems, both of which are diagnostic characters of section Densiflora. Consequently, it is reasonable to transfer D. microglaphys to section Densiflora. Meanwhile, the members of the basal subclade of Clade C, which includes Dendrobium melanostictum, the type species of section Amblyanthus, share features such as scales outside the perianth lobes, short racemes, and a peculiar scaly covering of the flowers and a backward-pointing lip appendage. Therefore, this subclade corresponds to section Amblyanthus and the rest of Clade C can be defined as section Densiflora.
Clade D comprises sections Pedilonum, Calcarifera, Oxyglossum, Calyptrochilus and Dolichocentrum. Since sections Pedilonum, Calcarifera, Oxyglossum and Dolichocentrum are not monophyletic, redefinition of these sections is necessary. If we conserve these sections, establishment of a large number of new sections is required. To avoid this complexity, we suggest designating Clade D as a single section. For this purpose, the section names Pedilonum and Calcarifera are not appropriate since their type species belong to Clade F. Therefore, the oldest available section name for Clade D is Calyptrochilus. An elongated mentum characterizes all species in this clade, while this character also defines Clade F and several species in other clades.
Clade E comprises sections Aporum, Crumenata and Bolbodium, the first two of which were found to not be monophyletic. As in the case of Clade D, conserving these two section names requires the establishment of multiple new sections. Thus, it is better to treat Clade E as a single section. The oldest available name corresponding to this clade is section Aporum. Clade E species have the following synapomorphies: a thick outer wall of epidermal cells on the stem surface and a thick sclerenchymatous cap on vascular bundles in the stem (Yukawa and Uehara 1996), and repeated flowering behaviour from the same reproductive shoot. Schuiteman (2011) also suggested that sections Aporum, Crumenata and Bolbodium should be treated as a single section.
Clade F comprises sections Pedilonum, Calcarifera and Platycaulon, the first two of which were found to not be monophyletic. Again, to avoid complexity by naming many new sections, it is reasonable to designate Clade F as a single section. The oldest available name corresponding to this clade is section Pedilonum. A shared character for Clade F is an elongated mentum, but this character also appeared in other clades, as mentioned previously.
The three species constituting Clade G have previously been treated as section Densiflora. Since we redefine section Densiflora as a subclade in Clade C where the type species is included, this section name cannot be used in Clade G. While the monophyly of this clade is strongly supported (BP100, PP100), its relationship to other clades remains to be resolved. Consequently, it is appropriate to deal with Clade G as an independent section. Given that no name corresponding to the three species constituting Clade G currently exists, a new section name will have to be assigned. Xiang et al. (2013) also suggested a separate status for a clade comprising D. jenkinsii and D. lindleyi. Members of this clade share the following combination of characters: pseudobulbous, heteroblastic, succulent stem, single leaf on the stem, and a few long flowering stems from the apical part of the vegetative stem. Among these characters, a single leaf represents the synapomorphy of this clade.
Clade H consists of members of sections Distichophyllae and Conostalix. As the latter section is nested within the first, the first section becomes paraphyletic. Except for thinner vegetative stems in section Conostalix, there is no obvious character to distinguish the two sections. Therefore, we deem it appropriate to redefine section Distichophyllae as the range of Clade H.
Clades I and J as well as unplaced species D. jerdonianum and D. trigonopus compose section Formosae. In addition to the definite monophyletic status of Clades I and J, it cannot be ruled out that some or all clades of this section along with section Distichophyllae form a monophyletic group. Since further analyses may resolve the ambiguous relationships of these clades, we suspend taxonomic treatment of section Formosae for the moment.
Clade K comprises section Stachyobium and D. oligophyllum of section Distichophyllae, which is the earliest divergent lineage in this clade. Re-examination of D. oligophyllum showed that the morphological characters of this species are consistent with the definition of section Stachyobium except for the long life-span of the leaf in this species. We thus transfer D. oligophyllum to section Stachyobium, and by this treatment, Clade K coincides with section Stachyobium. While the species of section Stachyobium share terminal inflorescences, we did not identify any synapomorphies of this section.
Clades L and M correspond to sections Herbaceae and Fytchianthe, respectively, endorsing the validity of the current definition of these sections. The species of section Herbaceae are characterized by ramified stems, terminal inflorescences and deciduous leaves. The species of section Fytchianthe are characterized by terminal inflorescences and deciduous leaves.
The scope of this study did not include the Australasian clade, the other major clade in genus Dendrobium. While a number of molecular phylogenetic studies have been conducted for this clade as well (Y. T. unpubl. res.; Yukawa et al. 1993, 1996, 2000; Yukawa 2001; Clements 2003, 2006; Burke et al. 2008), no attempt has been made to comprehensively analyse this clade as a whole. A consistent infrageneric classification of this genus should be proposed after combining the results of a phylogenetic analysis of both Asian and Australasian clades.
Sources of Funding
This work was partially supported by a Grant-in-Aid for Scientific Research (24370040 to T.Y.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Contributions by the Authors
T.Y., T.T., T.H., S.K. and H.I. conceived and designed the study. T.Y., T.T., P.W., A.S., T.N., S.S., S.K. and N.S. performed sequence and phylogenetic analyses. T.Y. and T.T. primarily wrote the manuscript with contributions from all the authors.
Conflicts of Interest Statement
None declared.
Supporting Information
The following Supporting Information is available in the online version of this article –
Figure S1. Consensus phylogram obtained from 100 754 Bayesian trees based on ITS sequences. Values below and above branches indicate percentage bootstrap values from maximum parsimony analysis and Bayesian posterior probabilities, respectively.
Figure S2. Consensus phylogram obtained from 119 935 Bayesian trees based on matK sequences. Values below and above branches indicate percentage bootstrap values from maximum parsimony analysis and Bayesian posterior probabilities, respectively.
Acknowledgements
We thank H. Okuizumi and D. Hirose for critical comments, K. Suzuki and K. Komura for plant cultivation, and T. Fujimoto, K. Kita, T. Sobu, K. Murase, Y. Matsuo and T. Yamada for technical support.
Appendix 1.
List of Dendrobium taxa and GenBank IDs.
| Species | Section | Voucher | ITS | matK |
|---|---|---|---|---|
| D. acerosum | Aporum | Yukawa04-32 | AB847639 | AB847678 |
| D. acinaciforme | Crumenata | TBG120483 | AB847640 | AB847679 |
| D. aduncum | Breviflores | TBG134842 | AB593484 | AB847680 |
| D. aff. anosmum | Dendrobium | Ting H19 | AB593490 | AB847686 |
| D. aff. densiflorum | Densiflora | TBG159449 | AB593485 | AB847681 |
| D. aff. infundibulum | Formosae | TBG128905 | AB593486 | AB847682 |
| D. aff. modestum | Crumenata | TBG119268 | AB847642 | AB847683 |
| D. aff. moniliforme | Dendrobium | TBG124453 | AB593488 | AB847684 |
| D. aff. ramosii | Pedilonum | HBG4335 | AB593492 | AB847687 |
| D. aff. signatum | Dendrobium | TBG118279 | AB593489 | AB847685 |
| D. alaticaulinum | Pedilonum | TBG135586 | AB593493 | AB847688 |
| D. albosanguineum | Dendrobium | TBG122507 | AB593494 | AB847689 |
| D. amabile | Densiflora | TBG111421 | AB593495 | AB847690 |
| D. amethystoglossum | Calcarifera | TBG120501 | AB593496 | AB847691 |
| D. amoenum | Dendrobium | TBG159411 | AB593497 | AB847692 |
| D. annae | Calcarifera | TBG111422 | AB593498 | AB847693 |
| D. anosmum | Dendrobium | TBG118511 | AB593499 | AB847694 |
| D. aphanochilum | Calyptrochilus | TBG142252 | AB593500 | AB847695 |
| D. aphrodite | Dendrobium | TBG122797 | AB593501 | AB847696 |
| D. aphyllum | Dendrobium | TBG122508 | AB593539 | AB847736 |
| D. atavus | Holochrysa | TBG159413 | AB593502 | AB847697 |
| D. auriculatum | Dolichocentrum | TBG120492 | AB593503 | AB847698 |
| D. austrocaledonicum | Distichophyllae | TBG130055 | AB593504 | AB847699 |
| D. auyongii | Aporum | TBG123341 | AB847643 | AB847700 |
| D. bellatulum | Formosae | TBG133256 | AB847644 | AB847701 |
| D. bensoniae | Dendrobium | TBG128899 | AB593505 | AB847702 |
| D. bifarium | Distichophyllae | TBG159414 | AB593506 | AB847703 |
| D. bifurcatum | Breviflores | TBG157293 | AB593507 | AB847704 |
| D. bracteosum | Pedilonum | TBG102743 | AB593509 | AB847705 |
| D. braianense | Holochrysa | TBG159424 | AB593510 | AB847706 |
| D. brymerianum | Dendrobium | TBG118826 | AB593511 | AB847707 |
| D. calcaratum | Pedilonum | TBG125164 | AB593512 | AB847708 |
| D. calceolum | Aporum | TBG122494 | AB593513 | AB847709 |
| D. caliculi-mentum | Pedilonum | TBG118610 | AB593514 | AB847710 |
| D. capillipes | Holochrysa | TBG128878 | AB593515 | AB847711 |
| D. cariniferum | Formosae | TBG128900 | AB847645 | AB847712 |
| D. catenatum | Dendrobium | TBG159450 | AB593517 | AB847713 |
| D. catillare | Pedilonum | TBG124769 | AB847646 | AB847714 |
| D. cf. bensoniae | Dendrobium | 036193-B | AB847647 | AB847715 |
| D. cf. candidum | Dendrobium | 29746 | AB593519 | AB847716 |
| D. cf. gratiosissimum | Dendrobium | TBG126713 | AB593522 | AB847719 |
| D. cf. griffithianum | Densiflora | TBG128876 | AB593523 | AB847720 |
| D. cf. hancockii | Dendrobium | TBG142251 | AB593524 | AB847721 |
| D. cf. jenkinsii | Densiflora | TBG129904 | AB593525 | AB847722 |
| D. cf. microglaphys | Amblyanthus | TBG116027 | AB593526 | AB847723 |
| D. cf. primulinum | Dendrobium | TBG118293 | AB593521 | AB847718 |
| D. cf. roseipes | Pedilonum | TBG159460 | AB847648 | AB847717 |
| D. chameleon | Calcarifera | TBG135506 | AB593527 | AB847724 |
| D. chrysanthum | Dendrobium | TBG124319 | AB593529 | AB847725 |
| D. chryseum | Holochrysa | TBG127382 | AB593530 | AB847726 |
| D. chrysocrepis | Dendrobium | TBG128879 | AB593531 | AB847727 |
| D. chrysotoxum | Densiflora | TBG118321 | AB593533 | AB847728 |
| D. codonosepalum | Calyptrochilus | TBG137050 | AB847649 | AB847729 |
| D. compactum | Stachyobium | TBG133060 | AB847650 | AB847730 |
| D. crepidatum | Dendrobium | TBG128871 | AB593534 | AB847731 |
| D. cruentum | Formosae | TBG134572 | AB593536 | AB847733 |
| D. crumenatum | Crumenata | TBG115833 | AB593537 | AB847734 |
| D. crystallinum | Dendrobium | TNS8502972 | AB593538 | AB847735 |
| D. cumulatum | Calcarifera | TBG159418 | AB593541 | AB847737 |
| D. cuthbertsonii | Oxyglossum | TBG116108 | AB593542 | AB847738 |
| D. cyanocentrum | Oxyglossum | TBG159420 | AB593543 | AB847739 |
| D. dearei | Formosae | TBG144587 | AB847651 | AB847740 |
| D. delacourii | Stachyobium | TBG100237 | AB593545 | AB847741 |
| D. densiflorum | Densiflora | TBG159421 | AB593546 | AB847742 |
| D. denudans | Stachyobium | TBG132760 | AB593547 | AB847743 |
| D. devonianum | Dendrobium | TBG124383 | AB593548 | AB847744 |
| D. diodon | Stachyobium | TBG116089 | AB593550 | AB847745 |
| D. distichum | Aporum | TBG137051 | AB593551 | AB847746 |
| D. dixanthum | Holochrysa | TBG128877 | AB593552 | AB847747 |
| D. draconis | Formosae | TBG118276 | AB593553 | AB847748 |
| D. ellipsophyllum | Distichophyllae | TBG126635 | AB593554 | AB847749 |
| D. ephemerum | Crumenata | TBG159426 | AB593555 | AB847750 |
| D. eriiflorum | Stachyobium | TBG140557 | AB593556 | AB847751 |
| D. erosum | Calyptrochilus | TBG159429 | AB593557 | AB847752 |
| D. eserre | Stachyobium | TBG133168 | AB593558 | AB847753 |
| D. faciferum | Crumenata | YukawaDNA2134 | AB847652 | AB847754 |
| D. fairchildiae | Calcarifera | TBG120506 | AB593559 | AB847755 |
| D. falconeri | Dendrobium | TBG128903 | AB593560 | AB847756 |
| D. farmeri | Densiflora | TBG159430 | AB593561 | AB847757 |
| D. fimbriatum | Holochrysa | 84386 | AB593562 | AB847758 |
| D. findlayanum | Dendrobium | TBG128904 | AB593563 | AB847759 |
| D. formosum | Formosae | TBG128902 | AB593564 | AB847760 |
| D. friedericksianum | Dendrobium | TBG124322 | AB593565 | AB847761 |
| D. furcatum | Dolichocentrum | TBG116070 | AB593566 | AB847762 |
| D. fytchianum | Fytchianthe | TBG128868 | AB593567 | AB847763 |
| D. gibsonii | Holochrysa | TBG129876 | AB593568 | AB847764 |
| D. glomeratum | Calyptrochilus | TBG138022 | AB593535 | AB847732 |
| D. goldfinchii | Crumenata | Yukawa 97-2008 | AB593569 | AB847765 |
| D. goldschmidtianum | Pedilonum | TBG159434 | AB593570 | AB847766 |
| D. gratiosissimum | Dendrobium | TBG126651 | AB593571 | AB847767 |
| D. gregulus | Stachyobium | TBG132862 | AB593572 | AB847768 |
| D. griffithianum | Densiflora | TBG138067 | AB593573 | AB847769 |
| D. guibertii | Densiflora | TBG132542 | AB593574 | AB847770 |
| D. hancockii | Dendrobium | TBG122506 | AB593575 | AB847771 |
| D. harveyanum | Dendrobium | TBG133184 | AB593576 | AB847772 |
| D. hasseltii | Pedilonum | TBG136022 | AB593577 | AB847773 |
| D. hemimelanoglossum | Stachyobium | TBG133257 | AB593578 | AB847774 |
| D. henryi | Holochrysa | TBG127415 | AB593579 | AB847775 |
| D. herbaceum | Herbaceae | TBG156719 | AB847654 | AB847776 |
| D. hercoglossum_1 | Breviflores | TBG118850 | AB593580 | AB847777 |
| D. hercoglossum_2 | Breviflores | TBG124432 | AB593581 | AB847778 |
| D. heterocarpum | Dendrobium | TBG116506 | AB593582 | AB847779 |
| D. hookerianum | Dendrobium | TBG133007 | AB593584 | AB847780 |
| D. hymenophyllum | Calcarifera | TBG140599 | AB847655 | AB847781 |
| D. intricatum | Calcarifera | TBG137294 | AB593586 | AB847782 |
| D. jacobsonii | Pedilonum | TBG133080 | AB593587 | AB847783 |
| D. jenkinsii | Densiflora | TBG129880 | AB593589 | AB847784 |
| D. jerdonianum | Formosae | TBG156717 | AB847656 | AB847785 |
| D. junceum | Crumenata | TBG119079 | AB593590 | AB847786 |
| D. kentrophyllum | Aporum | TBG102615 | AB593591 | AB847787 |
| D. kontumense | Formosae | TBG126642 | AB593592 | AB847788 |
| D. kraemeri | Pedilonum | TBG123234 | AB847657 | AB847789 |
| D. kratense | Stachyobium | TBG126712 | AB847658 | AB847790 |
| D. laevifolium | Oxyglossum | TBG142428 | AB593593 | AB847791 |
| D. lamyaiae | Dendrobium | TBG116046 | AB593595 | AB847792 |
| D. lancifolium | Calcarifera | TBG142427 | AB847659 | AB847793 |
| D. laterale | Stachyobium | TNS8501719 | AB847660 | AB847794 |
| D. lawesii | Calyptrochilus | TBG140275 | AB593596 | AB847795 |
| D. leonis | Aporum | 84206 | AB593597 | AB847796 |
| D. leptocladum | Dendrobium | TNS8502939 | AB593598 | AB847797 |
| D. linawianum | Dendrobium | TBG142429 | AB593599 | AB847798 |
| D. lindleyi | Densiflora | TBG115834 | AB593600 | AB847799 |
| D. linguella | Breviflores | TBG115831 | AB593601 | AB847800 |
| D. lituiflorum | Dendrobium | TBG128908 | AB593602 | AB847801 |
| D. lobbii | Conostalix | NSW426014 | AB593603 | AB847802 |
| D. loddigesii | Dendrobium | 74531 | AB593604 | AB847803 |
| D. lohohense | Holochrysa | TBG132863 | AB593605 | AB847804 |
| D. longicornu | Formosae | TBG122802 | AB847661 | AB847805 |
| D. lowii | Formosae | TBG134848 | AB593606 | AB847806 |
| D. luteolum | Dendrobium | TBG122798 | AB593607 | AB847807 |
| D. maccarthiae | Dendrobium | TBG128803 | AB593608 | AB847808 |
| D. macfarlanei | Crumenata | TBG122496 | AB847662 | AB847809 |
| D. macrostachyum | Dendrobium | TBG123340 | AB847663 | AB847810 |
| D. masarangense subsp. theionanthum | Oxyglossum | Tajima 27 | AB593609 | AB847811 |
| D. melanostictum | Amblyanthus | TBG123873 | AB593610 | AB847812 |
| D. melinanthum | Calyptrochilus | TBG159436 | AB593611 | AB847813 |
| D. microglaphys | Amblyanthus | TBG135481 | AB593612 | AB847814 |
| D. mohlianum | Calyptrochilus | TBG124783 | AB593613 | AB847815 |
| D. moniliforme | Dendrobium | TBG115845 | AB593614 | AB847816 |
| D. morrisonii | Pedilonum | Yukawa 97-2045 | AB847664 | AB847817 |
| D. moschatum | Holochrysa | 56113 | AB593616 | AB847818 |
| D. mutabile | Calcarifera | TBG159437 | AB593617 | AB847819 |
| D. nemorale | Distichophyllae | TBG159440 | AB593618 | AB847820 |
| D. nobile | Dendrobium | TBG128809 | AB593619 | AB847821 |
| D. ochreatum | Dendrobium | TBG128865 | AB593621 | AB847822 |
| D. okinawense | Dendrobium | TBG115938 | AB593622 | AB847823 |
| D. oligophyllum | Distichophyllae | TBG128728 | AB847665 | AB847824 |
| D. pachyglossum | Conostalix | TBG124380 | AB593623 | AB847825 |
| D. pachyphyllum | Bolbodium | TBG126623 | AB593624 | AB847826 |
| D. palpebrae_1 | Densiflora | TBG159441 | AB593625 | AB847827 |
| D. palpebrae_2 | Densiflora | TBG159442 | AB593626 | AB847828 |
| D. palpebrae_3 | Densiflora | TBG118313 | AB593627 | AB847829 |
| D. panduriferum | Calcarifera | TBG144531 | AB847666 | AB847830 |
| D. papilio | Dolichocentrum | TBG120493 | AB847667 | AB847831 |
| D. parciflorum | Aporum | TBG118261 | AB593628 | AB847832 |
| D. parcum | Herbaceae | TBG129884 | AB593629 | AB847833 |
| D. parishii | Dendrobium | TBG159443 | AB593630 | AB847834 |
| D. parthenium | Formosae | TBG136015 | AB847668 | AB847835 |
| D. patentilobum | Aporum | TBG137060 | AB847669 | AB847836 |
| D. pendulum | Dendrobium | TBG128867 | AB593633 | AB847837 |
| D. philippinense | Crumenata | TBG142250 | AB593634 | AB847838 |
| D. platygastrium | Platycaulon | TBG142432 | AB593635 | AB847839 |
| D. polyanthum | Dendrobium | TBG159417 | AB593636 | AB847840 |
| D. porphyrochilum | Stachyobium | TBG141468 | AB593637 | AB847841 |
| D. praecinctum | Stuposa | TBG116030 | AB593638 | AB847842 |
| D. prasinum | Oxyglossum | TBG119108 | AB593639 | AB847843 |
| D. prianganense | Calcarifera | TBG134835 | AB593640 | AB847844 |
| D. primulinum | Dendrobium | TBG159445 | AB593641 | AB847845 |
| D. profusum | Calcarifera | TBG133266 | AB593642 | AB847846 |
| D. pulchellum | Holochrysa | TBG118088 | AB593643 | AB847847 |
| D. rarum | Pedilonum | TBG124851 | AB593644 | AB847848 |
| D. regium | Dendrobium | TNS8501314 | AB593645 | AB847849 |
| D. rhombeum_1 | Dendrobium | TBG135976 | AB593646 | AB847850 |
| D. rhombeum_2 | Dendrobium | TBG142436 | AB593647 | AB847851 |
| D. roseiodorum | Formosae | TBG118270 | AB593648 | AB847852 |
| D. roseipes | Pedilonum | TBG142437 | AB593649 | AB847853 |
| D. rosellum | Aporum | TBG133320 | AB593650 | AB847854 |
| D. ruckeri | Dendrobium | TBG159446 | AB593651 | AB847855 |
| D. sanderae | Formosae | TBG120503 | AB593654 | AB847856 |
| D. sanguinolentum | Calcarifera | TBG159447 | AB593655 | AB847857 |
| D. scabrilingue | Formosae | TBG119087 | AB593656 | AB847858 |
| D. schuetzei | Formosae | TBG119088 | AB593658 | AB847859 |
| D. scoriarum | Dendrobium | YukawaDNA0381 | AB593659 | AB847860 |
| D. secundum | Pedilonum | TBG100260 | AB593660 | AB847862 |
| D. senile | Dendrobium | TBG119090 | AB593661 | AB847863 |
| D. signatum | Dendrobium | TBG133598 | AB593662 | AB847864 |
| D. singkawangense | Formosae | TBG141081 | AB593663 | AB847865 |
| D. smillieae | Pedilonum | TBG1402699 | AB593664 | AB847866 |
| D. sp. | Stachyobium | YukawaDNA0749 | AB847670 | AB847861 |
| D. spatella | Aporum | TBG84203 | AB847671 | AB847867 |
| D. speckmaieri | Platycaulon | TBG136027 | AB593665 | AB847868 |
| D. spectatissimum | Formosae | TBG133877 | AB593666 | AB847869 |
| D. stockeri | Amblyanthus | TBG130082 | AB593667 | AB847870 |
| D. stuposum | Stuposa | TBG134843 | AB593668 | AB847871 |
| D. sulcatum | Densiflora | TBG159451 | AB593670 | AB847872 |
| D. sutepense | Formosae | TNS8503543 | AB593671 | AB847873 |
| D. suzukii | Formosae | TNS9518319 | AB593672 | AB847874 |
| D. tetrachromum | Dendrobium | TBG137068 | AB847672 | AB847875 |
| D. thyrsiflorum | Densiflora | TBG159453 | AB593674 | AB847876 |
| D. tobaense | Formosae | TBG126681 | AB593677 | AB847877 |
| D. tortile | Dendrobium | TBG119099 | AB593678 | AB847878 |
| D. trankimianum | Formosae | TBG127512 | AB847673 | AB847879 |
| D. transparens | Dendrobium | TBG126731 | AB593679 | AB847880 |
| D. trantuanii | Breviflores | TBG157296 | AB847674 | AB847881 |
| D. trichostomum | Calyptrochilus | TBG142265 | AB593680 | AB847882 |
| D. trigonopus | Formosae | TBG119100 | AB847675 | AB847883 |
| D. unicum | Dendrobium | TBG119101 | AB593682 | AB847884 |
| D. uniflorum | Distichophyllae | TBG120499 | AB593683 | AB847885 |
| D. venustum | Stachyobium | TBG129885 | AB847676 | AB847886 |
| D. victoriae-reginae | Calcarifera | TBG141156 | AB593684 | AB847887 |
| D. violaceum | Oxyglossum | TBG100203 | AB593685 | AB847888 |
| D. wardianum | Dendrobium | TBG126641 | AB593686 | AB847889 |
| D. ypsilon | Platycaulon | TBG118514 | AB593688 | AB847890 |
Literature Cited
- Adams PB. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Botanical Journal of the Linnean Society. 2011;166:105–126. [Google Scholar]
- Barkman TJ, Lim SH, Salleh KM, Nais J. Mitochondrial DNA sequences reveal the photosynthetic relatives of Rafflesia, the world's largest flower. Proceedings of the National Academy of Sciences of the USA. 2004;101:787–792. doi: 10.1073/pnas.0305562101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham G, Hooker JD. Genera plantarum 3. London: Reeve and Co; 1883. [Google Scholar]
- Brieger FG. Subtribus Dendrobiinae. In: Brieger FG, Maatsch R, Sanghas K, editors. R. Schlechter Die orchideen. 3rd edn. Berlin: Parey; 1981. pp. 636–752. (11–12) [Google Scholar]
- Burke JM, Bayly MJ, Adams PB, Ladiges PY. Molecular phylogenetic analysis of Dendrobium (Orchidaceae), with emphasis on the Australian section Dendrocoryne, and implications for generic classification. Australian Systematic Botany. 2008;21:1–14. [Google Scholar]
- Burkill IH. Some notes on the pollination of flowers in the Botanic Gardens, Singapore, and other parts of the Malay Peninsula. Gardens’ Bulletin of the Straits Settlements. 1919;2:165–176. [Google Scholar]
- Chase MW, de Bruijn AY, Cox AV, Reeves G, Rudall PJ, Johnson MAT, Eguiarte LE. Phylogenetics of Asphodelaceae (Asparagales): an analysis of plastid rbcL and trnL-F DNA sequences. Annals of Botany. 2000;86:935–951. [Google Scholar]
- Clements MA. Molecular phylogenetic systematics in the Dendrobiinae (Orchidaceae), with emphasis on Dendrobium section Pedilonum. Telopea. 2003;10:247–298. [Google Scholar]
- Clements MA. Molecular phylogenetic systematics in Dendrobieae (Orchidaceae) Aliso. 2006;22:465–480. [Google Scholar]
- Coster L. Periodische Blutenerscheinungen in der Tropen. Annales du Jardin Botanique Buitenzorg. 1925;35:125–162. [Google Scholar]
- DeBry RW, Olmstead RG. A simulation study of reduced tree-search effort in bootstrap resampling analysis. Systematic Biology. 2000;49:171–179. doi: 10.1080/10635150050207465. [DOI] [PubMed] [Google Scholar]
- Dressler RL. The orchids: natural history and classification. Cambridge: Harvard University Press; 1981. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. Constructing a significance test for incongruence. Systematic Biology. 1995;44:570–572. [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Gerlach WWP. Flowering behaviour of ephemeral orchids of Western Samoa. 1. Flowering periodicity. Gartenbauwissenschaft. 1992;57:219–222. [Google Scholar]
- Hidayat T, Yukawa T, Ito M. Molecular phylogenetics of subtribe Aeridinae (Orchidaceae): insights from plastid matK and nuclear ribosomal ITS sequences. Journal of Plant Research. 2005;118:271–284. doi: 10.1007/s10265-005-0217-3. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Indsto JO, Weston PH. Near-ultraviolet reflectance in Dendrobium (Orchidaceae) In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Collingwood: CSIRO; 2000. pp. 326–334. [Google Scholar]
- Johnson LA, Soltis DE. Assessing congruence: empirical examples from molecular data. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants II: DNA sequencing. Boston: Kluwer Academic Publishers; 1998. pp. 297–348. [Google Scholar]
- Kim JH, Kim DK, Forest F, Fay MF, Chase MW. Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Annals of Botany. 2010;106:775–790. doi: 10.1093/aob/mcq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kränzlin F. Orchidaceae–Monandrae–Dendrobiinae I. In: Engler A, editor. Das Pflanzenreich. Heft 45. Leipzing: Wilhelm Engelmann; 1910. [Google Scholar]
- Lindley J. The genera and species of orchidaceous plants. XLVII. Dendrobium. London: Ridgeways; 1830. pp. 75–92. [Google Scholar]
- Maddison WP, Maddison DR. 2011. Mesquite 2.75: a modular system for evolutionary analysis http://mesquiteproject.org .
- Martínez-Azorín M, Crespo MB, Juan A, Fay MF. Molecular phylogenetics of subfamily Ornithogaloideae (Hyacinthaceae) based on nuclear and plastid DNA regions, including a new taxonomic arrangement. Annals of Botany. 2011;107:1–37. doi: 10.1093/aob/mcq207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MW, Stern WL, Judd WS. Vegetative anatomy and systematic of subtribe Dendrobiinae (Orchidaceae) Botanical Journal of the Linnean Society. 1996;120:89–144. [Google Scholar]
- Mort ME, Soltis PS, Soltis DE, Mabry ML. Comparison of three methods for estimating internal support on phylogenetic trees. Systematic Biology. 2000;49:160–171. doi: 10.1080/10635150050207456. [DOI] [PubMed] [Google Scholar]
- Müller L. Zur Kenntnis der Ölausscheidungen bei Orchideenblüten. Berichte der Deutschen Botanischen Gesellschaft. 1935;53:349–358. [Google Scholar]
- Namba T, Lin CC. Pharmacognostical studies on the crude drugs of Orchidaceae from Taiwan: 4. On ‘Chioh-hak’ (1) Shoyakugaku Zasshi. 1981a;35:221–232. [Google Scholar]
- Namba T, Lin CC. Pharmacognostical studies on the crude drugs of Orchidaceae from Taiwan: 5. On ‘Chioh-hak’ (2) Shoyakugaku Zasshi. 1981b;35:233–250. [Google Scholar]
- Nylander JAA. MRMODELTEST v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schlechter FRR. Die Orchidaceen von Deutsch-Neu-Guinea. Repertorium Specierum Novarum Regni Vegetabilis, Beihefte 1; , Dahlem bei Berlin, Germany. Koeltz; 1912. [Google Scholar]
- Schuiteman A. Dendrobium (Orchidaceae): to split or not to split? Gardens’ Bulletin Singapore. 2011;63:245–257. [Google Scholar]
- Schuiteman A, Adams PB. New combinations in Dendrobium (Orchidaceae) Muelleria. 2011;29:62–68. [Google Scholar]
- Seidenfaden G. Orchid genera in Thailand: 12. Dendrobium Sw. Opera Botanica. 1985;83:1–296. [Google Scholar]
- Seifriz W. The gregarious flowering of the orchid Dendrobium crumenatum. American Journal of Botany. 1923;10:32–37. [Google Scholar]
- Singh H. Anatomy of root in some Orchidaceae. Acta Botanica Indica. 1986;14:24–32. [Google Scholar]
- Soltis DE, Soltis PS, Doyle JJ. Molecular systematics of plants II: DNA sequencing. Boston: Kluwer Academic Publishers; 1998. [Google Scholar]
- Stern WL, Curry KJ, Whitten WM. Staining fragrance glands in orchid flowers. Bulletin of the Torrey Botanical Club. 1986;113:288–297. [Google Scholar]
- Sun Y, Skinner DZ, Liang GH, Hulbert SH. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics. 1994;89:26–32. doi: 10.1007/BF00226978. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4.0b10. Sunderland: Sinauer Associates, Inc; 2002. [Google Scholar]
- Takamiya T, Wongsawad P, Tajima N, Shioda N, Lu JF, Wen CL, Wu JB, Handa T, Iijima H, Kitanaka S, Yukawa T. Identification of Dendrobium species used for herbal medicines based on ribosomal DNA internal transcribed spacer sequence. Biological and Pharmaceutical Bulletin. 2011;34:779–782. doi: 10.1248/bpb.34.779. [DOI] [PubMed] [Google Scholar]
- Takamiya T, Kitamura S, Suzuki S, Shioda N, Furukawa M, Makino M, Matsuzaki K, Kitanaka S, Yukawa T, Iijima H. Search for potential medicinal resources in genus Dendrobium by analyses of genetic and chemical similarity. In: Ikeda T, ed. Proceedings of 11th Asia Pacific Orchid Conference; 2013. pp. 256–262. Okinawa, Japan. [Google Scholar]
- Wongsawad P, Handa T, Yukawa T. Molecular phylogeny of Dendrobium section Dendrobium. In: Nagata H, Ichihashi S, editors. Proceedings of Seventh Asia Pacific Orchid Conference. 2001. pp. 209–210. Nagoya, Japan. [Google Scholar]
- Wongsawad P, Handa T, Yukawa T. Molecular phylogeny of Dendrobium Callista-Dendrobium complex. In: Nair H, Arditti J, editors. Proceedings of the 17th World Orchid Conference, Shah Alam 2001. Kota Kinabalu: Natural History Publications (Borneo); 2005. pp. 131–133. [Google Scholar]
- Wood HP. The Dendrobium. Liechtenstein: A. R. G. Gantner Ruggell; 2006. [Google Scholar]
- Xiang XG, Schuiteman A, Li DZ, Huang WC, Chung SW, Li JW, Zhou HL, Jin WT, Lai YJ, Li ZY, Jin XH. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Molecular Phylogenetics and Evolution. 2013;69:950–960. doi: 10.1016/j.ympev.2013.06.009. [DOI] [PubMed] [Google Scholar]
- Yukawa T. Japan: Chiba University; 1993. Chloroplast DNA phylogeny and character evolution of the subtribe Dendrobiinae (Orchidaceae) PhD Thesis. [Google Scholar]
- Yukawa T. Molecular phylogeny of Dendrobium. In: Nagata H, Ichihashi S, eds. Proceedings of Seventh Asia Pacific Orchid Conference; Japan. Nagoya: 2001. pp. 69–71. [Google Scholar]
- Yukawa T, Uehara K. Vegetative diversification and radiation in subtribe Dendrobiinae (Orchidaceae): evidence from chloroplast DNA phylogeny and anatomical characters. Plant Systematics and Evolution. 1996;201:1–14. [Google Scholar]
- Yukawa T, Kurita S, Nishida M, Hasebe M. Phylogenetic implications of chloroplast DNA restriction site variation in subtribe Dendrobiinae (Orchidacaeae) Lindleyana. 1993;8:211–221. [Google Scholar]
- Yukawa T, Ohba H, Cameron KM, Chase MW. Chloroplast DNA phylogeny of subtribe Dendrobiinae (Orchidaceae): insights from a combined analysis based on rbcL sequences and restriction site variation. Journal of Plant Research. 1996;109:169–176. [Google Scholar]
- Yukawa T, Koga S, Handa T. DNA uncovers paraphyly of Dendrobium (Orchidaceae) In: Andrews S, Leslie L, Alexander C, editors. Taxonomy of cultivated plants, Third International Symposium. Kew: Royal Botanic Gardens; 1999. pp. 351–354. [Google Scholar]
- Yukawa T, Kita K, Handa T. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae) In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne: CSIRO; 2000. pp. 465–471. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.