Abstract
Iron–sulfur (Fe–S) cluster metalloproteins conduct essential functions in nearly all contemporary forms of life. The nearly ubiquitous presence of Fe–S clusters and the fundamental requirement for Fe–S clusters in both aerobic and anaerobic Archaea, Bacteria, and Eukarya suggest that these clusters were likely integrated into central metabolic pathways early in the evolution of life prior to the widespread oxidation of Earth’s atmosphere. Intriguingly, Fe–S cluster-dependent metabolism is sensitive to disruption by oxygen because of the decreased bioavailability of ferric iron as well as direct oxidation of sulfur trafficking intermediates and Fe–S clusters by reactive oxygen species. This fact, coupled with the ubiquity of Fe–S clusters in aerobic organisms, suggests that organisms evolved with mechanisms that facilitate the biogenesis and use of these essential cofactors in the presence of oxygen, which gradually began to accumulate around 2.5 billion years ago as oxygenic photosynthesis proliferated and reduced minerals that buffered against oxidation were depleted. This review highlights the most ancient of the Fe–S cluster biogenesis pathways, the Suf system, which likely was present in early anaerobic forms of life. Herein, we use the evolution of the Suf pathway to assess the relationships between the biochemical functions and physiological roles of Suf proteins, with an emphasis on the selective pressure of oxygen toxicity. Our analysis suggests that diversification into oxygen-containing environments disrupted iron and sulfur metabolism and was a main driving force in the acquisition of accessory Suf proteins (such as SufD, SufE, and SufS) by the core SufB–SufC scaffold complex. This analysis provides a new framework for the study of Fe–S cluster biogenesis pathways and Fe–S cluster-containing metalloenzymes and their complicated patterns of divergence in response to oxygen.
Metals and Evolution of the Biosphere
The bioavailability of essential transition metals such as copper (Cu), iron (Fe), molybdenum (Mo), nickel (Ni), and zinc (Zn) has varied substantially over geological and evolutionary time.1,2 Most researchers agree that the variation in metal availability through geological history has been driven largely by the advent of oxygenic photosynthesis >2.45 billion years ago (Ga)3−5 and concomitant increases in the oxidative weathering of continental sulfide minerals and delivery of these weathering products to ocean basins.2,6,7 Importantly, on geological time scales, the bulk oxidation state of oceans is determined by the ratio of photosynthetic oxygen production to the sum of biological respiration and chemical reducing equivalents.8 The oxidation state of oceans was critical to the bioavailability of metals given the differential behavior of metals in their oxidized and reduced states. While some metals such as Mo have decreased solubility under anoxic conditions, in particular when in complex with sulfide, other metals such as Fe are less soluble when they are oxidized and rapidly precipitate because of hydrolysis and formation of iron hydroxo(oxo) compounds. Thus, changes in the oxidation state of oceans and the differential behavior of metals in their oxidized and reduced forms have together impacted metal availability over geological history.
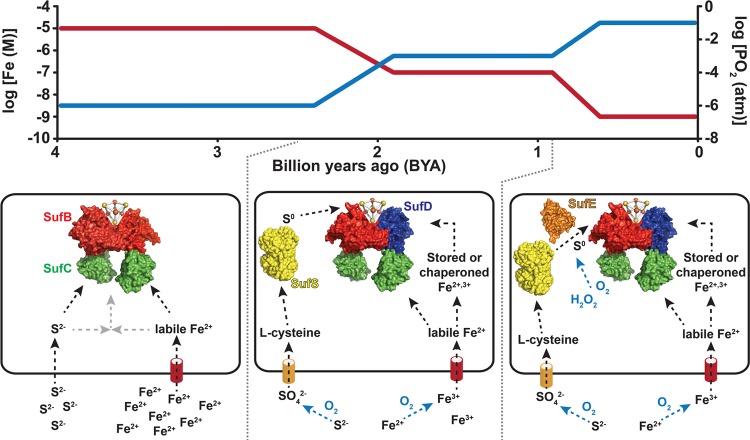
The bioavailability of metals strongly influenced early biological evolution and the metabolic strategies that sustained life during this time.1,9 Phylogenetic analysis of inferred proteomes from available sequenced genomes that span all domains of life indicates that selection for specific metalloprotein folds generally reflects the availability of metals through geological time.10,11 As previously mentioned, during the Archean eon (>2.5 Ga), Fe ions were bioavailable because of an absence or very low concentration of O2 and the solubility of Fe2+ that is greater than that of Fe3+. The delivery of Fe2+ from hydrothermal waters circulating through midocean ridge basalts is thought to be the predominant source of Fe for anoxic Archean ocean basins. The flux of Fe2+ to oceans from this source is thought to have greatly exceeded that of H2S or O2,12 leading to estimated free Fe2+ concentrations that range from 0.05 to 0.5 mM (Figure 1).13 In contrast, soluble Fe concentrations in modern oceans are far lower and rarely exceed several nanomolar (Figure 1).14 Consequently, protein folds that specifically bind Fe are well-represented in early evolving lineages, and their presence in biology can be mapped back to the Last Universal Common Ancestor (LUCA), indicating their early emergence. In contrast, protein folds specific for Cu and Zn ions are enriched in the genomes of organisms that emerged after the Great Oxidation Event (GOE) ∼2.45 Ga,10,11 likely because of the increased rate of delivery of these metals to ocean basins through greater oxidative weathering of continental Cu or Zn sulfide minerals as O2 became more available with the proliferation of oxygenic phototrophs.1
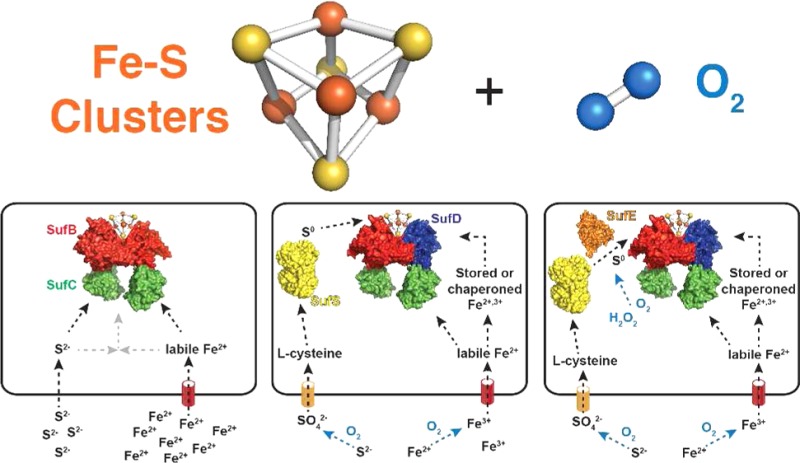
Figure 1.
Model of core Suf pathway evolution in response to increasing oxygen concentrations in the biosphere. In the top graph, iron (red) and oxygen (blue) concentrations refer to those in seawater as adapted from published estimates.1 The bottom panels show the addition of biogenesis components to the core SufBC scaffold complex in response to decreased iron bioavailability (SufD), altered sulfur metabolism (SufS), and oxidation of sulfur transfer intermediates (SufE). Surface representation structures are of SufB (red), SufC (green), SufD (blue), SufS homodimer (yellow), and SufE (orange). The model shows SufBC performing its proposed role as an Fe–S cluster scaffold. A proposed role for SufD in iron acquisition is also shown. SufS and SufE are shown mobilizing sulfide (as persulfide) from l-cysteine for cluster biogenesis in some species. See Table 1 for a summary of proposed Suf protein functions and the text for more details.
Biogenesis of Iron–Sulfur (Fe–S) Clusters
Fe2+ is common in ocean spreading centers and hydrothermal discharge where it often forms complexes with sulfide, resulting in iron–sulfur (Fe–S) mineral phases. Fe–S minerals, in particular pyrrhotite (FeS), have been shown to catalyze numerous small molecule interconversions, including the reduction of N2, reduction of CO2, and production of H2 using H2S as a reductant under high-pressure and high-temperature conditions mimicking those of hydrothermal vents.15−17 In modern biology, enzymes with Fe–S centers are widely distributed and catalyze a functionally diverse array of chemistry, including the reduction of N2/CO2 and production of H2. The strong parallels between the reactivity and structure of Fe–S minerals and Fe–S cluster-containing metalloenzymes, coupled with the presence of modern ancestors of these enzymes in early evolving lineages, represent one of the primary arguments in support of an “Fe–S World” theory for the origin of life.18 More recent phylogenetic analyses are consistent with this theory and suggest Fe–S cluster metalloprotein folds are among the most ancient and widely distributed, indicating a selective advantage for fine-tuning Fe–S cluster stability and chemical reactivity through ligation to a protein framework.11,19−21 The chemical reactivity of Fe–S clusters makes them versatile cofactors for electron transfer and substrate binding/activation reactions. Other functional folds for Fe–S proteins mediate their roles in catalysis and redox sensing.
In the laboratory, Fe–S clusters can be synthesized from iron and sulfide salts using low-molecular weight compounds containing suitable ligands such as sulfur (S) or nitrogen (N).22−25 Fe–S metalloproteins can also be chemically reconstituted using similar Fe and S sources or using purified physiological Fe and S donor proteins.26−29 However, in vitro reconstitution of Fe–S metalloproteins usually requires strict anoxic conditions and often generates polymeric Fe–S species not normally found in the cellular environment. While in vitro reconstitution is an important tool for studying active Fe–S metalloenzymes and for dissecting stepwise Fe–S cluster assembly, it is important to remember that the reconstitution procedure may not accurately reflect the biogenesis pathway in vivo.
Numerous genetic studies, starting with those focused on the maturation of the nitrogenase enzyme in Azotobacter vinelandii, have shown that in vivo Fe–S cluster biogenesis requires multiple protein components.30,31In vivo Fe–S cluster biogenesis is conducted by a somewhat bewildering array of proteins whose numbers seem to expand with each new organelle, organ system, and/or model organism under study.32,33 The basic process of Fe–S cluster biogenesis requires donation of iron (Fe2+ and/or Fe3+ depending on cluster type) and sulfide (S2–), which serves as a bridging ligand for the iron ions. In every system studied to date, these two components are first combined on a protein that serves as a “scaffold” for cluster assembly (Figure 1). The Fe–S cluster that is bound to the scaffold protein is often labile, which facilitates release of the cluster after assembly. The scaffold protein could theoretically transfer the assembled cluster to the appropriate apo form of the desired Fe–S metalloprotein. In practice, it appears that cells use a series of Fe–S cluster carrier proteins to mediate downstream trafficking and targeting of the mature Fe–S cluster. Some of the Fe–S carrier proteins may interact with additional targeting proteins that help dictate the final destination of the Fe–S cluster, providing an additional layer of control (and complexity) to the system.
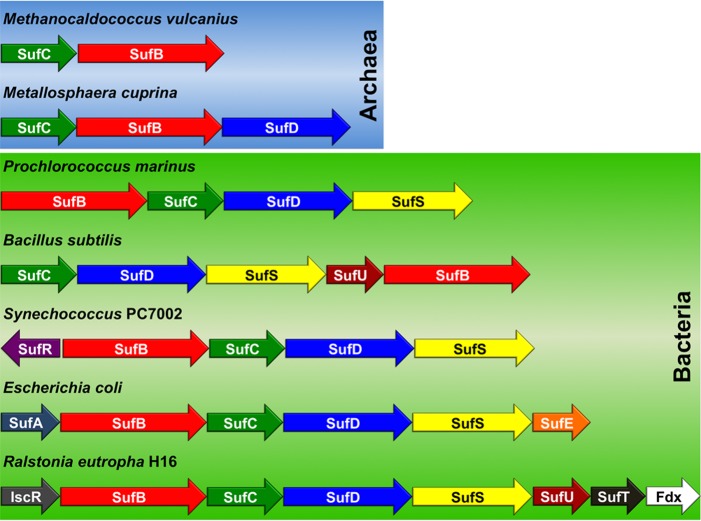
The Suf system is the most ancient of the currently identified Fe–S cluster biogenesis systems.34 The widespread taxonomic distribution of Suf and its presence in both aerobes and anaerobes suggest this system evolved prior to the widespread oxygenation of the biosphere and thus likely evolved mechanisms or strategies for maintaining functionality in an oxidizing environment (Figure 1 and Table 1). The suf operon is diverse and can contain from two to more than six genes organized as (presumed) single polycistronic transcriptional units (Figures 1 and 2). Furthermore, the physiological role of Suf appears to have diverged over evolutionary time. In some organisms, the Suf pathway for Fe–S cluster biogenesis is the only system present and is therefore essential for viability. In other organisms, Suf is one of multiple Fe–S cluster biogenesis systems and operates in parallel with the Isc (iron–sulfur cluster) and/or Nif (nitrogen fixation) pathways.33,35 While only a few Suf pathways have been extensively studied, it appears that in organisms that contain multiple Fe–S cluster biogenesis pathways, Suf functions as a stress-response cluster assembly system that operates under conditions of oxidative stress and iron starvation to augment other “housekeeping” biogenesis pathways like Isc.36−40 It is likely that much of the functional divergence of Suf across multiple phyla stems from changes in the gene composition of the suf operon as well as modifications to Suf regulation in response to the availability of iron and oxygen, which are interlinked as discussed above.
Table 1. Proposed Functions of Suf Proteins in the Fe–S Cluster Assembly Process.
| Suf protein | proposed function | refs |
|---|---|---|
| SufB | Fe–S scaffold protein | (41−45), (47) |
| SufC | ATPase | (36), (37), (44), (46), (48−52) |
| SufD | iron trafficking | (44), (60) |
| SufS | cysteine desulfurase | (42), (47), (60−63, 65−67, 81−83) |
| SufE | sulfur transfer shuttle | (42), (47), (61−67) |
| SufA | Fe–S carrier protein | (41), (45) |
| SufU | sulfur transfer or Fe-S scaffold protein | (80), (82), (83), (ref84), (102) |
Figure 2.
Select examples of suf operon diversity among Archaea (top) and Bacteria (bottom). Genes for sufA, sufB, sufC, sufD, sufS, and sufU are color-coded to reflect their homology in different organisms.
Over the past decade, we have studied the physiological role of one of the most complex suf operons, sufABCDSE, in the gamma proteobacterium Escherichia coli (Figure 2 and Table 1). The E. coli Suf system is an example of the stress-responsive class of suf operons as it is primarily used to augment the Isc housekeeping pathway under conditions of oxidative stress and iron starvation. In this review, we will attempt to correlate the results of biochemical and physiological studies with the taxonomic distribution of the core scaffold complex and sulfur delivery system of the Suf system. We then examine the evolutionary history of Suf to provide new insight into the influence of oxygen and its metabolism on the evolution of Fe–S cluster biogenesis.
SufBC Scaffold Complex
The simplest suf operon and that which contains the minimal functional core is comprised solely of sufBC (Figures 1 and 2).34 Despite their widespread occurrence in many Archaea and Bacteria, SufB and SufC have been most extensively studied at the genetic and biochemical levels in Bacteria, specifically E. coli and Erwinia chrysanthemi. The E. coli SufB protein stimulates Fe–S cluster assembly and insertion of a [2Fe-2S]2+ cluster into the adrenodoxin-like ferredoxin protein Fdx when iron and sulfide salts are provided as starting materials.41 SufB also assembles a stable [4Fe-4S]2+ cluster during in vitro reconstitution.42,43 (His)6-SufB is purified with both [4Fe-4S]2+ and linear [3Fe-4S] clusters after in vivo co-expression with the sufCDSE genes.44 Holo-SufB is competent to transfer intact Fe–S clusters to native E. coli target proteins such as SufA, Fdx, and aconitase (AcnA).41,43,45 On the basis of these results, we and others have proposed that E. coli SufB is an Fe–S scaffold protein in which the nascent Fe–S cluster assembles prior to its transfer to Fe–S metalloproteins. As illustrated in Figure 1, early during the evolution of life the SufB scaffold may have been selected on the basis of its ability to isolate reactive intermediates that form during “spontaneous” cluster assembly, to prevent polymerization of iron sulfide species during cluster assembly, and/or to provide some specificity for downstream targeting of mature Fe–S clusters to their respective metalloproteins. Scaffold proteins like SufB may have represented the simplest biogenesis system for capturing abiological Fe–S compounds from the environment and incorporating them into proteins as cofactors. Support for this notion, as discussed in more detail below, is suggested by phylogenetic analysis that indicates Suf likely emerged in the Archaea but after the divergence of Bacteria and Archaea from the LUCA, which indicates the presence of other more primitive mechanisms for Fe–S cluster biosynthesis in earlier evolving but likely extinct life forms. Thus, the ancestral suf operon likely consisted of only sufBC, with subsequent acquisition of sufD through a duplication of sufB (Figures 1 and 2).
SufC is encoded along with the SufB scaffold protein in all suf operons identified in sequenced genomes (Table 1 of the Supporting Information), which is consistent with biochemical evidence indicating that the two proteins physically interact to form a SufBC complex (Figure 1).37,42,43,46−49 Primary sequence analysis of SufC reveals the presence of signature motifs normally found in the ATPase subunit of ATP-binding cassette (ABC) transporters.37,50−52 SufC has an overall L-shaped structure consisting of two domains, a RecA-like catalytic domain containing the Walker A (GxxxxGKT/S) (Figure 3A, yellow) and Walker B (hhhhD) (Figure 3A, yellow) motifs (where h denotes a hydrophobic residue) and a helical domain that contains the ABC signature motif (L/FSGGQ/E) (Figure 3A, yellow) that is strictly conserved among ABC ATPases. Upon dimerization, the Walker motifs of one SufC monomer should orient with the ABC signature motif of the other monomer to create two ATP-binding sites, both buried at the dimer interface (Figure 1 of the Supporting Information). A highly conserved glutamate residue immediately following the Walker B motif is the proposed catalytic residue for ATP hydrolysis in ABC ATPases (although this has not been conclusively shown for SufC). In structures of other ABC ATPases, this glutamate residue interacts with ATP in the nucleotide-binding site via a water molecule. In the structure of the SufC monomer, this glutamate residue (Glu171) is positioned away from the active site and forms a salt bridge with Lys152 (Figure 3A,B).50−52 Lys152 is adjacent to the ABC signature sequence, so the ionic bond with Glu171 also serves to link the helical and catalytic domains. Residues Lys152 and Glu171 are conserved in all SufC proteins, and the Lys152–Glu171 salt bridge is observed in SufC structures from E. coli and Thermus thermophilus. The SufC primary sequence also contains two other motifs peculiar to ABC ATPases. The D-loop (Figure 3) is a flexible structure that seems to alter its position to facilitate ATP hydrolysis. A putative dimer model of SufC, generated by superimposing the structure of monomeric SufC onto the structure of an ATP-bound form of the paralogous HlyB (H662A) dimer from E. coli,53 reveals possible steric hindrance of SufC dimer formation due to protrusion of the D-loop into the putative dimer interface. This feature distinguishes SufC from other ABC transporter ATPases.54 The Q-loop (Figure 3) connects the catalytic and helical domains in ABC ATPases and mediates the interaction between the ATPase and transmembrane domains of ABC transporters.54−57 SufC instead forms soluble cytoplasmic complexes with SufB (and SufD in some organisms).55,57 The Q-loop putatively contributes to the interaction of SufC with its partner proteins.
Figure 3.
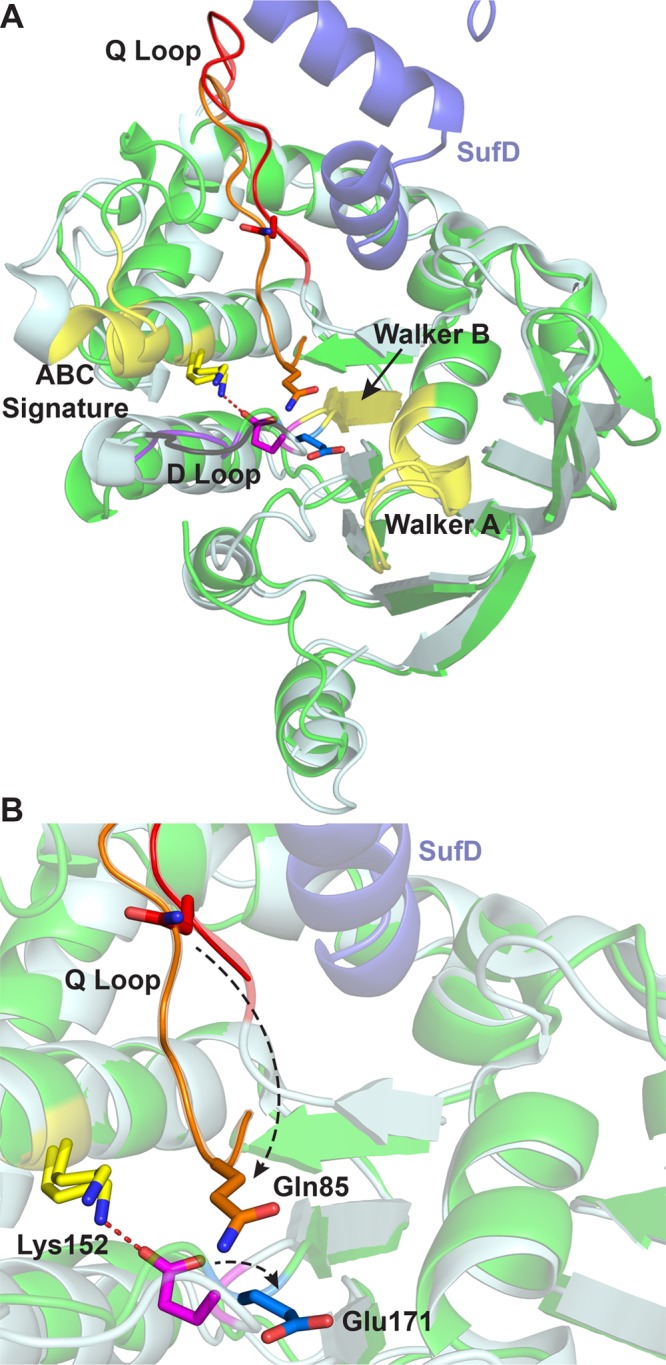
(A) Structural alignment of the nucleotide-free SufC monomer (light blue, PDB entry 2D3W) and one nucleotide-free SufC monomer from the SufC2D2 complex (green, chain C of PDB entry 2ZU0). C-Terminal helices 6 and 7 of SufD from SufC2D2 are shown at the top (blue). The Walker A, Walker B, and ABC signature motifs are colored yellow. The Q-loop is colored red (SufC monomer) or orange (SufC2D2). The D-loop is colored black (SufC monomer) or purple (SufC2D2). (B) Close-up view of panel A. Black dashed arrows indicate positional changes of those residues going from the structure of SufC alone to the SufC from SufC2D2. Gln85 (red to orange), Lys152 (yellow), and Glu171 (pink to cyan) are shown as sticks, and the Lys152–Glu171 salt bridge is shown as a red dashed line.
The interaction of SufC with both SufB and SufD is consistent with the considerable sequence homology between SufB and SufD (Figure 4), the latter of which evolved from a duplication of a sufB gene (discussed in more detail below). The region of shared homology largely covers the β-helix structure of SufD and SufB as well as α-helices shown to interact with SufC. In E. coli, SufC can form three stable complexes. SufB and SufC can interact to form a stable SufB2C2 heterotetramer in vivo.41,42,44 While it is not clear if SufB2C2 is physiologically relevant in E. coli, this complex may reflect the active SufBC complex in organisms that lack SufD and have only the minimal sufBC operon (Figure 1). If the entire sufABCDSE operon is co-expressed in E. coli, the SufBC2D heterotetramer is the primary complex purified, which may suggest that it is the most stable form of SufB, SufC, and SufD (Figures 1 and 4).43,47 In addition, a SufC2D2 complex can form if SufC and SufD are co-expressed in the absence of SufB.49,51 A crystal structure of SufC2D2 from E. coli has been determined.51 The quaternary structure of the E. coli SufC2D2 complex revealed that the SufC catalytic site is remodeled by its interactions with SufD. The Glu171–Lys152 salt bridge is broken, and Glu171 is rotated toward the ATP-binding pocket (Figure 3B).51 The D-loop is also rotated away from the dimer interface, making the nucleotide-binding site of SufC more accessible and facilitating the dimerization of SufC for ATP binding and hydrolysis. These structural observations are supported by studies that show that the basal ATPase activity of SufC alone is quite low but is significantly enhanced when SufC forms a complex with either SufB or SufD.48,49 It is proposed that the salt bridge in the monomeric SufC structure is used to downregulate ATP hydrolysis when SufC is not bound to SufB or SufD (and perhaps in response to ATP occupancy of the active site).50,51 All of these changes appear to be controlled by interaction of SufC with its partner protein(s), indicating an appreciable degree of coordinated regulation of SufC ATPase activity. At present, the exact function of SufC in the Fe–S cluster biogenesis cycle is unknown.
Figure 4.
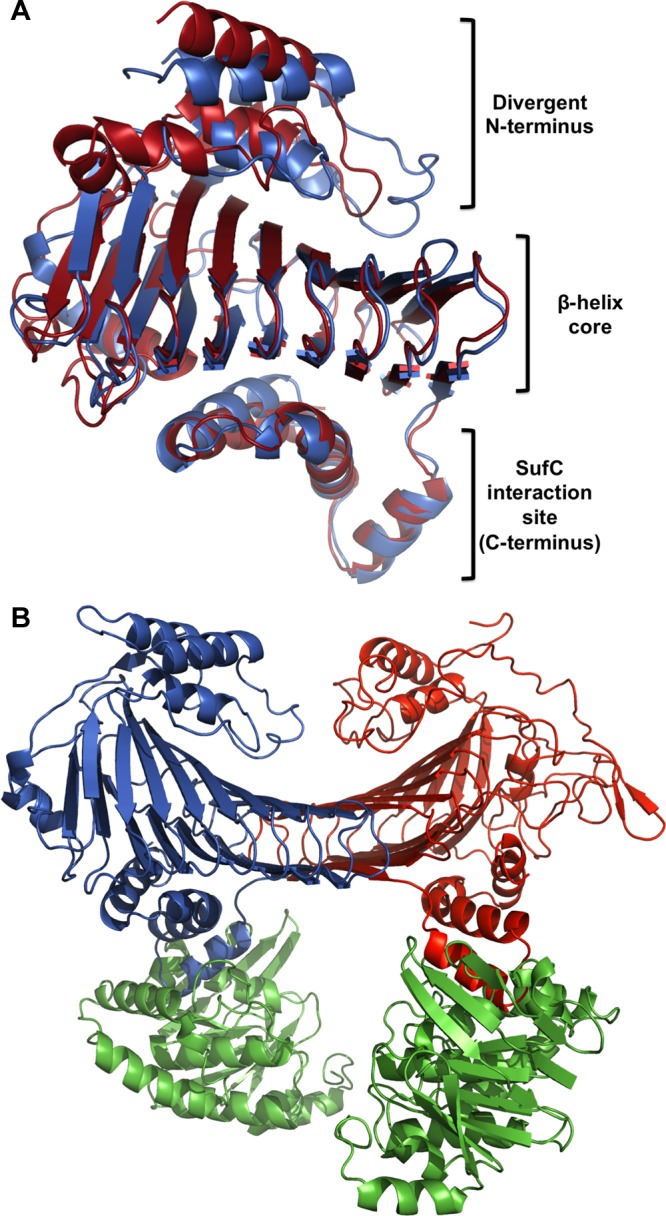
(A) Structural alignment of E. coli SufD (PDB entry 1VH4) and Methanosarcina mazei Go1 SufB (PDB entry 4DN7). The alignment was generated using the FATCAT Pairwise Alignment tool. (B) Model structure of the E. coli SufBC2D complex generated by modeling E. coli SufB on one chain of the SufC2D2 structure (PDB entry 2ZU0). SufB is colored red; SufC monomers are colored green, and SufD is colored blue. The alignment was generated using the FATCAT Pairwise Alignment tool.
There is currently no direct structural characterization of SufB2C2 or SufBC2D. Presumably, similar interactions occur between SufC and the SufB or SufD partner protein (Figures 3A and 4). It is possible the SufB2C2 and SufC2D2 complexes conduct discrete steps in cluster assembly, but this has not been conclusively shown in vivo or in vitro. A number of organisms contain the sufBC genes and lack sufD (Figure 5, discussed below); however, there is currently no evidence of sufCD being found in the absence of sufB in any genome (Table 1 of the Supporting Information). This finding coupled with the substantial sequence and structural homology between SufB and SufD suggests that sufD results from a duplication of sufB.
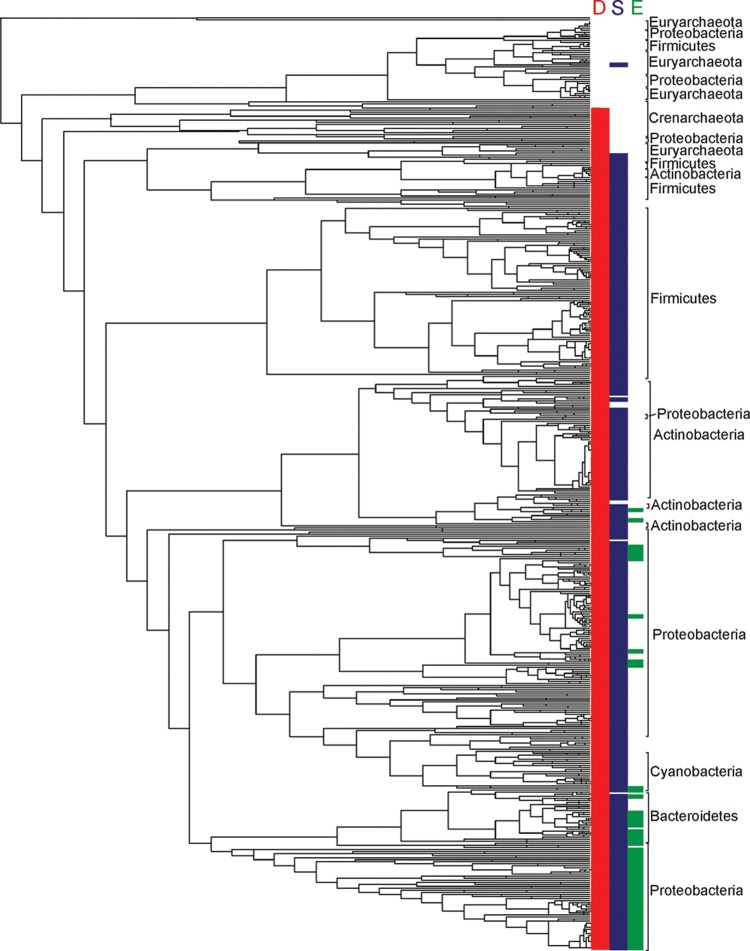
Figure 5.
Rate-smoothed phylogenetic reconstruction of concatenated SufBC alignment blocks from 1094 bacterial and archaeal taxa (eukarya excluded). Representative SufB and SufC were identified in available genome sequences in October 2011 and were aligned individually with ClustalW103 specifying default alignment parameters. Paralogs of SufB (i.e., SufD) and SufC (i.e., other members of the ABC transporter ATP-binding protein family) were included in each alignment, respectively, and were later used to root the phylogeny. SufB and SufC alignment blocks were concatenated with PAUP (version 4.0, Sinauer Associates, Inc.). The maximum likelihood phylogenetic reconstruction was inferred using PhyML version 3.0 specifying the LG substitution matrix and a four-category gamma substitution model.104 The concatenated SufBC phylogeny was rate-smoothed using the penalized-likelihood approach105 as implemented by the “chronopl” command in the R statistical package (version 2.15.0) Picante (version 1.3)106 specifying a smoothing parameter of 1.0. The distribution of SufD (red), SufS (green), and SufE (blue) in the suf operon in each taxon was mapped on the phylogeny using base commands in the Picante package within R. The primary taxonomic rank at the phylum level is indicated for major clusters.
Taxonomic Distribution and Phylogeny of the Core sufB and sufC Genes
The taxonomic distribution of Suf was determined via BLASTp analysis of publically available genome sequences in October 2011 (full results presented in Table 1 of the Supporting Information). The genetic composition of operons was characterized using the KEGG gene viewer, with manual verification using BLASTp or sequence alignments. We found that 1094 genomes of a total of 1667 genome sequences (65.6% of the total) were found to encode sufB and sufC. The sufB and sufC loci were almost exclusively found adjacent to one another in an apparent operon leading us to define sufBC as the minimal suf operon, with the only exceptions being a single species of cyanobacterium (Cyanothece sp. PCC 7425) and two clostridial species (Clostridium difficile CD196 and Clostridium tetani E88). As a percent of available genomes, the sufBC genes were more prevalent among archaeal genomes than bacterial genomes, with only a few eukaryal genomes containing sufBC. Among the Archaea, sufBC was identified in nearly all of the available genomes, including those affiliated with the Crenarchaeota (35 of 35 genomes), Euryarchaeota (75 of 77 genomes), Nanoarchaeota (1 of 1 genome), and Thaumarchaeota (2 of 2 genomes). Among the bacteria, sufBC was prevalent among phyla that are traditionally characterized as anaerobes or facultative anaerobes, including the Bacteroidetes (60 of 62 genomes), Firmicutes (248 of 299 genomes), green-nonsulfur bacteria (15 of 15 genomes), and Spirochaetes (14 of 31 genomes). In addition, sufBC was prevalent among Acidobacteria (147 of 151 genomes), Chlamydiae (20 of 21 genomes), and Cyanobacteria (40 of 40 genomes) but exhibited an uneven distribution among Proteobacteria. A higher percentage of taxa affiliated with the alphaproteobacteria (121 of 155 genomes, or 78.1% of the total), deltaproteobacteria (26 of 42 genomes, or 61.9% of the total), and gammaproteobacteria (185 of 308, or 60.1% of the total) were found to encode sufBC compared to the percentage in betaproteobacteria (29 of 105 genomes, or 27.6% of the total) or epsilonproteobacteria (5 of 37 genomes, or 13.5% of the total); both of the latter phyla tend to harbor aerobic members. Other lineages in which sufBC was commonly identified include the Aquificae, Dictyoglomi, Fibrobacteres, Fusobacteria, Nitrospirae, Planctomycetes, Thermotogae, and Verrucomicrobia.
The sufBC genes are also present among several eukaryal genomes, most notably in the chloroplast or mitochondrial genomes of constituent taxa. sufBC was identified among the chlorophytes (e.g., green algae; 3 of 4 genomes), rhodophytes (e.g., red algae; 1 of 1 genome), and streptophytes (e.g., eudicots, monocots, ferns, and mosses; 8 of 14 genomes). Among protists, sufBC was identified among taxa affiliated with stramenophiles (e.g., diatoms; 1 of 2 genomes) and alveolates (1 of 16 genomes).
We determined the phylogeny of a concatenation of SufBC for use in defining patterns in the evolution of the suf operon and for predicting the composition of the ancestral suf operon (Figure 5). Methods used to concatenate SufBC, reconstruct the SufBC phylogeny, rate-smooth the phylogeny, and map the distribution of ancillary genes on the resultant rate-smoothed phylogeny are included in the figure legend. The earliest branching lineage associated with the SufBC phylogeny is comprised of two sublineages, one of which is comprised exclusively of Crenarchaeota and the other of which is comprised of Euryarcheota, Proteobacteria, and Firmicutes (Figure 5). In the second sublineage, sequences affiliated with the Euryarchaea nest the bacterial lineages (Figure 5). Together, these observations may suggest that suf first emerged among the Archaea. This hypothesis is supported by the observation that SufBC associated with Candidatus Korarchaeota (i.e., unclassified Archaea) also cluster within this second sublineage (data not shown). Branching after the large crenarchaeal/euryarchaeal/firmicute/proteobacterial SufBC lineage is a cluster of sequences comprised of a second monophyletic lineage of crenarchaea, adding further support for an archaeal origin for suf. Importantly, the finding that euryarchaeal SufBC nest SufBC sequences affiliated with bacteria in the earliest branching lineage strongly suggests a role for lateral gene transfer (LGT) during the early evolution of the suf operon. Additional evidence supporting a role for LGT in the evolution of suf derives from the observation that proteobacterial SufBC form a number of large polyphyletic clades (Figure 5), which generally correspond to the phylum level (alpha, beta, delta, epsilon, and gamma) classification of this lineage (data not shown). This indicates that suf was likely acquired in these lineages early, but after the primary divergence among proteobacteria had taken place.
Consistent with previous reports indicating that chloroplast suf genes associated with eukaryote plants likely are the result of the primary endosymbiosis of a cyanobacterium, SufBC proteins from the red alga Cyanidioschyzon merolae and the green alga Chlamydomonas reinhardtii cluster with cyanobacterial SufBC (E. S. Boyd, unpublished data). Likewise, the complements of genes encoded by plant chloroplast and cyanobacterial suf operons are similar, adding further support to this hypothesis. Interestingly, the suf proteins encoded by mitochondrian genomes in taxa such as Toxoplasma gondii are also very similar to cyanobacterial sequences (data not shown). Recently, a chimeric fusion protein of SufB and SufC was discovered in the human protozoan parasite Blastocystis.58 The Bh-SufCB fusion protein was likely acquired by lateral gene transfer from an archaeon from the Methanomicrobiales and now plays a role in cytosolic Fe–S cluster metabolism in Blastocystis.58
Diversification of the Core suf Operon
A number of accessory genes are found encoded with sufBC in many organisms (Figure 2). A qualitative examination of the operon composition (Figure 5) in early evolving taxa when compared with more recently evolved taxa suggests that the operon has grown in complexity with time, similar to what has been observed in other operons, including that which encodes the mercury detoxification systems (mer).59 Consistent with previous work, our analysis clearly shows that suf operons associated with early evolving taxa, such as members of the Euryarchaeota and Crenarchaeota, as well as early evolving members of the Proteobacteria and Firmicutes, all harbor operons that are simpler than those harbored by more recently evolved taxa (e.g., most other Proteobacteria).34 This suggests that natural selection has promoted the recruitment of new gene functions in the suf operon to optimize the efficiency of Fe–S cluster biogenesis in the various ecological niches inhabited by these taxa. Collectively, our analysis suggests an evolutionary trajectory in which suf grew in complexity from an operon encoding only sufBC through the sequential recruitment of other genes such as sufD, sufS, and sufE (Figures 1 and 2). Importantly, when the distribution of accessory genes is mapped on the SufBC tree, numerous examples of gene loss are also evident as evinced by large gaps in the phylogenetic distribution of these genes. Thus, both gene recruitment and loss are likely to have shaped the functional and evolutionary history of suf.
For our initial analysis of suf operon evolution and diversity, we chose to focus primarily on the recruitment of sufD, sufE, and sufS to the operon (Figure 5). Previous studies have suggested a possible connection between SufD and the iron acquisition step of Fe–S cluster assembly by SufBC.44 Similarly, extensive biochemical and genetic experiments have shown that SufS and SufE act as a concerted sulfur transfer pathway for donation of sulfide during cluster assembly.42,47,60−67 Thus, our phylogenetic analysis of these genes can be used in conjunction with extensive biochemical and genetic data to formulate new hypotheses about their functional roles in the E. coli Suf system.
To predict the composition of the ancestral suf operon, as well as to define the trajectory of recruitment of genes to the operon, we mapped the distribution of individual gene functions on the SufBC tree (Figure 5). The sufD gene appears earliest in evolutionary time among the suf genes when mapped on the SufBC tree. Sequence homology suggests the sufD gene derives from a duplication of an ancestral sufB sequence. The earliest evolving sufD proteins are not yet present in the earliest evolved SufBC lineage from the Euryarchaeota, Crenarchaeota, and several Firmicutes and Proteobacteria. Rather, the earliest evolving sufD genes likely emerged as the crenarchaeal lineage diversified at a time that likely predated the divergence of the Sulfolobales and the Desulfurococcales (E. S. Boyd, unpublished data). Thus, the ancestral suf operon likely consisted of only sufBC, with acquisition of sufD through gene duplication occurring early during the evolution of suf. Shortly after the acquisition of sufD, the gene encoding the cysteine desulfurase (sufS) was recruited to the operon (Figure 5). The SufS cysteine desulfurase is likely to have predated in evolutionary time the emergence of the ancillary sulfurtransferase component encoded by sufE (Figure 5). Following the recruitment of the cysteine desulfurase, other Fe–S cluster biogenesis proteins were recruited to the operon, including the U-type scaffold or sulfur trafficking protein SufU, the A-type cluster trafficking protein SufA, and others. A detailed analysis of the evolution and complexity of the complete array of suf operon genes is underway but is beyond the scope of this review (E. S. Boyd, J. M. Boyd, and F. W. Outten, manuscript in preparation).
Phylogenetic Distribution and Function of sufD
The sufD gene was identified in 85% of the suf operons examined. In general, suf operons associated with Archaea appeared to have fewer gene complements than those associated with bacteria. The majority of taxa that encode only sufBC (and do not include sufD) were affiliated with the Euryarcheota (only 23.7% of genomes encode sufD), Crenarchaeota (74.3% of genomes encode sufD), and Deltaproteobacteria (42.3% of genomes encode sufD) (Table 1 of the Supporting Information). Close examination of the sufD distribution pattern shows that 91.5% of Archaeal genomes that contain sufBC but lack sufD are obligate anaerobes or microaerobes. A similar trend was observed among the Bacteria in which 86.5% of the genomes that contain sufBC but lack sufD are also anaerobes or microaerobes. These observations suggest that recruitment and selection for sufD in the core sufBC operon conveys an advantage for aerobic prokaryotes but not necessarily for anaerobic or microaerobic Archaea and Bacteria (Figure 1).
As previously mentioned, SufD is a paralog of SufB and evolved from gene duplication. SufD and SufB harbor significant sequence homology throughout their C-terminal domains but considerable divergence in the N-terminal region, suggesting that the duplication of these genes occurred prior to differentiation in the N-terminus. In E. coli, SufD interacts with the SufC ATPase and SufB to form the SufBC2D complex but can also form the SufC2D2 complex if SufB is absent.36,37,46,47,49,51 Thus, it appears that duplication of SufB allowed for new functionality of the core scaffold complex, replacing (or perhaps expanding) the SufB2C2 stoichiometry with a new SufBC2D complex.
While studying the E. coli Suf system, we discovered that if SufD is absent, in vivo incorporation of iron on SufB during Fe–S cluster assembly is abolished while the acquisition of sulfide is only modestly decreased.44 Similar results were obtained if the ATPase activity of SufC is altered by introduction of a single point mutation into the Walker A motif (K40R).44 On the basis of these results, we proposed that SufC ATPase activity works in concert with SufD to mediate the delivery of iron into the Suf pathway. The iron donation step may require ATPase activity to provide energy to mobilize iron for cluster assembly from an inert or inaccessible form (such as an iron storage protein or ferric siderophore). Barras and Expert discovered a number of intriguing genetic defects linked to iron metabolism that are caused by deletion of the suf genes.36,37,68 In Er. chrysanthemi grown under iron-replete conditions, deletion of the individual suf genes led to an increased sensitivity to the iron-activated antibiotic streptonigrin, with the sufC deletion causing the greatest sensitivity.36 These results suggest that the size of the intracellular labile iron pool increases in most suf deletion strains (leading to an increase in the extent of streptonigrin activation). Interestingly, a sufD mutant did not cause increased sensitivity to streptonigrin, suggesting that sufD may be required for the phenotype observed in the other suf deletion strains. It was later found in Er. chrysanthemi that deletion of sufA, sufB, or sufD leads to a decrease in the iron-loaded form of the iron storage protein bacterioferritin (Bfr) under iron-limited conditions.68 This result suggests that in the absence of some suf genes, the size of the labile iron pool is increased under iron-limited conditions because of a decrease in iron storage capacity. That same work also showed that sufC deletion actually causes the opposite phenotype, an increase in the iron-loaded form of Bfr. Finally, it was observed that deletion of sufC or sufD weakened the ability of Er. chrysanthemi to utilize its native siderophore chrysobactin as an iron source when the cells were stressed with the iron chelator 2,2′-dipyridyl.37 Together, these genetic studies suggest that the suf system, especially sufC and sufD, plays an active role in modulating the intracellular iron distribution to direct iron into the Fe–S cluster assembly pathway. It remains to be seen if Suf plays this role through direct interaction with the iron homeostasis system or if the cell indirectly channels iron to Suf through altered expression of iron homeostasis components by regulators like Fur and IscR.
The SufBC2D complex isolated from E. coli is copurified with 1 equiv of FADH2 per complex.43,44 All three Suf proteins are required for stoichiometric binding of FADH2, which has a dissociation constant (Kd) of 12 μM.43 When the bound FADH2 is oxidized to FAD by oxygen, the affinity of SufBC2D for the flavin is decreased and FAD dissociates from the complex. Reduced flavins are efficient ferric iron-reducing agents, and SufBC2D-FADH2 was shown to mobilize iron from ferric citrate or the ferric-loaded form of bacterial frataxin (CyaY), presumably via a reductive mechanism.43 It was proposed that SufBC2D uses FADH2 as a redox cofactor to mobilize iron for the Suf pathway, although the physiological in vivo iron donor remains unclear.
The distribution of sufD in aerobic microorganisms encoding suf is consistent with our hypothesized role for SufD in iron trafficking during Fe–S cluster biogenesis (Figure 1 and Table 1). Iron bioavailability is more restricted in aerobic habitats because of the insolubility of ferric iron, in particular in neutral to alkaline environments such as the cytoplasm of most cells. Intracellular iron metabolism (including Fe–S cluster metabolism) is also perturbed by oxygen and reactive oxygen species, which can lead to increased iron demand for metalloprotein maturation. The oxygen-dependent disruption of iron metabolism likely led to selection for a multitude of adaptive measures to acquire iron and protect it from spurious chemistry. It is unclear how SufD may directly or indirectly mediate iron acquisition for Fe–S cluster biogenesis, but part of the answer may lie in the creation of a composite FADH2-binding site when SufD joins the SufBC2D complex. This new functionality may have provided SufBC with a way to funnel reducing equivalents into ferric iron reduction to acquire iron by reductive release from ferric siderophores, ferric iron storage proteins, or another unknown ferric chelate.
Recruitment of SufS by SufBC Parallels the Use of l-Cysteine as the Sulfur Source for Cluster Biogenesis
l-Cysteine is a major physiological sulfur source for Fe–S cluster biosynthesis in bacteria as well as eukaryotic mitochondria and chloroplasts. A family of cysteine desulfurases is responsible for the mobilization of the sulfur atom via a pyridoxal 5′-phosphate (PLP) enzymatic mechanism first characterized for the NifS cysteine desulfurase.69,70 In this mechanism, substrate l-cysteine binds to PLP and forms a PLP–cysteine adduct as a Schiff base (sometimes termed the external aldimine). Next, a catalytic cysteine residue acts as a nucleophile to attack the sulfhydryl group of the substrate cysteine, which has been activated by binding to PLP. The nucleophilic attack results in formation of an enzyme-bound persulfide (R-S-SH) and a PLP-bound enamine that is ultimately released as l-alanine. The reactive enzyme-bound persulfide group can then be transferred to cysteine residues on the final scaffold protein directly or via a sulfur shuttle protein using a mechanism similar to protein disulfide bond exchange. Despite some disagreement about nomenclature, the persulfide (or sulfane sulfur) species has a formal oxidation state of zero and is often termed S0 in the biochemical literature (even though it is not technically zerovalent sulfur).71,72 At some point, the persulfide (S0) is reduced to sulfide (S2–) for incorporation into the Fe–S cluster, although this likely occurs on the scaffold protein (Figure 1).
On the basis of sequence similarity, the cysteine desulfurases can be subdivided into group I (NifS and IscS) and group II (SufS and CsdA) enzymes.73 Key differences between the groups are observed in the structure around the catalytic cysteine residue. The catalytic cysteine occurs as part of a short, more rigid loop with a more hydrophobic environment in group II cysteine desulfurases than in group I enzymes.74 On the basis of multiple three-dimensional structures of SufS, the shortness and decreased flexibility of the active site loop containing the catalytic Cys364 likely explain the catalytic inefficiency of group II desulfurase enzymes compared to group I enzymes, which have a flexible catalytic cysteine loop that is 11 amino acids longer than the group II enzymes.75−79 However, we now know that accessory proteins can enhance the activity of SufS and other group II enzymes to a level comparable to that of group I enzymes. For SufS in E. coli and Er. chrysanthemi, the accessory protein is SufE, while SufS in other organisms, such as Bacillus subtilis, utilizes SufU.47,61,63,64,66,67,80−83,ref84
Despite its broad use in bacteria, the SufS cysteine desulfurase exhibited a patchy taxonomic distribution in the Archaea, with 0.0 and 3.9% of total crenarchaeal and euryarchaeal suf operons encoding sufS, respectively (Table 1 of the Supporting Information). Even though l-cysteine is the sulfur source for Fe–S cluster biogenesis in many organisms, most of the Archaea analyzed inhabit environments characterized as sulfur- and/or sulfide-rich, which may explain the lack of cysteine desulfurase homologues in these organisms (Figure 1). The observed taxonomic distribution of SufS may have resulted from evolutionary changes in l-cysteine biosynthesis and metabolism in response to the advent of oxygenic photosynthesis. In the absence of significant concentrations of atmospheric O2, the primary source of delivery of sulfate to ocean basins was deposition of volcanogenic sulfur species (e.g., SO2) and more reduced species that had been photochemically oxidized in the Archean atmosphere.12 Once the O2 concentration increased in the atmosphere, sulfate was delivered to the oceans through continental weathering and oxidation of mineral sulfides.4,7 The selection for dissimilatory and assimilatory sulfate reduction pathways due to the greater availability of sulfate in anerobes and microaerophiles likely had an impact on organisms that eventually used SufS to synthesize Fe–S clusters. In many organisms, sulfur is assimilated via sulfate reduction to sulfide and incorporation into O-acetylserine for synthesis of free l-cysteine. Free l-cysteine then serves as the primary sulfur donor for sulfur-containing metabolites, including Fe–S clusters (Figure 1). In contrast, methanogenic Archaea such as Methanococcus maripaludis use environmental sulfide, which is abundant in those organisms’ anaerobic habitats, to generate an uncharacterized protein persulfide, which then donates sulfur to directly generate Cys-tRNACys in a reaction dependent on SepCysS.84−86 Free cysteine is not generated by this tRNA-dependent pathway, and free cysteine pools in methanogens (arising primarily from protein turnover) can be 5–10-fold smaller than cysteine pools in bacteria.87 Furthermore, it has been shown that free cysteine is not the in vivo sulfur source for Fe–S cluster biogenesis in M. maripaludis where the sulfur donor is likely an unknown compound derived from exogenous sulfide.87 Thus, SufS may have been incorporated into Suf to better interface with the changing availability of sulfur because of the accumulation of oxygen and with new sulfur metabolic pathways that arose in response to such changes.
SufE and the Functional Divergence of the Stress-Response suf Operon in Gammaproteobacteria
A SufE family member is found to be encoded with a group II cysteine desulfurase in many Gram-negative bacterial genomes. However, suf operons from the Crenarcheota and Euryarcheota do not encode sufE, and a lower percentage of bacterial suf operons encode sufE (16.8% of the total) compared to sufS (88.4% of he total). Indeed, all suf operons from members of the Betaproteobacteria and Firmicutes analyzed here lack homologues of sufE. A sufE gene is more commonly found in the suf operons of Bacteriodetes and Gammaproteobacteria. With the exception of Salmonella enterica subsp. enterica serovar Paratyphi A AKU12601 and Shigella flexneri 301 (serotype 2a), all bacterial suf operons that encode sufE also encode sufS, suggesting an interaction between these proteins.
Early studies from Er. chrysanthemi showed that SufS and SufE interact in a complex and that SufE increases the desulfurase activity of SufS by ∼50-fold.61 The highest specific activity was obtained upon addition of 1 equiv of SufE. Preliminary steady-state kinetic results indicated that SufS from Er. chrysanthemi, alone or in complex with SufE, seems to display Michaelis–Menten behavior using cysteine as a substrate. Binding of SufE to SufS had no effect on the KM value for cysteine (500 μM) but had a large effect on Vmax (0.9 unit/mg compared to 0.019 unit/mg). Fontecave and co-workers subsequently demonstrated transfer of the sulfur from SufS to SufE via a SufS-bound persulfide intermediate and suggested that the acceleration of persulfide cleavage by SufE is primarily responsible for the observed activation of desulfurase activity.62 However, a possible conformational change in SufS upon SufE binding that may enhance substrate cysteine binding has not been excluded.
We reported that E. coli SufE can stimulate the cysteine desulfurase activity of the E. coli SufS enzyme up to 8-fold and that SufE Cys51 accepts sulfane sulfur from SufS.47 This sulfur transfer process from SufS to SufE is sheltered from the environment on the basis of its resistance to added reductants and the analysis of available crystal structures of the proteins.47,75,77 We also found that in the presence of SufE, the SufBC2D or SufB2C2 complexes further stimulate SufS activity up to 32-fold.42,44,47 The cysteine desulfurase SufS donates sulfur to the sulfur transfer protein SufE, and then SufE in turn interacts with the SufB protein for sulfur transfer to SufB.42 The interaction occurs only if SufC is also present. On the basis of protein–protein interaction and sulfur transfer experiments, the proposed route for the persulfide intermediate is from the catalytic Cys364 of SufS to the active site Cys51 of SufE, and finally transfer to SufB as part of SufBC2D for cluster assembly. SufBC2D likely enhances SufS–SufE activity by removal of the SufE persulfide via sulfur transfer to SufB, which allows SufE to recycle faster.65,66
Approximately 87% of Gammaproteobacteria that contain sufE (as part of the suf operon) also contain a second, independent Fe–S cluster biogenesis machinery, the Isc (iron–sulfur cluster) operon (E. S. Boyd, unpublished data). Those Gammabacteria that contain Suf and Isc are predominantly facultative anaerobes from the order Enterobacteriales. In most organisms in which both systems are present and have been characterized, the Isc system is the basal, housekeeping Fe–S cluster biogenesis pathway used under optimal growth conditions while Suf has been adapted to function as a stress-responsive cluster biogenesis pathway used under conditions that perturb Fe–S cluster metabolism.36,37,39,40,60,88 In E. coli and other related Gammaproteobacteria, the sufABCDSE operon is induced by oxidative or nitrosative stress and iron deprivation.36,38,39,60,89,90 Under normal growth conditions, these organisms express the iscRSUA-hscBA-fdx-iscX operon for basal Fe–S cluster assembly. However, via intricate regulatory circuits involving the Fur iron metalloregulatory protein, the IscR Fe–S cluster sensor, the OxyR hydrogen peroxide sensor, and the Fur-regulated small RNA rhyB, the Isc system is downregulated under stress while the level of Suf expression increases to allow it to play a more critical role in maintaining Fe–S cluster assembly until the stress is removed.39,60,91−95In vivo studies in E. coli indicate that the Suf pathway works better under oxidative stress than Isc.88,96 As little as 1 μM H2O2in vivo can deactivate the Isc machinery and lead to a growth requirement for the Suf pathway.
The inclusion of sufE in the subset of suf operons that are utilized for stress-responsive Fe–S cluster biogenesis suggests that SufE provides some protection against disruption of cluster assembly by stress (Figure 1 and Table 1). Fe–S cluster biogenesis is sensitive to oxygen because of the proclivity of iron, sulfide, and protein sulfhydryl groups to be modified by oxygen or reactive oxygen species. The transfer of sulfur from a cysteine desulfurase enzyme to other proteins is a key step in Fe–S cluster assembly. Sulfur is transferred between active site cysteine residues as a highly reactive S-sulfanyl cysteine moiety. The sulfanyl cysteine species is sensitive to reduction or oxidation if exposed to the environment. Because of the reactivity of both the persulfide intermediate and active site sulfhydryl groups on the enzymes, oxidative stress may block the sulfur donation step of Fe–S cluster biogenesis (Figure 1). To test if sulfur trafficking by the Suf pathway may be more resistant to disruption than the Isc system, we characterized the sulfurtransferase reaction of E. coli SufS and SufE and compared its kinetic features to those of the E. coli IscS–IscU system.65 IscU is the scaffold protein for the Isc system that accepts sulfane sulfur from IscS. Surprisingly, we found that the SufS–SufE system is more active than the IscS–IscU system at low but physiologically relevant concentrations of l-cysteine. The enhanced activity at low l-cysteine concentrations may allow Suf to function better than Isc under conditions of oxidative stress when cellular l-cysteine pools might be depleted by oxidation or by use in repair or new synthesis of damaged protein thiols.
We also directly compared the oxidative stress resistance of the E. coli SufS–SufE sulfur transfer pathway to that of the IscS–IscU system during the cysteine desulfurase reaction cycle.65 The results indicated that SufS–SufE sulfurtransferase activity is more resistant to H2O2 exposure than that of the IscS–IscU system. The active site Cys328 residue of IscS is more easily oxidized by H2O2 than the SufS active site when the enzymes are turning over. IscU provided some protection to the IscS active site but was itself sensitive to oxidation at the critical Cys63 and Cys106 residues, which would presumably disrupt sulfur transfer between IscS and IscU.97−100 In contrast, SufE active site Cys51 showed only mild sensitivity to oxidation by H2O2 and was able to continue to enhance SufS activity in the presence of oxidative stress. These results coupled with the phylogenetic distribution of SufE suggest that the addition of SufE transforms the Suf system from a housekeeping system to a stress-response system in gammaproteobacteria.
Conclusions and Unanswered Questions
Our phylogenetic analysis of the core Suf system supports the biochemical and physiological studies of the Suf system in E. coli. The recruitment of SufD and the SufS–SufE sulfurtransferase pair to the core SufBC system parallels the extensive metabolic remodeling necessary for early life forms to adapt to progressive increases in the atmospheric oxygen level (Figure 1). Fe–S cluster metabolism can be perturbed by oxygen in multiple ways, including decreased iron bioavailability, altered sulfur metabolism, disruption of sulfur trafficking, and direct damage to Fe–S metalloproteins (including Fe–S scaffold proteins). These stresses may have selected for components to coordinate iron mobilization for cluster assembly (SufD) and to protect sulfur donation steps (SufS–SufE). Despite the genetic evidence of a connection among SufD, SufC ATPase activity, and iron donation, there is still no clear iron donor protein or chelate identified in E. coli for the Suf system. It is not clear if flavin may be involved in this iron donation process or if it plays a role in a different redox step of cluster assembly. Further biochemical and physiological studies are necessary to fully understand the drivers in the evolutionary diversification of the Suf system.
These initial studies open the door to a much deeper phylogenetic analysis of Suf throughout the three domains of life. Although we focus here on the SufD, SufE, and SufS accessory proteins, suf operons often contain a number of other genes, including those encoding alternate scaffolds (SufU), Fe–S carrier proteins (SufA), ferredoxins, iron storage proteins, iron transporters, and iron metalloenzymes. For example, most Gram-positive bacteria, including B. subtilis, do not encode a SufE homologue. Instead, they often contain a gene encoding SufU located adjacent to a group II cysteine desulfurase gene sufS.80−83,101,102 SufU is a homologue of IscU, but there are significant sequence differences between the two genes. In B. subtilis SufU is essential for viability.80 SufU has been shown to enhance SufS cysteine desulfurase activity in a manner similar to that of SufE in other organisms.81,82 SufU works as the second substrate in the catalytic ping-pong mechanism of the overall sulfurtransferase reaction of the SufS cysteine desulfurase. The mechanism of SufU enhancement may be due to the acceleration of persulfide cleavage to recycle the catalytic cysteine on SufS.81,82 The exact role of SufU in these Suf systems is still an area of active study. We do note, however, that the distribution of sufE in individual operons shows a significant negative covariance with sufU, indicating those suf operons with sufU rarely encode sufE (E. S. Boyd, J. M. Boyd, and F. W. Outten, manuscript in preparation). However, in many organisms, sufE homologues are separately encoded at locations outside of the suf operon, so their presence cannot be excluded by our analysis. While not homologous at the primary sequence level, SufE and SufU proteins are structurally related, including similar positioning of the Cys residues used to accept persulfide from SufS (Figure 6). They may therefore have been separately selected to play similar roles in the mobilization of sulfur from SufS-type cysteine desulfurases through convergent evolution.ref84 This intriguing gene covariance and the recruitment of genes to the suf operon await further phylogenetic and biochemical analysis.
Figure 6.
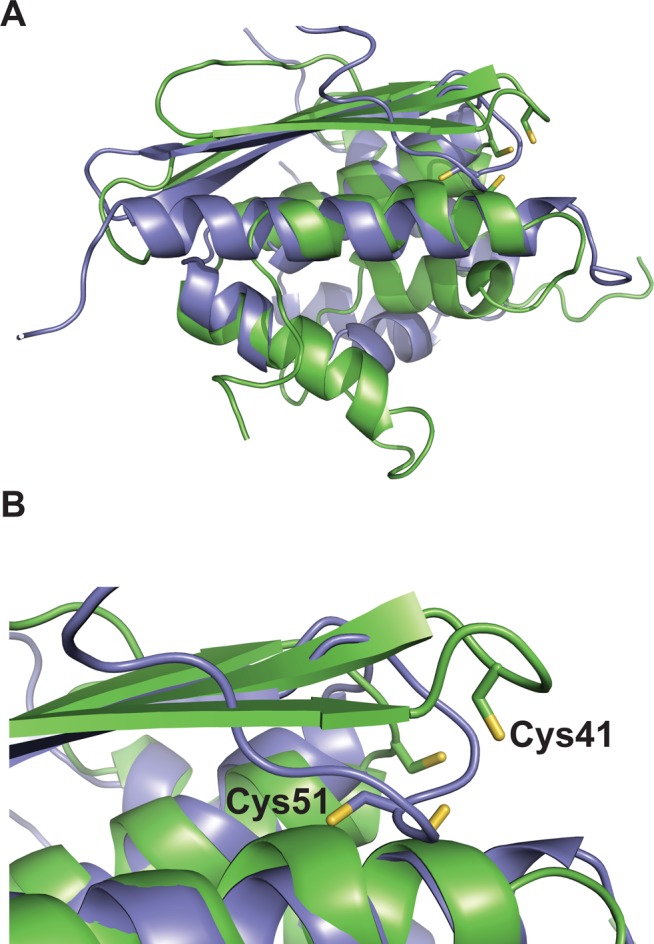
(A) Structural alignment of E. coli SufE (purple, PDB entry 1MZG) and B. subtilis SufU (green, PDB entry 2AZH). Active site Cys residues are show as sticks. (B) Enlarged view of the alignment showing the relative orientation of SufE Cys51 and SufU Cys41. The alignment was generated using the FATCAT Pairwise Alignment tool.
Acknowledgments
We thank Dr. Lesa Offermann and Dr. Leslie Lovelace for assistance with structural analysis software and the members of the broader Fe–S cluster biogenesis community for many insightful discussions.
Glossary
Abbreviations
- Fe–S
iron–sulfur
- Ga
billion years ago
- GOE
great oxidation event
- isc
iron–sulfur cluster
- nif
nitrogen fixation
- PDB
Protein Data Bank
- suf
sulfur formation
- LUCA
Last Universal Common Ancestor.
Supporting Information Available
Locus tags used for bioinformatics analysis provided as a Microsoft Excel file (Table 1) and a model structure of the ATP-bound SufC dimer (Figure 1). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
⊥ Y.D.: Department of Surgery, School of Medicine, University of California—San Francisco, 2340 Sutter St., San Francisco, CA 94109.
This work was supported by grants from NASA Exobiology and Evolutionary Biology (NNX13AI11G) and NASA Astrobiology Institute (NNA13AA94A) to E.S.B., National Institutes of Health Grant GM81706 to F.W.O., and a United Negro College Fund-Merck Graduate Science Research Dissertation Fellowship to K.M.T. J.M.B. was supported by startup funds from Rutgers University, the Charles and Johanna Busch Biomedical Foundation, and U.S. Department of Agriculture Hatch Grant NE-1028.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Anbar A. D. (2008) Oceans, Elements, and Evolution. Science 322, 1481–1483. [DOI] [PubMed] [Google Scholar]
- Anbar A. D.; Duan Y.; Lyons T. W.; Arnold G. L.; Kendall B.; Creaser R. A.; Kaufman A. J.; Gordon G. W.; Scott C.; Garvin J.; Buick R. (2007) A whiff of oxygen before the Great Oxidation Event?. Science 317, 1903–1906. [DOI] [PubMed] [Google Scholar]
- Bekker A.; Holland H. D.; Rumble D.; Yang W.; Wang P. L.; Coetzee L. L. (2002) MIF of S, oolitic ironstones, redox sensitive elements in shales, and the rise of atmospheric oxygen. Geochim. Cosmochim. Acta 66, A64. [Google Scholar]
- Farquhar J.; Bao H. M.; Thiemens M. (2000) Atmospheric influence of Earth’s earliest sulfur cycle. Science 289, 756–758. [DOI] [PubMed] [Google Scholar]
- Canfield D. E. (2005) The early history of atmospheric oxygen: Homage to Robert A. Garrels. Annu. Rev. Earth Planet. Sci. 33, 1–36. [Google Scholar]
- Scott C.; Lyons T. W.; Bekker A.; Shen Y.; Poulton S. W.; Chu X.; Anbar A. D. (2008) Tracing the stepwise oxygenation of the Proterozoic ocean. Nature 452, 456–459. [DOI] [PubMed] [Google Scholar]
- Reinhard C. T.; Raiswell R.; Scott C.; Anbar A. D.; Lyons T. W. (2009) A late Archean sulfidic sea stimulated by early oxidative weathering of the continents. Science 326, 713–716. [DOI] [PubMed] [Google Scholar]
- Falkowski P. G. (1997) Photosynthesis: The paradox carbon dioxide efflux. Curr. Biol. 7, R637–R639. [DOI] [PubMed] [Google Scholar]
- Konhauser K. O.; Pecoits E.; Lalonde S. V.; Papineau D.; Nisbet E. G.; Barley M. E.; Arndt N. T.; Zahnle K.; Kamber B. S. (2009) Oceanic nickel depletion and a methanogen famine before the Great Oxidation Event. Geochim. Cosmochim. Acta 73, A678. [DOI] [PubMed] [Google Scholar]
- David L. A.; Alm E. J. (2011) Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469, 93–96. [DOI] [PubMed] [Google Scholar]
- Dupont C. L.; Butcher A.; Valas R. E.; Bourne P. E.; Caetano-Anolles G. (2010) History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U.S.A. 107, 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. C.; Brimblecombe P. (1985) Iron and sulfur in the pre-biologic ocean. Precambrian Res. 28, 205–222. [DOI] [PubMed] [Google Scholar]
- Holland H. D. (1973) Oceans: Possible source of iron in iron formations. Econ. Geol. 68, 1169–1172. [Google Scholar]
- Falkowski P. G.; Barber R. T.; Smetacek V. V. (1998) Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–207. [DOI] [PubMed] [Google Scholar]
- Drobner E.; Huber H.; Wachtershauser G.; Rose D.; Stetter K. O. (1990) Pyrite formation linked with hydrogen evolution under anaerobic conditions. Nature 346, 742–744. [Google Scholar]
- Huber C.; Wachtershauser G. (1997) Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science 276, 245–247. [DOI] [PubMed] [Google Scholar]
- Schoonen M. A.; Xu Y. (2001) Nitrogen reduction under hydrothermal vent conditions: Implications for the prebiotic synthesis of C-H-O-N compounds. Astrobiology 1, 133–142. [DOI] [PubMed] [Google Scholar]
- Wachtershauser G. (1988) Pyrite Formation, the 1st Energy-Source for Life: A Hypothesis. Syst. Appl. Microbiol. 10, 207–210. [Google Scholar]
- Mulder D. W.; Boyd E. S.; Sarma R.; Lange R. K.; Endrizzi J. A.; Broderick J. B.; Peters J. W. (2010) Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(ΔEFG). Nature 465, 248–251. [DOI] [PubMed] [Google Scholar]
- Boyd E. S.; Anbar A. D.; Miller S.; Hamilton T. L.; Lavin M.; Peters J. W. (2011) A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9, 221–232. [DOI] [PubMed] [Google Scholar]
- Boyd E. S.; Peters J. W. (2013) New insights into the evolutionary history of biological nitrogen fixation. Front. Microbiol. 4, 00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H.; Holm R. H.; Munck E. (1997) Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277, 653–659. [DOI] [PubMed] [Google Scholar]
- Hagen K. S.; Reynolds J. G.; Holm R. H. (1981) Definition of Reaction Sequences Resulting in Self-Assembly of [Fe4S4(Sr)4]2– Clusters from Simple Reactants. J. Am. Chem. Soc. 103, 4054–4063. [Google Scholar]
- Hagen K. S.; Watson A. D.; Holm R. H. (1983) Synthetic routes to Fe2S2, Fe3S4, Fe4S4, and Fe6S9 clusters from the common precursor [Fe(Sc2H5)4]2–: Structures and properties of [Fe3S4(Sr)4]3– and [Fe6S9(Sc2H5)2]4–, examples of the newest types of Fe-S-Sr clusters. J. Am. Chem. Soc. 105, 3905–3913. [Google Scholar]
- Qi W. B.; Li J. W.; Chain C. Y.; Pasquevich G. A.; Pasquevich A. F.; Cowan J. A. (2012) Glutathione Complexed Fe-S Centers. J. Am. Chem. Soc. 134, 10745–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. C.; Beinert H. (1988) The state of cluster SH and S2– of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J. Biol. Chem. 263, 8194–8198. [PubMed] [Google Scholar]
- Kennedy M. C.; Emptage M. H.; Dreyer J. L.; Beinert H. (1983) The role of iron in the activation-inactivation of aconitase. J. Biol. Chem. 258, 11098–11105. [PubMed] [Google Scholar]
- Malkin R.; Rabinowitz J. C. (1966) The reconstitution of clostridial ferredoxin. Biochem. Biophys. Res. Commun. 23, 822–827. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama P.; Agar J. N.; Cash V. L.; Johnson M. K.; Dean D. R. (2000) NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. U.S.A. 97, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R.; Brigle K. E.; Bennett L. T.; Setterquist R. A.; Wilson M. S.; Cash V. L.; Beynon J.; Newton W. E.; Dean D. R. (1989) Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J. Bacteriol. 171, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. R.; Cash V. L.; Weiss M. C.; Laird N. F.; Newton W. E.; Dean D. R. (1989) Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet. 219, 49–57. [DOI] [PubMed] [Google Scholar]
- Lill R.; Muhlenhoff U. (2008) Maturation of iron-sulfur proteins in eukaryotes: Mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77, 669–700. [DOI] [PubMed] [Google Scholar]
- Roche B.; Aussel L.; Ezraty B.; Mandin P.; Py B.; Barras F. (2013) Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. [DOI] [PubMed] [Google Scholar]
- Takahashi Y.; Tokumoto U. (2002) A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383. [DOI] [PubMed] [Google Scholar]
- Johnson D. C.; Dean D. R.; Smith A. D.; Johnson M. K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. [DOI] [PubMed] [Google Scholar]
- Nachin L.; El Hassouni M.; Loiseau L.; Expert D.; Barras F. (2001) SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: The key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39, 960–972. [DOI] [PubMed] [Google Scholar]
- Nachin L.; Loiseau L.; Expert D.; Barras F. (2003) SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H.; Yeo W. S.; Roe J. H. (2004) Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: Involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51, 1745–1755. [DOI] [PubMed] [Google Scholar]
- Outten F. W.; Djaman O.; Storz G. (2004) A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52, 861–872. [DOI] [PubMed] [Google Scholar]
- Tokumoto U.; Kitamura S.; Fukuyama K.; Takahashi Y. (2004) Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 136, 199–209. [DOI] [PubMed] [Google Scholar]
- Chahal H. K.; Outten F. W. (2012) Separate Fe-S scaffold and carrier functions for SufB2C2 and SufA during in vitro maturation of [2Fe-2S] Fdx. J. Inorg. Biochem. 116, 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G.; Gaddam S. A.; Ayala-Castro C. N.; Ollagnier-de Choudens S.; Lascoux D.; Fontecave M.; Outten F. W. (2007) SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350. [DOI] [PubMed] [Google Scholar]
- Wollers S.; Layer G.; Garcia-Serres R.; Signor L.; Clemancey M.; Latour J. M.; Fontecave M.; Ollagnier de Choudens S. (2010) Iron-sulfur (Fe-S) cluster assembly: The SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini A.; Mapolelo D. T.; Chahal H. K.; Johnson M. K.; Outten F. W. (2010) SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry 49, 9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahal H. K.; Dai Y.; Saini A.; Ayala-Castro C.; Outten F. W. (2009) The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48, 10644–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangachari K.; Davis C. T.; Eccleston J. F.; Hirst E. M.; Saldanha J. W.; Strath M.; Wilson R. J. (2002) SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 514, 225–228. [DOI] [PubMed] [Google Scholar]
- Outten F. W.; Wood M. J.; Munoz F. M.; Storz G. (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278, 45713–45719. [DOI] [PubMed] [Google Scholar]
- Eccleston J. F.; Petrovic A.; Davis C. T.; Rangachari K.; Wilson R. J. (2006) The kinetic mechanism of the SufC ATPase: The cleavage step is accelerated by SufB. J. Biol. Chem. 281, 8371–8378. [DOI] [PubMed] [Google Scholar]
- Petrovic A.; Davis C. T.; Rangachari K.; Clough B.; Wilson R. J.; Eccleston J. F. (2008) Hydrodynamic characterization of the SufBC and SufCD complexes and their interaction with fluorescent adenosine nucleotides. Protein Sci. 17, 1264–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka S.; Wada K.; Hasegawa Y.; Minami Y.; Fukuyama K.; Takahashi Y. (2006) Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 580, 137–143. [DOI] [PubMed] [Google Scholar]
- Wada K.; Sumi N.; Nagai R.; Iwasaki K.; Sato T.; Suzuki K.; Hasegawa Y.; Kitaoka S.; Minami Y.; Outten F. W.; Takahashi Y.; Fukuyama K. (2009) Molecular dynamism of Fe-S cluster biosynthesis implicated by the structure of SufC2-SufD2 complex. J. Mol. Biol. 387, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S.; Kita A.; Miki K. (2005) Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8. J. Mol. Biol. 353, 1043–1054. [DOI] [PubMed] [Google Scholar]
- Zaitseva J.; Jenewein S.; Jumpertz T.; Holland I. B.; Schmitt L. (2005) H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24, 1901–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. M.; George A. M. (2012) Role of the D-loops in allosteric control of ATP hydrolysis in an ABC transporter. J. Phys. Chem. A 116, 3004–3013. [DOI] [PubMed] [Google Scholar]
- Higgins C. F.; Linton K. J. (2004) The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11, 918–926. [DOI] [PubMed] [Google Scholar]
- Linton K. J.; Higgins C. F. (1998) The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28, 5–13. [DOI] [PubMed] [Google Scholar]
- Linton K. J.; Higgins C. F. (2007) Structure and function of ABC transporters: The ATP switch provides flexible control. Pfluegers Arch. 453, 555–567. [DOI] [PubMed] [Google Scholar]
- Tsaousis A. D.; Ollagnier de Choudens S.; Gentekaki E.; Long S.; Gaston D.; Stechmann A.; Vinella D.; Py B.; Fontecave M.; Barras F.; Lukes J.; Roger A. J. (2012) Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc. Natl. Acad. Sci. U.S.A. 109, 10426–10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. S.; Barkay T. (2012) The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 3, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer S. I.; Hantke K. (1999) SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181, 3307–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau L.; Ollagnier-de-Choudens S.; Nachin L.; Fontecave M.; Barras F. (2003) Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 278, 38352–38359. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S.; Lascoux D.; Loiseau L.; Barras F.; Forest E.; Fontecave M. (2003) Mechanistic studies of the SufS-SufE cysteine desulfurase: Evidence for sulfur transfer from SufS to SufE. FEBS Lett. 555, 263–267. [DOI] [PubMed] [Google Scholar]
- Xu X. M.; Möller S. G. (2006) AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 25, 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Abdel-Ghany S. E.; Anderson T. D.; Pilon-Smits E. A.; Pilon M. (2006) CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 281, 8958–8969. [DOI] [PubMed] [Google Scholar]
- Dai Y.; Outten F. W. (2012) The E. coli SufS-SufE sulfur transfer system is more resistant to oxidative stress than IscS-IscU. FEBS Lett. 586, 4016–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach B. P.; Pradhan P. K.; Dos Santos P. C. (2013) Protected sulfur transfer reactions by the Escherichia coli Suf system. Biochemistry 52, 4089–4096. [DOI] [PubMed] [Google Scholar]
- Singh H.; Dai Y.; Outten F. W.; Busenlehner L. S. (2013) Escherichia coli SufE sulfur transfer protein modulates the SufS cysteine desulfurase through allosteric conformational dynamics. J. Biol. Chem. 288, 36189–36200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert D.; Boughammoura A.; Franza T. (2008) Siderophore-controlled iron assimilation in the enterobacterium Erwinia chrysanthemi: Evidence for the involvement of bacterioferritin and the Suf iron-sulfur cluster assembly machinery. J. Biol. Chem. 283, 36564–36572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.; White R. H.; Cash V. L.; Jack R. F.; Dean D. R. (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.; White R. H.; Cash V. L.; Dean D. R. (1994) Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720. [DOI] [PubMed] [Google Scholar]
- Beinert H. (2000) A tribute to sulfur. Eur. J. Biochem. 267, 5657–5664. [DOI] [PubMed] [Google Scholar]
- Toohey J. I. (1989) Sulfane sulfur in biological systems: A possible regulatory role. Biochem. J. 264, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara H.; Kurihara T.; Yoshimura T.; Soda K.; Esaki N. (1997) Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272, 22417–22424. [DOI] [PubMed] [Google Scholar]
- Mihara H.; Esaki N. (2002) Bacterial cysteine desulfurases: Their function and mechanisms. Appl. Microbiol. Biotechnol. 60, 12–23. [DOI] [PubMed] [Google Scholar]
- Fujii T.; Maeda M.; Mihara H.; Kurihara T.; Esaki N.; Hata Y. (2000) Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase. Biochemistry 39, 1263–1273. [DOI] [PubMed] [Google Scholar]
- Mihara H.; Kurihara T.; Yoshimura T.; Esaki N. (2000) Kinetic and mutational studies of three NifS homologs from Escherichia coli: Mechanistic difference between l-cysteine desulfurase and l-selenocysteine lyase reactions. J. Biochem. 127, 559–567. [DOI] [PubMed] [Google Scholar]
- Lima C. D. (2002) Analysis of the E. coli NifS CsdB protein at 2.0 Å reveals the structural basis for perselenide and persulfide intermediate formation. J. Mol. Biol. 315, 1199–1208. [DOI] [PubMed] [Google Scholar]
- Mihara H.; Fujii T.; Kato S.; Kurihara T.; Hata Y.; Esaki N. (2002) Structure of external aldimine of Escherichia coli CsdB, an IscS/NifS homolog: Implications for its specificity toward selenocysteine. J. Biochem. 131, 679–685. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery J. R.; Urbina H.; Vickery L. E. (2003) Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059. [DOI] [PubMed] [Google Scholar]
- Albrecht A. G.; Netz D. J.; Miethke M.; Pierik A. J.; Burghaus O.; Peuckert F.; Lill R.; Marahiel M. A. (2010) SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J. Bacteriol. 192, 1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach B.; Earles E.; Dos Santos P. C. (2010) Kinetic analysis of the bisubstrate cysteine desulfurase SufS from Bacillus subtilis. Biochemistry 49, 8794–8802. [DOI] [PubMed] [Google Scholar]
- Albrecht A. G.; Peuckert F.; Landmann H.; Miethke M.; Seubert A.; Marahiel M. A. (2011) Mechanistic characterization of sulfur transfer from cysteine desulfurase SufS to the iron-sulfur scaffold SufU in Bacillus subtilis. FEBS Lett. 585, 465–470. [DOI] [PubMed] [Google Scholar]
- Riboldi G. P.; de Oliveira J. S.; Frazzon J. (2011) Enterococcus faecalis SufU scaffold protein enhances SufS desulfurase activity by acquiring sulfur from its cysteine-153. Biochim. Biophys. Acta 1814, 1910–1918. [DOI] [PubMed] [Google Scholar]
- Selbach B. P.; Chung A. H.; Scott A. D.; George S. J.; Cramer S. P.; Dos Santos P. C. (2014) Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry 53, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein S. I.; Perona J. J. (2008) Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J. Biol. Chem. 283, 22007–22017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Dos Santos P. C.; Zhu X.; Orlando R.; Dean D. R.; Soll D.; Yuan J. (2012) Catalytic mechanism of Sep-tRNA:Cys-tRNA synthase: Sulfur transfer is mediated by disulfide and persulfide. J. Biol. Chem. 287, 5426–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A.; Zhu W.; Major T. A.; Roy H.; Palioura S.; Jahn D.; Whitman W. B.; Yates J. R. III; Ibba M.; Soll D. (2005) RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972. [DOI] [PubMed] [Google Scholar]
- Liu Y. C.; Sieprawska-Lupa M.; Whitman W. B.; White R. H. (2010) Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic Archaeon Methanococcus maripaludis. J. Biol. Chem. 285, 31923–31929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S.; Imlay J. A. (2010) Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 78, 1448–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay P.; Zheng M.; Bedzyk L. A.; LaRossa R. A.; Storz G. (2004) Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U.S.A. 101, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Wang X.; Templeton L. J.; Smulski D. R.; LaRossa R. A.; Storz G. (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183, 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G.; Morissette A.; Prevost K.; Masse E. (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 28, 1551–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel J. L.; Nesbit A. D.; Mettert E. L.; Fleischhacker A. S.; Wanta B. T.; Kiley P. J. (2013) Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol. Microbiol. 87, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel J. L.; Rodionov D.; Liu M.; Blattner F. R.; Kiley P. J. (2006) IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60, 1058–1075. [DOI] [PubMed] [Google Scholar]
- Lee K. C.; Yeo W. S.; Roe J. H. (2008) Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J. Bacteriol. 190, 8244–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo W. S.; Lee J. H.; Lee K. C.; Roe J. H. (2006) IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61, 206–218. [DOI] [PubMed] [Google Scholar]
- Jang S.; Imlay J. A. (2007) Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 282, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S.; Mihara H.; Kurihara T.; Takahashi Y.; Tokumoto U.; Yoshimura T.; Esaki N. (2002) Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: Implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 5948–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni E. N.; de Oliveira J. S.; Nicolet Y.; Raulfs E. C.; Amara P.; Dean D. R.; Fontecilla-Camps J. C. (2012) (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem., Int. Ed. 51, 5439–5442. [DOI] [PubMed] [Google Scholar]
- Smith A. D.; Frazzon J.; Dean D. R.; Johnson M. K. (2005) Role of conserved cysteines in mediating sulfur transfer from IscS to IscU. FEBS Lett. 579, 5236–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina H. D.; Silberg J. J.; Hoff K. G.; Vickery L. E. (2001) Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276, 44521–44526. [DOI] [PubMed] [Google Scholar]
- Shimomura Y.; Wada K.; Fukuyama K.; Takahashi Y. (2008) The asymmetric trimeric architecture of [2Fe-2S] IscU: Implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 383, 133–143. [DOI] [PubMed] [Google Scholar]
- Riboldi G. P.; Verli H.; Frazzon J. (2009) Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem. 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A.; Blackshields G.; Brown N. P.; Chenna R.; McGettigan P. A.; McWilliam H.; Valentin F.; Wallace I. M.; Wilm A.; Lopez R.; Thompson J. D.; Gibson T. J.; Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Guindon S.; Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Sanderson M. J. (2002) Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol. Biol. Evol. 19, 101–109. [DOI] [PubMed] [Google Scholar]
- Kembel S. W.; Cowan P. D.; Helmus M. R.; Cornwell W. K.; Morlon H.; Ackerly D. D.; Blomberg S. P.; Webb C. O. (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.